An Enzymatic and Proteomic Analysis of Panus lecomtei during Biodegradation of Gossypol in Cottonseed
Abstract
1. Introduction
2. Materials and Methods
2.1. Cottonseed and Fungal Strain
2.2. Cultivation of P. lecomtei on Crushed Whole Cottonseed
2.3. Quantification of Gossypol and Crude Enzyme Extracts
2.4. Global Secretome Analysis
2.5. Sample Preparation and Processing
2.6. LC-MS/MS Analysis
2.7. Chromatography
2.8. Mass Spectrometry
2.9. Data Analysis
2.10. Protein Grouping and Functional Inference
2.11. Partial Purification of the Crude Extract
3. Results
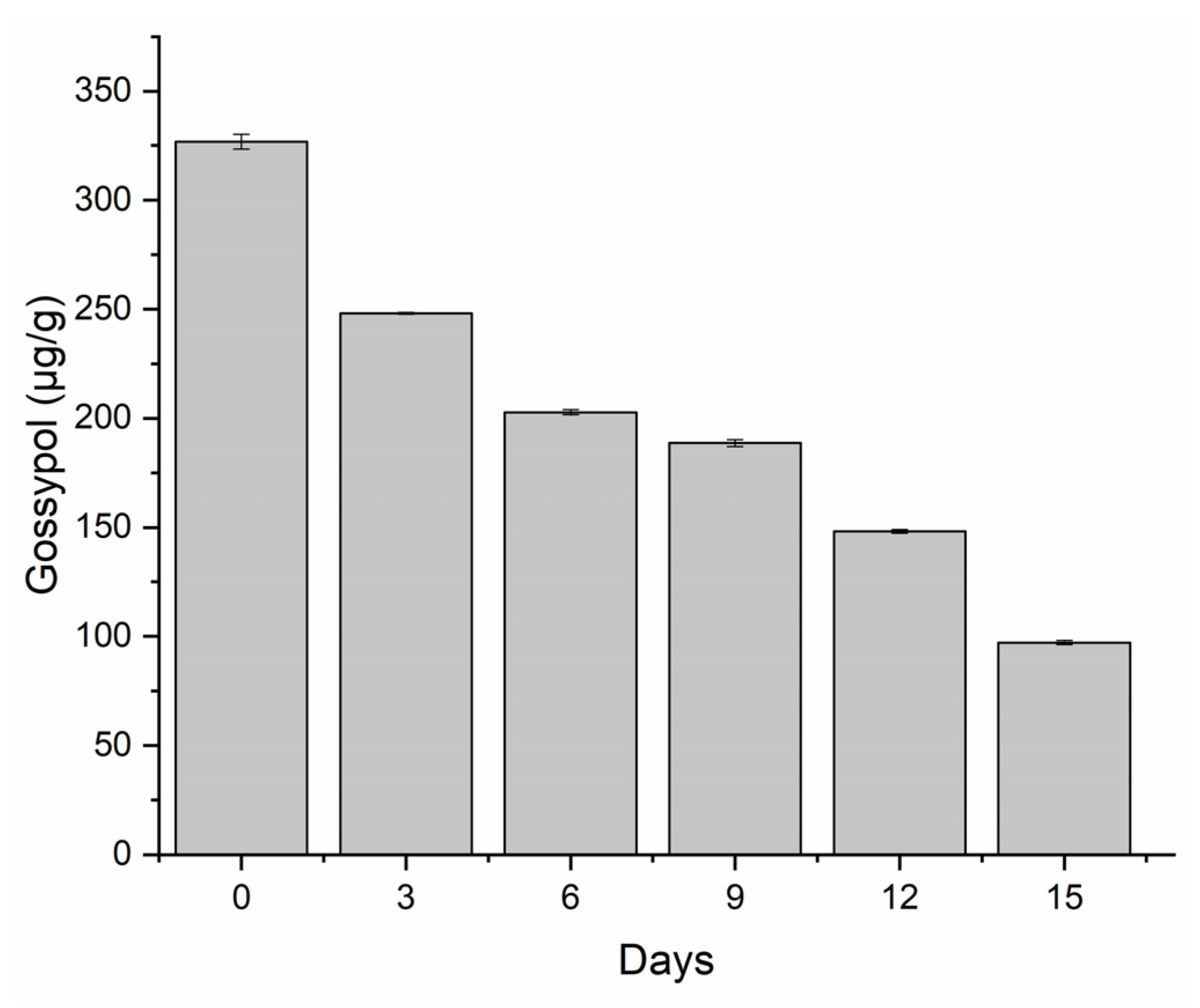
3.1. Degradation of Free Gossypol (FG)
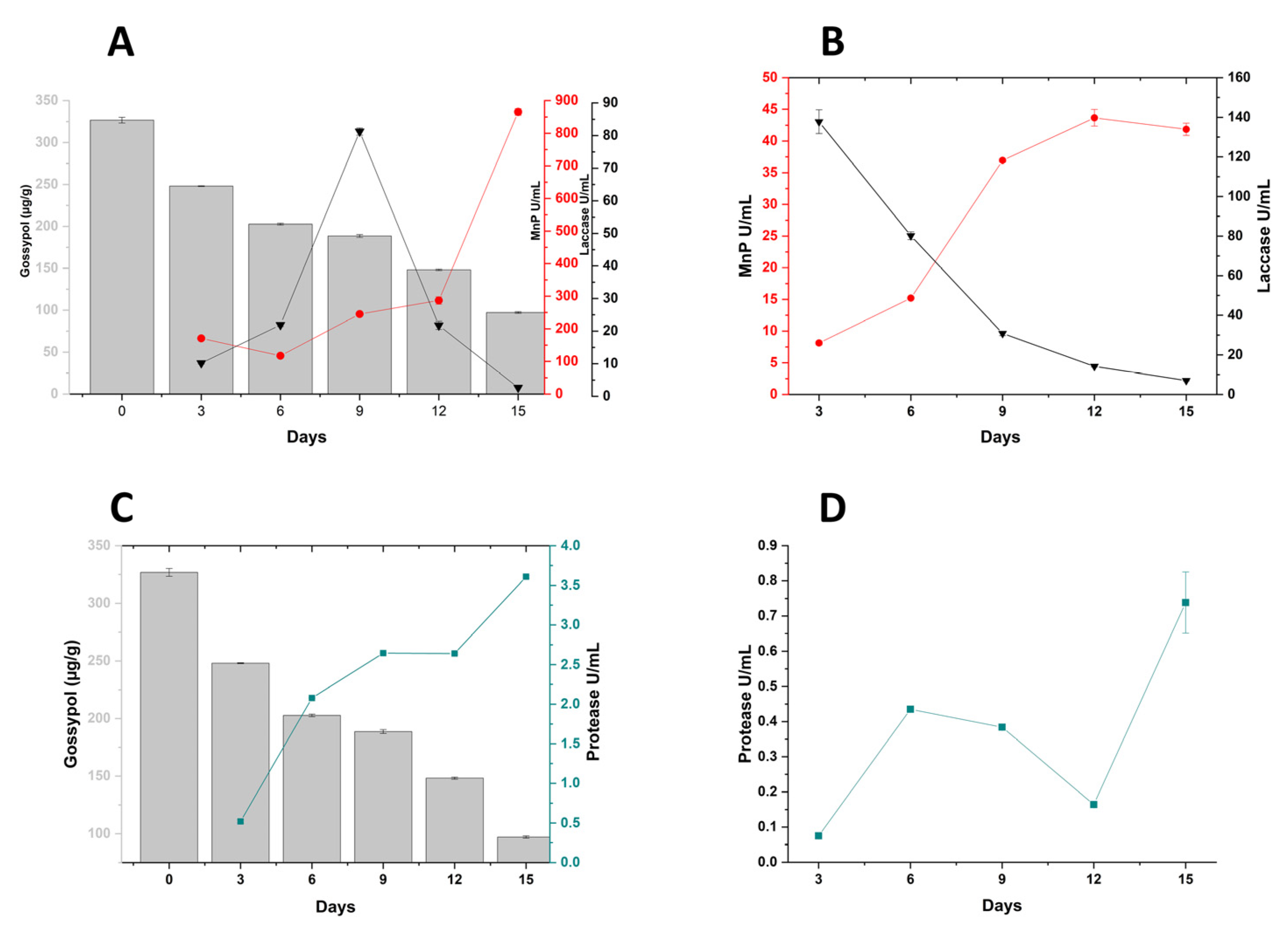
3.2. Enzyme Profiling during Cultivation of P. lecomtei BRM044603
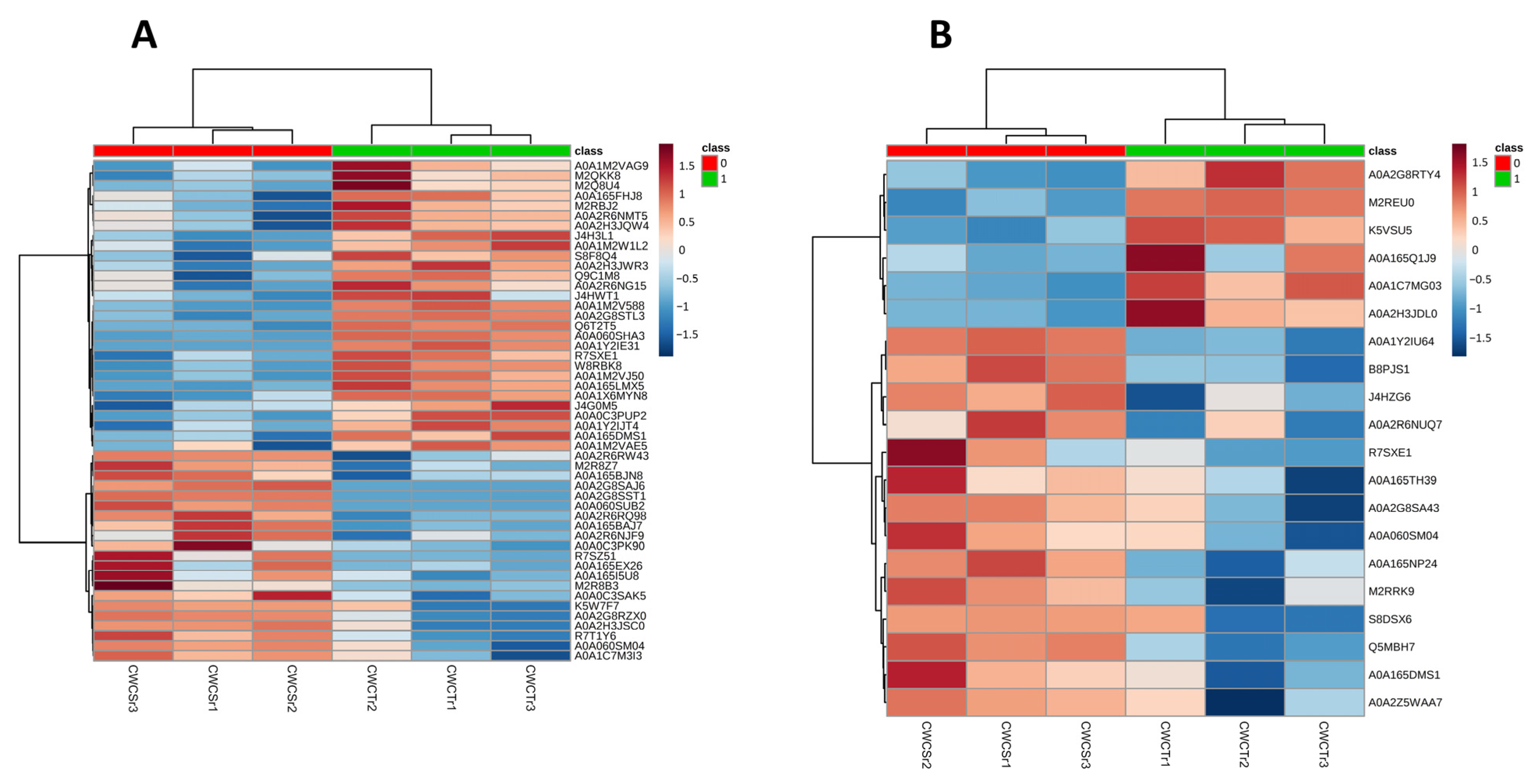
3.3. Proteomic Analysis of Crude Extracts from P. lecomtei BRM044603 Cultivated in the Presence of Gossypol (CWCS) and in the Absence of Gossypol (CWCT) at 6 DAI and 12 DAI
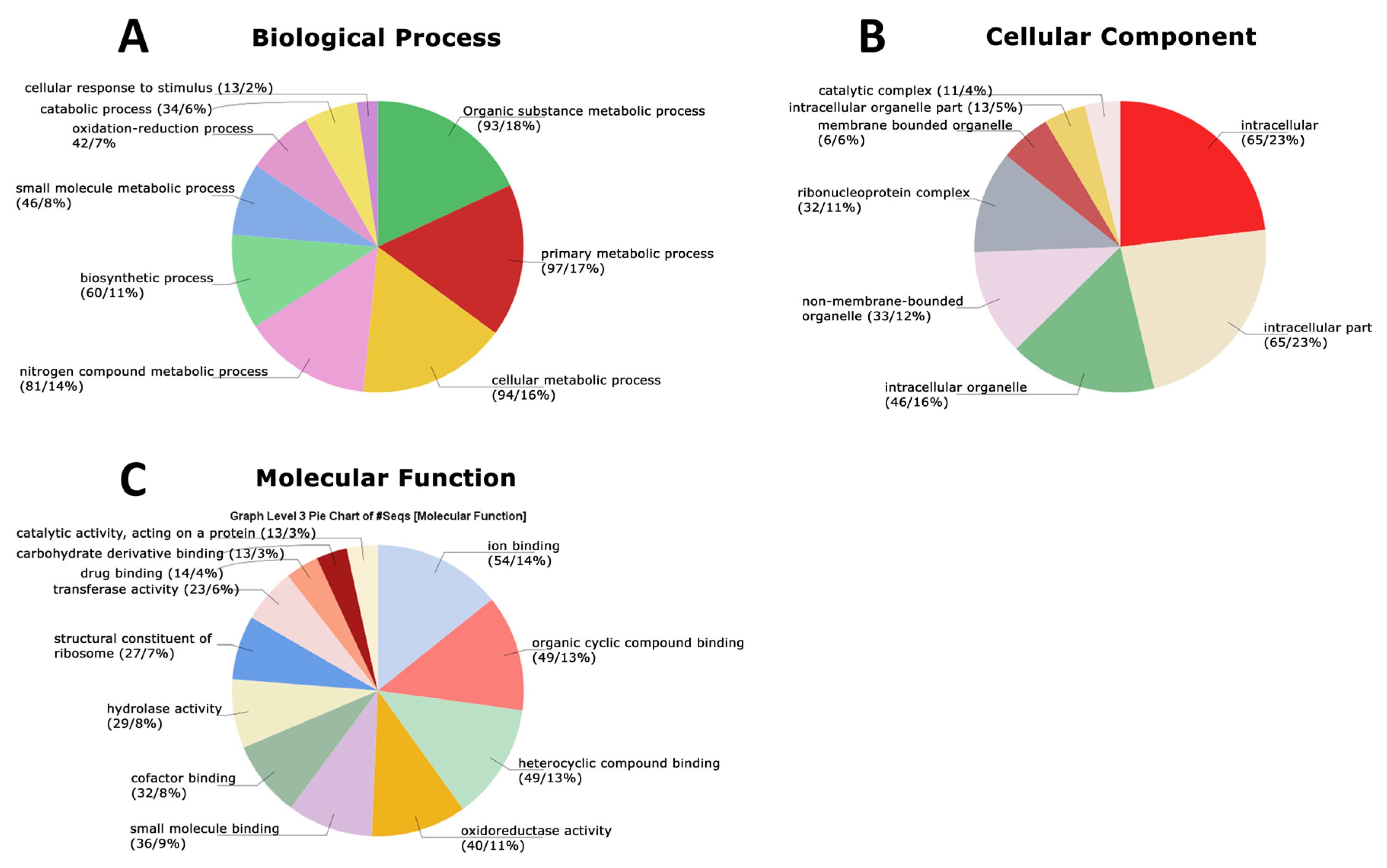
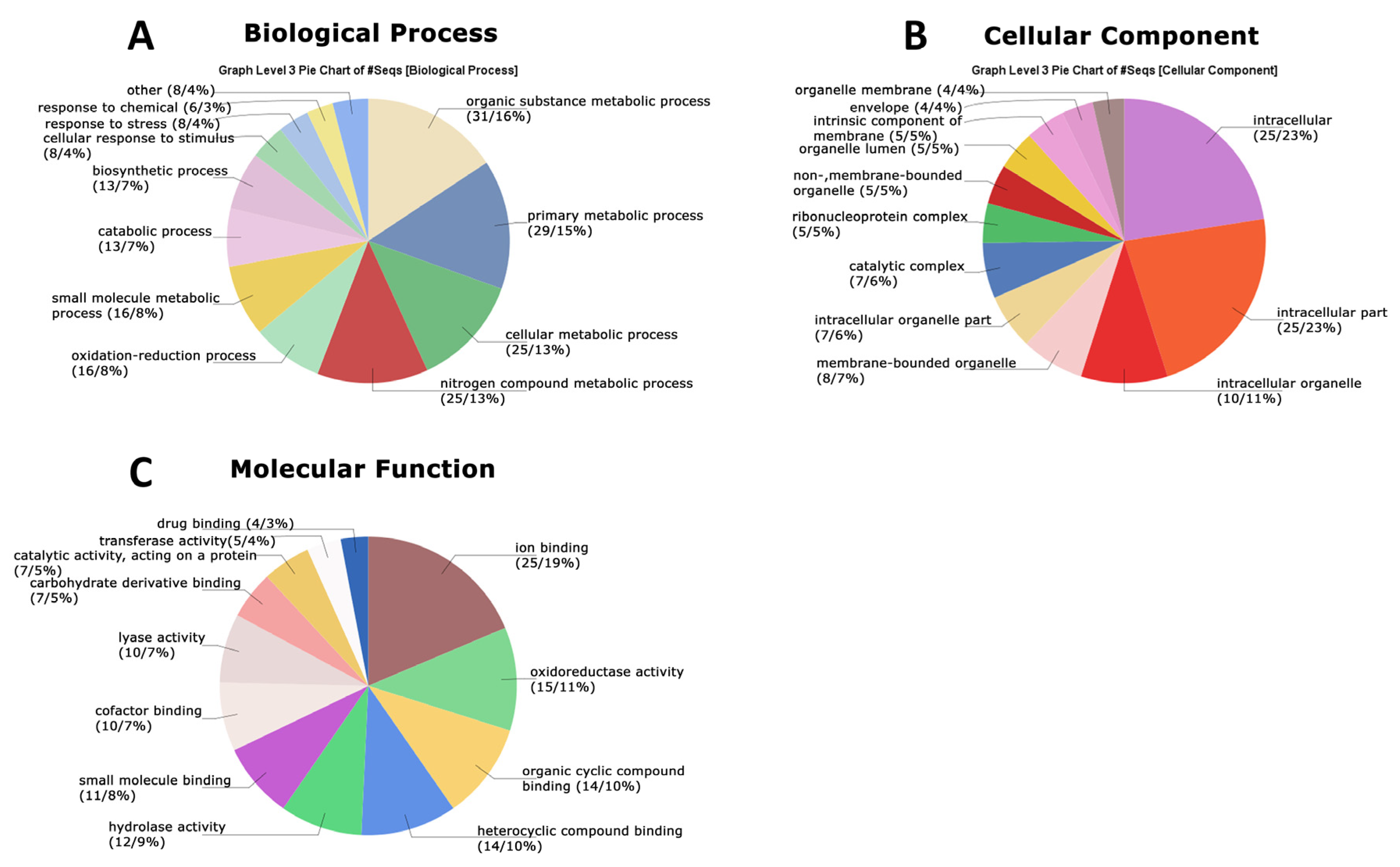
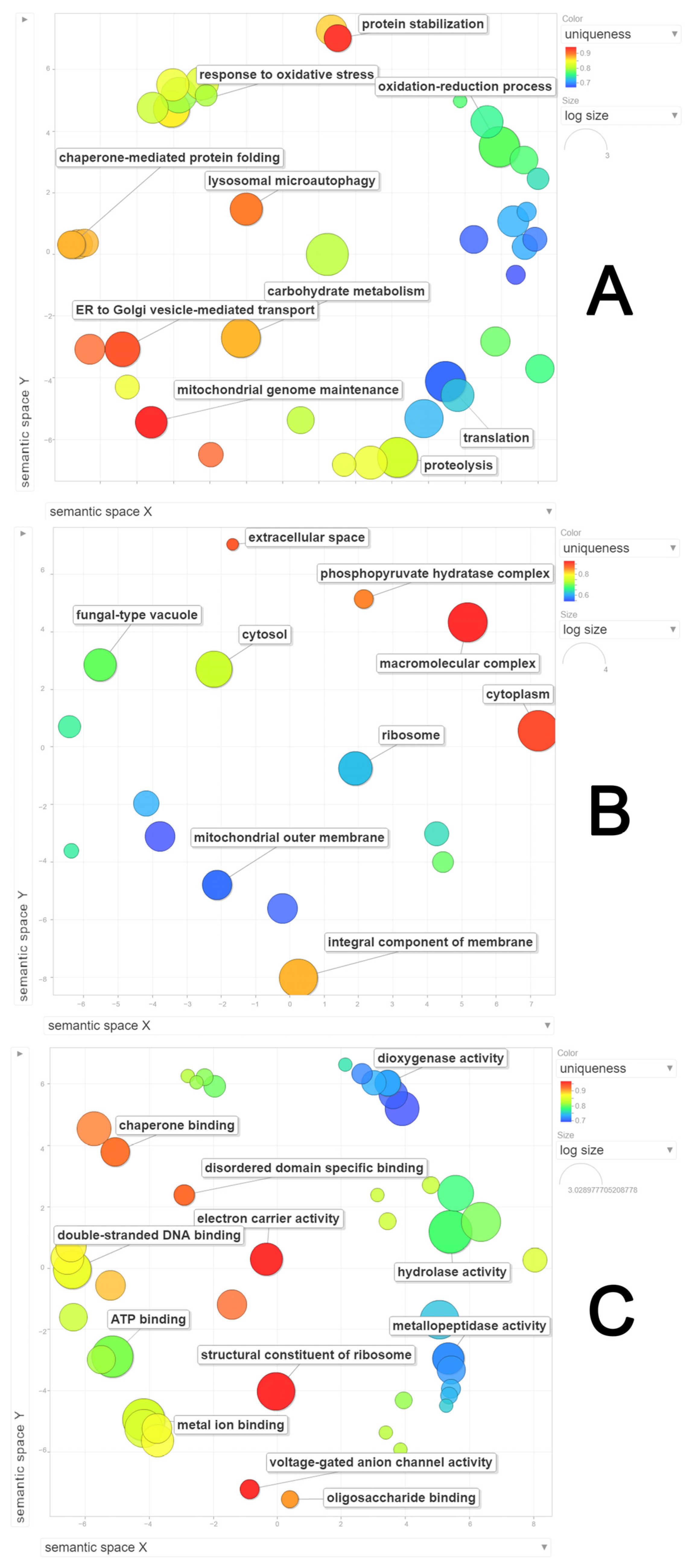
3.4. Functional Analysis of the Global Secretome of P. lecomtei BRM044603 via Gene Ontology (GO), String-Derived Protein–Protein Interaction Networks, and Kyoto Encyclopedia of Genes and Genomes (KEGG) Metabolic Pathway Mapping
3.5. Partial Purification of Laccase Enzyme Fractions and Evaluation of Free-Gossypol Degradation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. The State of Food Security and Nutrition in the World 2018: Building Climate Resilience for Food Security and Nutrition; Food & Agriculture Organization: Rome, Italy, 2018. [Google Scholar]
- Molotoks, A.; Smith, P.; Dawson, T.P. Impacts of land use, population, and climate change on global food security. Food Energy Secur. 2021, 10, e261. [Google Scholar] [CrossRef]
- Georganas, A.; Giamouri, E.; Pappas, A.C.; Papadomichelakis, G.; Fortatos, S.; Manios, T.; Lasaridi, K.; Fegeros, K.; Tsiplakou, E.; Zervas, G. Redefining the future of catering waste application in animal diets—A review on the minimization of potential hazards in catering waste prior to application in animal diets. Anim. Feed Sci. Technol. 2022, 289, 115334. [Google Scholar] [CrossRef]
- Paris Agreement. In Proceedings of the Report of the Conference of the Parties to the United Nations Framework Convention on Climate Change, 21st Session, Paris, France, 30 November–13 December 2015; p. 2017. Available online: https://heinonline.org/HOL/Page?handle=hein.journals/intlm55&div=46&g_sent=1&casa_token= (accessed on 1 February 2024).
- Welsby, D.; Price, J.; Pye, S.; Ekins, P. Unextractable fossil fuels in a 1.5 °C world. Nature 2021, 597, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, I.A.; Ötvös, T.; Gilmanova, A.; Rocca, E.; Ghanem, C.; Wanat, M. International Energy Agency; Kluwer Law International BV: Alphen aan den Rijn, The Netherlands, 2021. [Google Scholar]
- Ramos, M.D.N.; Milessi, T.S.; Candido, R.G.; Mendes, A.A.; Aguiar, A. Enzymatic catalysis as a tool in biofuels production in Brazil: Current status and perspectives. Energy Sustain. Dev. 2022, 68, 103–119. [Google Scholar] [CrossRef]
- Mahdi, H.I.; Ramlee, N.N.; da Silva Duarte, J.L.; Cheng, Y.-S.; Selvasembian, R.; Amir, F.; de Oliveira, L.H.; Azelee, N.I.W.; Meili, L.; Rangasamy, G. A comprehensive review on nanocatalysts and nanobiocatalysts for biodiesel production in Indonesia, Malaysia, Brazil and USA. Chemosphere 2023, 319, 138003. [Google Scholar] [CrossRef] [PubMed]
- Riayatsyah, T.; Sebayang, A.; Silitonga, A.; Padli, Y.; Fattah, I.; Kusumo, F.; Ong, H.; Mahlia, T. Current progress of Jatropha curcas commoditisation as biodiesel feedstock: A comprehensive review. Front. Energy Res. 2022, 9, 815416. [Google Scholar] [CrossRef]
- Babadi, A.A.; Rahmati, S.; Fakhlaei, R.; Barati, B.; Wang, S.; Doherty, W.; Ostrikov, K.K. Emerging technologies for biodiesel production: Processes, challenges, and opportunities. Biomass Bioenergy 2022, 163, 106521. [Google Scholar] [CrossRef]
- Milanez, A.Y.; Baptista da Silva Maia, G.; Guimarães, D.D.; Ferreira, C.L.A. Biodiesel e Diesel Verde no Brasil: Panorama Recente e Perspectivas; Banco Nacional de Desenvolvimento Econômico e Social: Rio de Janeiro, Brazil, 2022; Volume 6, pp. 41–71. [Google Scholar]
- Gaonkar, V.; Rosentrater, K.A. Soybean. In Integrated Processing Technologies for Food and Agricultural By-Products; Elsevier: Amsterdam, The Netherlands, 2019; pp. 73–104. [Google Scholar]
- Mirpoor, S.F.; Giosafatto, C.V.L.; Porta, R. Biorefining of seed oil cakes as industrial co-streams for production of innovative bioplastics. A review. Trends Food Sci. Technol. 2021, 109, 259–270. [Google Scholar] [CrossRef]
- Yesilyurt, M.K.; Aydin, M. Experimental investigation on the performance, combustion and exhaust emission characteristics of a compression-ignition engine fueled with cottonseed oil biodiesel/diethyl ether/diesel fuel blends. Energy Convers. Manag. 2020, 205, 112355. [Google Scholar] [CrossRef]
- Parkash, V.; Snider, J.L.; Bruce, A.; Ermanis, A.; Virk, G.; Kaur, N.; Collins, G.; Chapman, K.D. Effects of cultivar and nitrogen application rate on lint, seed, oil, and protein yields of field-grown cotton. Crop Sci. 2023, 63, 1541–1554. [Google Scholar] [CrossRef]
- Mizik, T.; Gyarmati, G. Economic and sustainability of biodiesel production—A systematic literature review. Clean Technol. 2021, 3, 19–36. [Google Scholar] [CrossRef]
- Świątkiewicz, S.; Arczewska-Włosek, A.; Józefiak, D. The use of cottonseed meal as a protein source for poultry: An updated review. Worlds Poult. Sci. J. 2016, 72, 473–484. [Google Scholar] [CrossRef]
- Gadelha, I.C.N.; Fonseca, N.B.S.; Oloris, S.C.S.; Melo, M.M.; Soto-Blanco, B. Gossypol toxicity from cottonseed products. Sci. World J. 2014, 2014, 231635. [Google Scholar] [CrossRef] [PubMed]
- Tiago Do Prado, P.; Louvandini, H.; Mcmanus, C.M.; Abdalla, A.L. Uso de subprodutos do algodão na nutrição de ruminantes. Ciênc. Vet. Tróp. 2010, 13, 24–37. [Google Scholar]
- Yu, F.; Barry, T.; Moughan, P.; Wilson, G. Condensed tannin and gossypol concentrations in cottonseed and in processed cottonseed meal. J. Sci. Food Agric. 1993, 63, 7–15. [Google Scholar] [CrossRef]
- Yu, F.; Moughan, P.; Barry, T. The effect of cottonseed condensed tannins on the ileal digestibility of amino acids in casein and cottonseed kernel. Br. J. Nutr. 1996, 75, 683–698. [Google Scholar] [CrossRef] [PubMed]
- Elkin, R.G. Influence of Plant Toxins on Laying Hen Performance and Egg Quality. In Egg Innovations and Strategies for Improvements; Elsevier: Amsterdam, The Netherlands, 2017; pp. 499–512. [Google Scholar]
- Kong, G.; Daud, M.K.; Zhu, S. Effects of pigment glands and gossypol on growth, development and insecticide-resistance of cotton bollworm (Heliothis armigera (Hübner). J. Crop Prot. 2010, 29, 813–819. [Google Scholar] [CrossRef]
- Benedict, C.R.; Martin, G.S.; Liu, J.; Puckhaber, L.; Magill, C.W. Terpenoid aldehyde formation and lysigenous gland storage sites in cotton: Variant with mature glands but suppressed levels of terpenoid aldehydes. Phytochemistry 2004, 65, 1351–1359. [Google Scholar] [CrossRef]
- Abdurakhimov, R.S.; Veshkurova, O.; Uzbekov, V.; Arzanova, I.; Sultanova, E.; Salikhov, S.I. Effect of cotton-seed biocidal peptides and gossypol on resistance to biotic factors. Chem. Nat. Compd. 2009, 45, 213–216. [Google Scholar] [CrossRef]
- Authority, E.F.S. Gossypol as undesirable substance in animal feed-Scientific Opinion of the Panel on Contaminants in the Food Chain. EFSA J. 2009, 7, 908. [Google Scholar]
- Beltrão, N.D.M.; de Araujo, A.E.; de Araujo, A.E. Algodão: O Produtor Pergunta, a Embrapa Responde; Embrapa Informação Tecnológica: Brasília, Brazil, 2004. [Google Scholar]
- Zhang, W.-J.; Xu, Z.-R.; Zhao, S.-H.; Sun, J.-Y.; Yang, X. Development of a microbial fermentation process for detoxification of gossypol in cottonseed meal. Anim. Feed Sci. Technol. 2007, 135, 176–186. [Google Scholar] [CrossRef]
- Gomes, T.G.; Hadi, S.m.I.; Costa Alves, G.S.; Mendonça, S.; De Siqueira, F.G.; Miller, R.N. Current strategies for the detoxification of Jatropha curcas seed cake: A review. J. Agric. Food Chem. 2018, 66, 2510–2522. [Google Scholar] [CrossRef] [PubMed]
- Das Graças Miranda Nunes, M.; Oliveira da Cruz, M.; Alves, M.H. Panus lecomtei (Fr.) Corner (Agaricomycetes, Polyporales, Panaceae): Primeira ocorrência no Estado do Piauí, Brasil. Hoehnea 2020, 47, e132020. [Google Scholar] [CrossRef]
- Gugliotta, A.; Gibertoni, T.; Drechsler-Santos, E.; Silveira, R.; Chikowski, R.; Pires, R.; Montoya, C.; Souza, J.; Palacio, M.; Rezende, D. Polyporales in Lista de Espécies da Flora do Brasil; Jardim Botânico do Rio de Janeiro: Rio de Janeiro, Brazil, 2015. [Google Scholar]
- Vargas-Isla, R.; Capelari, M.; Menolli, N.; Nagasawa, E.; Tokimoto, K.; Ishikawa, N.K. Relationship between Panus lecomtei and P. strigellus inferred from their morphological, molecular and biological characteristics. Mycoscience 2015, 56, 561–571. [Google Scholar] [CrossRef]
- Benti, N.E.; Aneseyee, A.B.; Geffe, C.A.; Woldegiyorgis, T.A.; Gurmesa, G.S.; Bibiso, M.; Asfaw, A.A.; Milki, A.W.; Mekonnen, Y.S. Biodiesel production in Ethiopia: Current status and future prospects. Sci. Afr. 2023, 19, e01531. [Google Scholar] [CrossRef]
- Soares Neto, C.B.; Conceicao, A.A.; Gomes, T.G.; de Aquino Ribeiro, J.A.; Campanha, R.B.; Barroso, P.A.V.; Machado, A.E.V.; Mendonca, S.; De Siqueira, F.G.; Miller, R.N.G. A comparison of physical, chemical, biological and combined treatments for detoxification of free gossypol in crushed whole cottonseed. Waste Biomass Valoriz. 2021, 12, 3965–3975. [Google Scholar] [CrossRef]
- Conceicao, A.A.; Soares Neto, C.B.; de Aquino Ribeiro, J.A.; Goncalves de Siqueira, F.; Miller, R.N.G.; Mendonca, S. Development of an RP-UHPLC-PDA method for quantification of free gossypol in cottonseed cake and fungal-treated cottonseed cake. PLoS ONE 2018, 13, e0196164. [Google Scholar] [CrossRef]
- Wolfenden, B.S.; Willson, R.L. Radical-cations as reference chromogens in kinetic studies of ono-electron transfer reactions: Pulse radiolysis studies of 2, 2′-azinobis-(3-ethylbenzthiazoline-6-sulphonate). J. Chem. Soc. Perkin Trans. 2 1982, 7, 805–812. [Google Scholar] [CrossRef]
- Kuwahara, M.; Glenn, J.K.; Morgan, M.A.; Gold, M.H. Separation and characterization of two extracelluar H2O2-dependent oxidases from ligninolytic cultures of Phanerochaete chrysosporium. FEBS Lett. 1984, 169, 247–250. [Google Scholar] [CrossRef]
- Tien, M.; Kirk, T.K. Lignin-degrading enzyme from Phanerochaete chrysosporium: Purification, characterization, and catalytic properties of a unique H2O2-requiring oxygenase. Proc. Natl. Acad. Sci. USA 1984, 81, 2280–2284. [Google Scholar] [CrossRef]
- Charney, J.; Tomarelli, R.M. A colonmetric method for the determination of the proteolytic activity of duodenal juice. J. Biol. Chem. 1947, 171, 501–505. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M. Analysis of the topology of protein complexes using cross-linking and mass spectrometry. Cold Spring Harb. Protoc. 2007, 2007, pdb.prot4594. [Google Scholar] [CrossRef]
- Crowell, A.M.; Wall, M.J.; Doucette, A.A. Maximizing recovery of water-soluble proteins through acetone precipitation. Anal. Chim. Acta. 2013, 796, 48–54. [Google Scholar] [CrossRef]
- Arshid, S.; Tahir, M.; Fontes, B.; de Souza Montero, E.F.; Castro, M.S.; Sidoli, S.; Roepstorff, P.; Fontes, W. High performance mass spectrometry based proteomics reveals enzyme and signaling pathway regulation in neutrophils during the early stage of surgical trauma. Proteom. Clin. Appl. 2017, 11, 1600001. [Google Scholar] [CrossRef]
- Kalli, A.; Smith, G.T.; Sweredoski, M.J.; Hess, S. Evaluation and optimization of mass spectrometric settings during data-dependent acquisition mode: Focus on LTQ-Orbitrap mass analyzers. J. Proteome Res. 2013, 12, 3071–3086. [Google Scholar] [CrossRef]
- Hawkridge, A.M. Practical considerations and current limitations in quantitative mass spectrometry-based proteomics. In Quantitative Proteomics; Eyers, C.E., Gaskell, S., Eds.; The Royal Society of Chemistry: Cambridge, UK, 2014; pp. 1–25. [Google Scholar]
- Andrews, G.L.; Simons, B.L.; Young, J.B.; Hawkridge, A.M.; Muddiman, D.C. Performance characteristics of a new hybrid quadrupole time-of-flight tandem mass spectrometer (TripleTOF 5600). Anal. Chem. 2011, 83, 5442–5446. [Google Scholar] [CrossRef] [PubMed]
- Välikangas, T.; Suomi, T.; Elo, L.L. A comprehensive evaluation of popular proteomics software workflows for label-free proteome quantification and imputation. Brief. Bioinform. 2018, 19, 1344–1355. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
- Mudunuri, U.; Che, A.; Yi, M.; Stephens, R.M. bioDBnet: The biological database network. Bioinform. 2009, 25, 555–556. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Biol. 2008, 2008, 619832. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, D183–D189. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2012, 41, D377–D386. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016, 45, gkw937. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Bjørkskov, F.B.; Krabbe, S.L.; Nurup, C.N.; Missel, J.W.; Spulber, M.; Bomholt, J.; Molbaek, K.; Helix-Nielsen, C.; Gotfryd, K.; Gourdon, P. Purification and functional comparison of nine human Aquaporins produced in Saccharomyces cerevisiae for the purpose of biophysical characterization. Sci. Rep. 2017, 7, 16899. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Mageshwaran, V.; Majee, S.; Pandiyan, K. Isolation and identification of potential Gossypol degrading fungal strains from cotton growing soil. Microbiol. Res. J. Int. 2017, 21, 35364. [Google Scholar] [CrossRef]
- Rajarathnam, S.; Shashirekha, M.; Bano, Z. Biodegradation of gossypol by the white oyster mushroom, Pleurotus florida, during culturing on rice straw growth substrate, supplemented with cottonseed powder. World J. Microbiol. Biotechnol. 2001, 17, 221–227. [Google Scholar] [CrossRef]
- Weng, X.-Y.; Sun, J.-Y. Biodegradation of free gossypol by a new strain of Candida tropicalis under solid state fermentation: Effects of fermentation parameters. Process Biochem. 2006, 41, 1663–1668. [Google Scholar] [CrossRef]
- Yang, X.; Guo, J.; Sun, J. Biodegradation of free-gossypol by a new fungus isolated from cotton planted soil. Afr. J. Microbiol Res. 2011, 5, 3066–3072. [Google Scholar]
- Noss, R.F. Indicators for monitoring biodiversity: A hierarchical approach. Biol. Conserv. 1990, 4, 355–364. [Google Scholar] [CrossRef]
- Valli, M.; Russo, H.M.; Bolzani, V.S. The potential contribution of the natural products from Brazilian biodiversity to bioeconomy. An. Acad. Bras. Cienc. 2018, 90, 763–778. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Sun, J.Y.; Guo, J.L.; Weng, X.Y. Identification and proteomic analysis of a novel gossypol-degrading fungal strain. J. Sci. Food Agric. 2012, 92, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Tilocca, B.; Balmas, V.; Hassan, Z.U.; Jaoua, S.; Migheli, Q. A proteomic investigation of Aspergillus carbonarius exposed to yeast volatilome or to its major component 2-phenylethanol reveals major shifts in fungal metabolism. Int. J. Food Microbiol. 2019, 306, 108265. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.-Z.; Wang, X.; Liu, W.; Cheng, J.-S.; Yang, Y.; Yuan, Y.-J. Proteomic research reveals the stress response and detoxification of yeast to combined inhibitors. PLoS ONE 2012, 7, e43474. [Google Scholar] [CrossRef] [PubMed]
- Dey, P.; Malik, A.; Singh, D.K.; Haange, S.-B.; von Bergen, M.; Jehmlich, N. Insight into the molecular mechanisms underpinning the mycoremediation of multiple metals by proteomic technique. Front. Microbiol. 2022, 13, 872576. [Google Scholar] [CrossRef]
- Khalaf, M.A.; Meleigy, S.A. Reduction of free gossypol levels in cottonseed meal by microbial treatment. Int. J. Agric. Biol. 2008, 10, 185–190. [Google Scholar]
- Nelson, D.L.; Cox, M.M. Princípios de Bioquímica de Lehninger; Artmed Editora: Porto Alegre, Brazil, 2022. [Google Scholar]
- Hamel, R.D.; Appanna, V.D. Modulation of TCA cycle enzymes and aluminum stress in Pseudomonas fluorescens. J. Inorg. Biochem. 2001, 87, 1–8. [Google Scholar] [CrossRef]
- Kovacic, P. Mechanism of drug and toxic actions of gossypol: Focus on reactive oxygen species and electron transfer. Curr. Med. Chem. 2003, 10, 2711–2718. [Google Scholar] [CrossRef] [PubMed]
- Santana, A.T.; Guelfi, M.; Medeiros, H.C.; Tavares, M.A.; Bizerra, P.F.; Mingatto, F.E. Mechanisms involved in reproductive damage caused by gossypol in rats and protective effects of vitamin E. Biol. Res. 2015, 48, 43. [Google Scholar] [CrossRef] [PubMed]
- Grewal, J.; Tiwari, R.; Khare, S. Secretome analysis and bioprospecting of lignocellulolytic fungal consortium for valorization of waste cottonseed cake by hydrolase production and simultaneous gossypol degradation. Waste Biomass Valoriz. 2020, 11, 2533–2548. [Google Scholar] [CrossRef]
- Leonowicz, A.; Cho, N.; Luterek, J.; Wilkolazka, A.; Wojtas-Wasilewska, M.; Matuszewska, A.; Hofrichter, M.; Wesenberg, D.; Rogalski, J. Fungal laccase: Properties and activity on lignin. J. Basic Microbiol. 2001, 41, 185–227. [Google Scholar] [CrossRef] [PubMed]
- Claus, H.; Filip, Z. Degradation and transformation of aquatic humic substances by laccase-producing fungi Cladosporium cladosporioides and Polyporus versicolor. Acta Hydrochim. Hydrobiol. 1998, 26, 180–185. [Google Scholar] [CrossRef]
- Strong, P.; Claus, H. Laccase: A review of its past and its future in bioremediation. Crit. Rev. Environ. Sci. Technol. 2011, 41, 373–434. [Google Scholar] [CrossRef]
- Wang, L.; Chen, M.; Luo, X.; Fan, Y.; Zheng, Z.; He, Z.; Yin, R.; Meng, T.; Xu, S.; Pan, Y. Intramolecular annulation of gossypol by laccase to produce safe cottonseed protein. Front. Chem. 2020, 8, 583176. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.-L.; Miao, H.-B.; Wu, Q.; Huang, Z.-X. Heterologous Expression, Enzymatic Characterization of Laccase BmLac and Degradation of Gossypol by It. Biotechnol. Bull. 2023, 39, 320. [Google Scholar]
- Zhang, L.; Zheng, H.; Zhang, X.; Chen, X.; Liu, Y.; Tang, Y.; Zhang, W.; Wang, Z.; Zhao, L.; Guo, Y. Effective Degradation of Free Gossypol in Defatted Cottonseed Meal by Bacterial Laccases: Performance and Toxicity Analysis. Foods 2024, 13, 566. [Google Scholar] [CrossRef]
- Denisov, I.G.; Makris, T.M.; Sligar, S.G.; Schlichting, I. Structure and chemistry of cytochrome P450. Chem. Rev. 2005, 105, 2253–2278. [Google Scholar] [CrossRef]
- Hafeez, M.; Liu, S.; Jan, S.; Shi, L.; Fernández-Grandon, G.M.; Gulzar, A.; Ali, B.; Rehman, M.; Wang, M. Knock-down of gossypol-inducing cytochrome P450 genes reduced deltamethrin sensitivity in Spodoptera exigua (Hübner). Int. J. Mol. Sci. 2019, 20, 2248. [Google Scholar] [CrossRef] [PubMed]
- Krempl, C.; Heidel-Fischer, H.M.; Jiménez-Alemán, G.H.; Reichelt, M.; Menezes, R.C.; Boland, W.; Vogel, H.; Heckel, D.G.; Joußen, N. Gossypol toxicity and detoxification in Helicoverpa armigera and Heliothis virescens. Insect Biochem. Mol. 2016, 78, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Song, Y.; Zeng, R. The role of cytochrome P450-mediated detoxification in insect adaptation to xenobiotics. Curr. Opin. Insect Sci. 2021, 43, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Tolleson, W.H.; Yu, D.; Chen, S.; Guo, L.; Xiao, W.; Tong, W.; Ning, B. Regulation of cytochrome P450 expression by microRNAs and long noncoding RNAs: Epigenetic mechanisms in environmental toxicology and carcinogenesis. J. Environ. Sci. Health C Toxicol. Carcinog. 2019, 37, 180–214. [Google Scholar] [CrossRef]
- Upadhyay, P.; Shrivastava, R.; Agrawal, P.K. Bioprospecting and biotechnological applications of fungal laccase. 3 Biotech 2016, 6, 15. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neto, C.B.S.; Gomes, T.G.; Filho, E.X.F.; Fontes, W.; Ricart, C.A.O.; de Almeida, J.R.M.; de Siqueira, F.G.; Miller, R.N.G. An Enzymatic and Proteomic Analysis of Panus lecomtei during Biodegradation of Gossypol in Cottonseed. J. Fungi 2024, 10, 321. https://doi.org/10.3390/jof10050321
Neto CBS, Gomes TG, Filho EXF, Fontes W, Ricart CAO, de Almeida JRM, de Siqueira FG, Miller RNG. An Enzymatic and Proteomic Analysis of Panus lecomtei during Biodegradation of Gossypol in Cottonseed. Journal of Fungi. 2024; 10(5):321. https://doi.org/10.3390/jof10050321
Chicago/Turabian StyleNeto, Clemente Batista Soares, Taísa Godoy Gomes, Edivaldo Ximenes Ferreira Filho, Wagner Fontes, Carlos André Ornelas Ricart, João Ricardo Moreira de Almeida, Félix Gonçalves de Siqueira, and Robert Neil Gerard Miller. 2024. "An Enzymatic and Proteomic Analysis of Panus lecomtei during Biodegradation of Gossypol in Cottonseed" Journal of Fungi 10, no. 5: 321. https://doi.org/10.3390/jof10050321
APA StyleNeto, C. B. S., Gomes, T. G., Filho, E. X. F., Fontes, W., Ricart, C. A. O., de Almeida, J. R. M., de Siqueira, F. G., & Miller, R. N. G. (2024). An Enzymatic and Proteomic Analysis of Panus lecomtei during Biodegradation of Gossypol in Cottonseed. Journal of Fungi, 10(5), 321. https://doi.org/10.3390/jof10050321







