Strategies for the Enhancement of Secondary Metabolite Production via Biosynthesis Gene Cluster Regulation in Aspergillus oryzae
Abstract
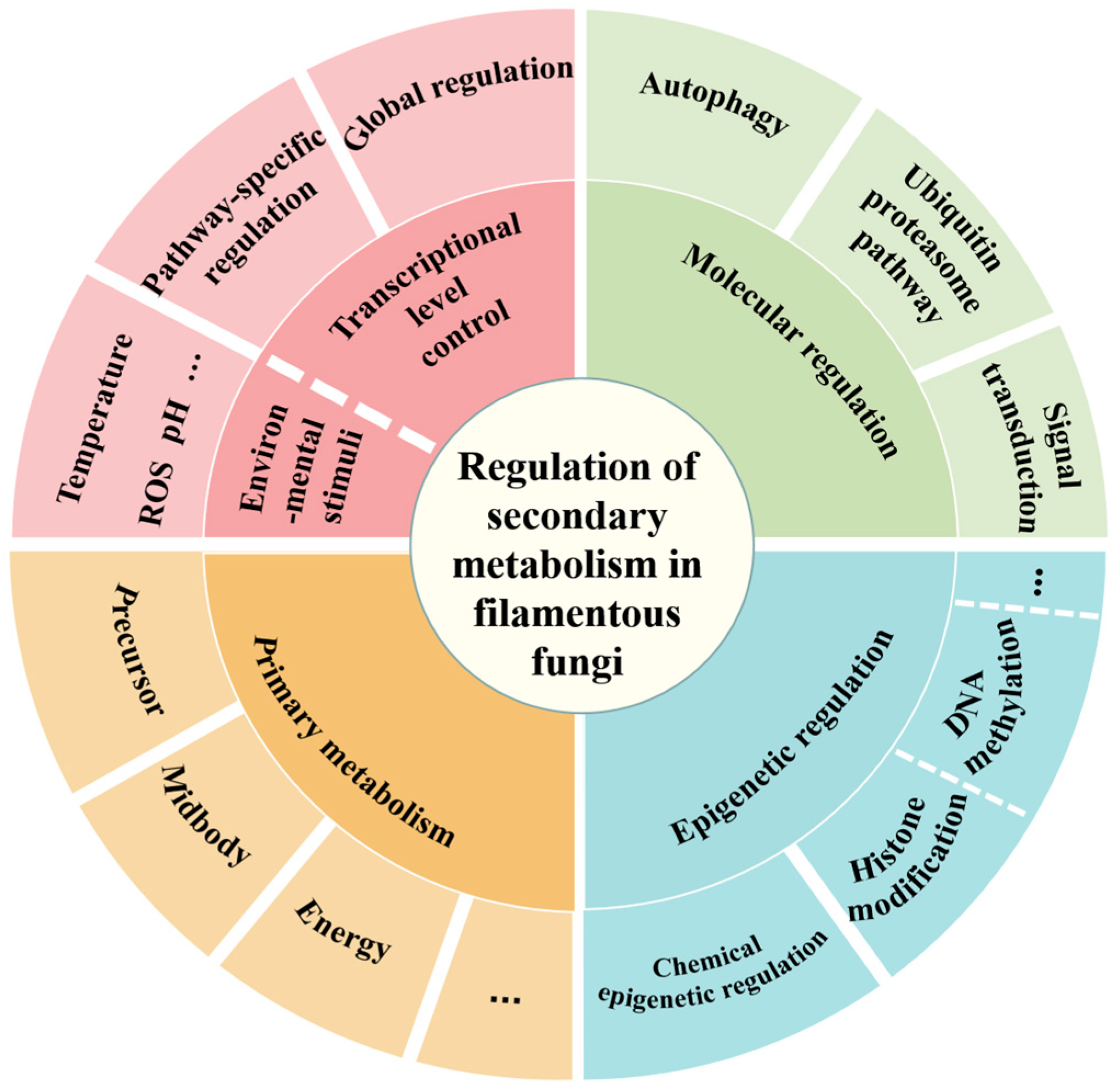
1. Introduction
| Species | Chromosome Number | Genome Size (Mb) | GC Content (%) | Gene Annotation | Reference |
|---|---|---|---|---|---|
| Aspergillus oryzae | 8 | 37.9 | 48.2 | 13120 | [7] |
| Aspergillus nidulans | 8 | 31 | 50.4 | 10560 | [8] |
| Aspergillus fumigatus | 8 | 29.4 | 49.9 | 9926 | [39] |
| Saccharomyces cerevisiae | 16 | 12.1 | 38.3 | 5885 | [40] |
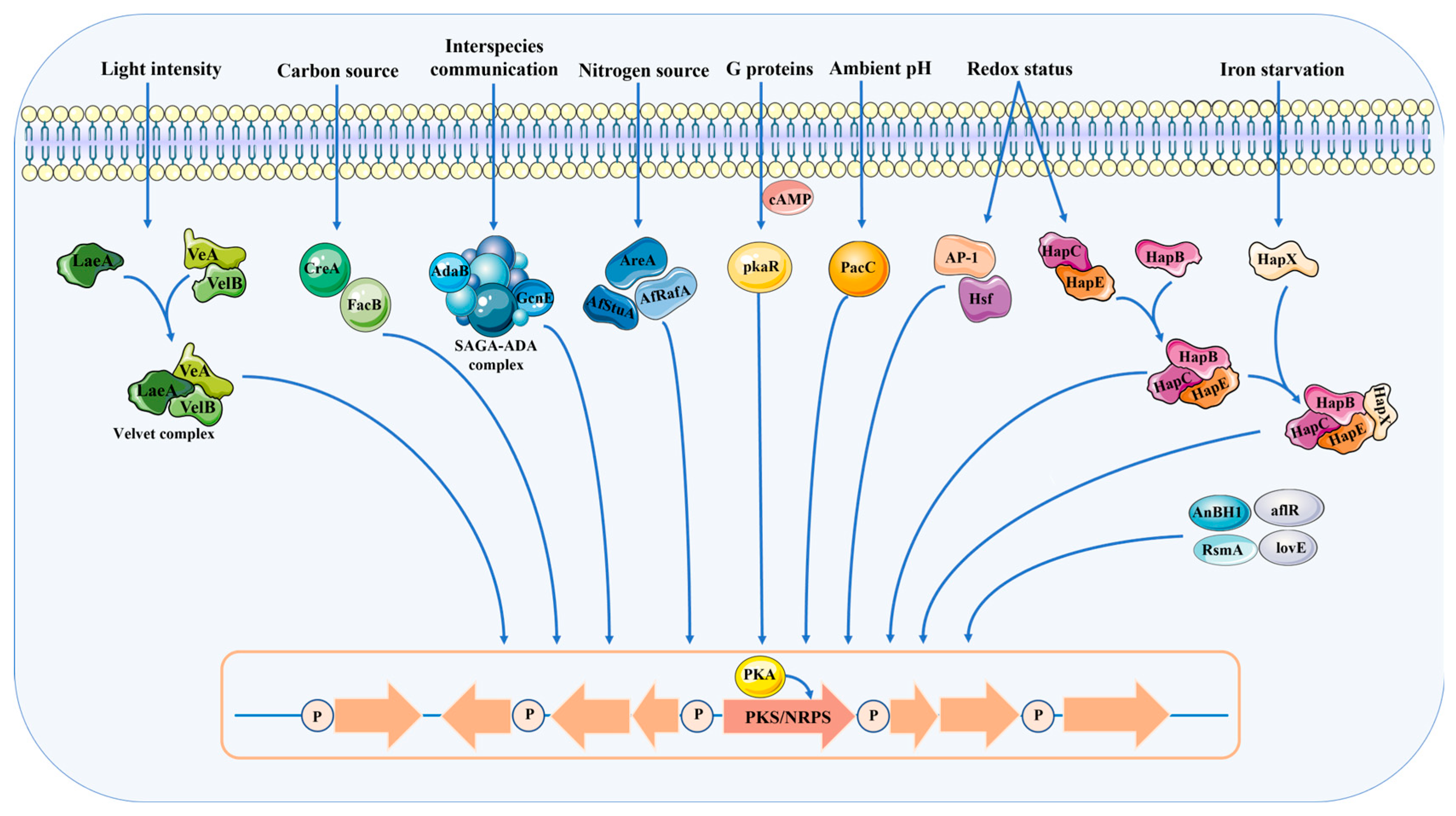
2. Transcriptional Regulation of Secondary Metabolism in A. oryzae
2.1. Global Regulatory Factors
2.2. Pathway-Specific Regulators
3. Epigenetic Regulations of Secondary Metabolism in A. oryzae
3.1. Effect of Epigenetic-Related Genes on Secondary Metabolism
3.2. Effect of Chemical Epigenetic Agents on Secondary Metabolism
4. Environmental Factor Regulation of Secondary Metabolism in A. oryzae
4.1. The Impact of Carbon Sources
4.2. The Impact of Nitrogen Sources
4.3. The Impact of Temperature
4.4. The Impact of Other Factors
5. Conclusions and Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Oda, K.; Kakizono, D.; Yamada, O.; Iefuji, H.; Akita, O.; Iwashita, K. Proteomic analysis of extracellular proteins from Aspergillus oryzae grown under submerged and solid-state culture conditions. Appl. Environ. Microbiol. 2006, 72, 3448–3457. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Abe, K.; Asai, K.; Gomi, K.; Juvvadi, P.R.; Kato, M.; Kitamoto, K.; Takeuchi, M.; Machida, M. Genomics of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2007, 71, 646–670. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Tu, Y.Y.; Jiang, C.M.; Zhang, Z.; Li, Y.K.; Zeng, B. Functional Genomics of Aspergillus oryzae: Strategies and Progress. Microorganisms 2019, 7, 103. [Google Scholar] [CrossRef] [PubMed]
- Lambre, C.; Baviera, J.M.B.; Bolognesi, C.; Cocconcelli, P.S.; Crebelli, R.; Gott, D.M.; Grob, K.; Lampi, E.; Mengelers, M.; Mortensen, A.; et al. Safety evaluation of the food enzyme phospholipase A1 from the genetically modified Aspergillus oryzae strain NZYM-LJ. Efsa J. 2022, 20, e07381. [Google Scholar] [PubMed]
- Lambree, C.; Baviera, J.M.B.; Bolognesi, C.; Cocconcelli, P.S.; Crebelli, R.; Gott, D.M.; Grob, K.; Lampi, E.; Mengelers, M.; Mortensen, A.; et al. Safety evaluation of the food enzyme phospholipase A1 from the genetically modified Aspergillus oryzae strain NZYM-PP. Efsa J. 2023, 21, e07835. [Google Scholar]
- Ichishima, E. Development of enzyme technology for Aspergillus oryzae, A. sojae, and A. luchuensis, the national microorganisms of Japan. Biosci. Biotechnol. Biochem. 2016, 80, 1681–1692. [Google Scholar] [CrossRef] [PubMed]
- Machida, M.; Asai, K.; Sano, M.; Tanaka, T.; Kumagai, T.; Terai, G.; Kusumoto, K.I.; Arima, T.; Akita, O.; Kashiwagi, Y.; et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 2005, 438, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Galagan, J.E.; Calvo, S.E.; Cuomo, C.; Ma, L.J.; Wortman, J.R.; Batzoglou, S.; Lee, S.I.; Basturkmen, M.; Spevak, C.C.; Clutterbuck, J.; et al. Sequencing of Aspergillus nidulans and comparative analysis with A-fumigatus and A-oryzae. Nature 2005, 438, 1105–1115. [Google Scholar] [CrossRef]
- Alberti, F.; Foster, G.D.; Bailey, A.M. Natural products from filamentous fungi and production by heterologous expression. Appl. Microbiol. Biotechnol. 2017, 101, 493–500. [Google Scholar] [CrossRef]
- Liu, L.; Feizi, A.; Osterlund, T.; Hjort, C.; Nielsen, J. Genome-scale analysis of the high-efficient protein secretion system of Aspergillus oryzae. BMC Syst. Biol. 2014, 8, 73. [Google Scholar] [CrossRef]
- Oikawa, H. Reconstitution of biosynthetic machinery of fungal natural products in heterologous hosts. Biosci. Biotechnol. Biochem. 2020, 84, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Jun, S.C.; Han, K.H.; Hong, S.B.; Yu, J.H. Diversity, Application, and Synthetic Biology of Industrially Important Aspergillus Fungi. In Advances in Applied Microbiology; Sariaslani, S., Gadd, G.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; Volume 100, pp. 161–202. [Google Scholar]
- Oikawa, H. Heterologous production of fungal natural products: Reconstitution of biosynthetic gene clusters in model host Aspergillus oryzae. Proc. Jpn. Acad. Ser. B-Phys. Biol. Sci. 2020, 96, 420–430. [Google Scholar] [CrossRef]
- Maruyama, J. Genome Editing Technology and Its Application Potentials in the Industrial Filamentous Fungus Aspergillus oryzae. J. Fungi 2021, 7, 638. [Google Scholar] [CrossRef]
- Yamashita, N.; Komori, Y.; Okumura, Y.; Uchiya, K.; Matsui, T.; Nishimura, A.; Ogawa, K.; Nikai, T. High-yields heterologous production of the novel Aspergillus fumigatus elastase inhibitor AFUEI in Aspergillus oryzae. J. Biosci. Bioeng. 2011, 112, 114–117. [Google Scholar] [CrossRef]
- Shenouda, M.L.; Ambilika, M.; Cox, R.J. Trichoderma reesei Contains a Biosynthetic Gene Cluster That Encodes the Antifungal Agent Ilicicolin H. J. Fungi 2021, 7, 1034. [Google Scholar] [CrossRef]
- Dufour, N.; Rao, R.P. Secondary metabolites and other small molecules as intercellular pathogenic signals. FEMS Microbiol. Lett. 2011, 314, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.P.D.; Carvalho, A.S.C.; Santos, A.S.; Alves, C.N.; do Nascimento, J.L.M.; Silva, E.O. Kojic acid, a secondary metabolite from Aspergillus sp., acts as an inducer of macrophage activation. Cell Biol. Int. 2011, 35, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Rohlfs, M.; Churchill, A.C.L. Fungal secondary metabolites as modulators of interactions with insects and other arthropods. Fungal Genet. Biol. 2011, 48, 23–34. [Google Scholar] [CrossRef]
- Vining, L.C. Secondary metabolism, inventive evolution and biochemical diversity—A review. Gene 1992, 115, 135–140. [Google Scholar] [CrossRef]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism—From biochemistry to genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar] [CrossRef]
- Shwab, E.K.; Keller, N.P. Regulation of secondary metabolite production in filamentous ascomycetes. Mycol. Res. 2008, 112, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Tokuoka, M.; Seshime, Y.; Fujii, I.; Kitamoto, K.; Takahashi, T.; Koyama, Y. Identification of a novel polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) gene required for the biosynthesis of cyclopiazonic acid in Aspergillus oryzae. Fungal Genet. Biol. 2008, 45, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Orth, R. Mycotoxins of Aspergillus oryzae strains for use in the food industry as starters and enzyme producing molds. Ann. Nutr. Aliment. 1977, 31, 617–624. [Google Scholar]
- Iizuka, H.; Iida, M. Maltoryzine, a new toxic metabolite produced by a strain of Aspergillus oryzae var. microsporus isolated from the poisonous malt sprout. Nature 1962, 196, 681–682. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, T.; Oka, H.; Goto, R.; Tsurigami, R.; Maruyama, J.; Shimizu, M.; Kato, M.; Nakano, H.; Kojima, T. The Identification of a Target Gene of the Transcription Factor KojR and Elucidation of Its Role in Carbon Metabolism for Kojic Acid Biosynthesis in Aspergillus oryzae. J. Fungi 2024, 10, 113. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, M.; Eslamifar, M.; Khezri, K. Kojic acid applications in cosmetic and pharmaceutical preparations. Biomed. Pharmacother. 2019, 110, 582–593. [Google Scholar] [CrossRef] [PubMed]
- Chib, S.; Jamwal, V.L.; Kumar, V.; Gandhi, S.G.; Saran, S. Fungal production of kojic acid and its industrial applications. Appl. Microbiol. Biotechnol. 2023, 107, 2111–2130. [Google Scholar] [CrossRef] [PubMed]
- Zilles, J.C.; dos Santos, F.L.; Kulkamp-Guerreiro, I.C.; Contri, R.V. Biological activities and safety data of kojic acid and its derivatives: A review. Exp. Dermatol. 2022, 31, 1500–1521. [Google Scholar] [CrossRef] [PubMed]
- Brtko, J. Biological functions of kojic acid and its derivatives in medicine, cosmetics, and food industry: Insights into health aspects. Arch. Pharm. 2022, 355, 2200215. [Google Scholar] [CrossRef]
- Zhou, M.; Zhou, K.; He, P.; Wang, K.M.; Zhu, R.Z.; Wang, Y.D.; Dong, W.; Li, G.P.; Yang, H.Y.; Ye, Y.Q.; et al. Antiviral and Cytotoxic Isocoumarin Derivatives from an Endophytic Fungus Aspergillus oryzae. Planta Medica 2016, 82, 414–417. [Google Scholar] [CrossRef]
- Usov, A.N.; Blanko, F.F.; Ivanova, V.S.; Bedrina, E.N.; Firsova, S.A.; Sedakova, L.A.; Funtikova, N.S. Structure and antitumor activity of polysaccharides from the micelles of Aspergillus oryzae. Bioorganicheskaia Khimiia 1991, 17, 121–125. [Google Scholar]
- Li, M.F.; Xiao, D.; Zhu, L.C.; Liu, L.; Zheng, J.N.; Gu, X.J.; Zhu, Y.N.; Xie, J.; Wang, X.; Dai, J.M.; et al. Indole Alkaloids from the Cigar Tobacco-Derived Endophytic Fungus Aspergillus oryzae and Their Antibacterial Activity. Chem. Nat. Compd. 2022, 58, 1093–1097. [Google Scholar] [CrossRef]
- Sakata, K.; Kuwatsuka, T.; Sakurai, A.; Takahashi, N.; Tamura, G. Isolation of Aspirochlorine (=Antibiotic A30641) as a True Antimicrobial Constituent of the Antibiotic, Oryzachlorin, from Aspergillus oryzae. J. Agric. Chem. Soc. Jpn. 1983, 47, 2673–2674. [Google Scholar] [CrossRef]
- Nonaka, N.; Asai, Y.; Nishio, M.; Takahashi, K.; Okuda, T.; Tanaka, S.; Sugita, T.; Ohnuki, T.; Komatsubara, S. TMC-2A, -2B and -2C, novel dipeptidyl peptidase IV inhibitors produced by Aspergillus oryzae A374. 1. Taxonomy of producing strain, fermentation, and biochemical properties. J. Antibiot. 1997, 50, 646–652. [Google Scholar] [CrossRef][Green Version]
- Li, Y.Z.; Zhang, H.X.; Chen, Z.M.; Fan, J.X.; Chen, T.M.; Xiao, Y.; Jie, J.Y.; Zeng, B.; Zhang, Z. Overexpression of a novel gene Aokap2 affects the growth and kojic acid production in Aspergillus oryzae. Mol. Biol. Rep. 2022, 49, 2745–2754. [Google Scholar] [CrossRef]
- Tamano, K.; Kuninaga, M.; Kojima, N.; Umemura, M.; Machida, M.; Koike, H. Use of the kojA promoter, involved in kojic acid biosynthesis, for polyketide production in Aspergillus oryzae: Implications for long-term production. BMC Biotechnol. 2019, 19, 70. [Google Scholar] [CrossRef]
- Marui, J.; Ohashi-Kunihiro, S.; Ando, T.; Nishimura, M.; Koike, H.; Machida, M. Penicillin biosynthesis in Aspergillus oryzae and its overproduction by genetic engineering. J. Biosci. Bioeng. 2010, 110, 8–11. [Google Scholar] [CrossRef]
- Nierman, W.C.; Pain, A.; Anderson, M.J.; Wortman, J.R.; Kim, H.S.; Arroyo, J.; Berriman, M.; Abe, K.; Archer, D.B.; Bermejo, C.; et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 2005, 438, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M.; et al. Life with 6000 genes. Science 1996, 274, 563–567. [Google Scholar] [CrossRef]
- Feng, J.; Hauser, M.; Cox, R.J.; Skellam, E. Engineering Aspergillus oryzae for the Heterologous Expression of a Bacterial Modular Polyketide Synthase. J. Fungi 2021, 7, 1085. [Google Scholar] [CrossRef]
- Zhang, W.Y.; Zhong, Y.; Yu, Y.; Shi, D.F.; Huang, H.Y.; Tang, X.L.; Wang, Y.H.; Chen, G.D.; Zhang, H.P.; Liu, C.L.; et al. 4-Hydroxy Pyridones from Heterologous Expression and Cultivation of the Native Host. J. Nat. Prod. 2020, 83, 3338–3346. [Google Scholar] [CrossRef]
- Guo, J.J.; Cai, Y.S.; Cheng, F.C.; Yang, C.J.; Zhang, W.Q.; Yu, W.L.; Yan, J.J.; Deng, Z.X.; Hong, K. Genome Mining Reveals a Multiproduct Sesterterpenoid Biosynthetic Gene Cluster in Aspergillus ustus. Org. Lett. 2021, 23, 1525–1529. [Google Scholar] [CrossRef]
- Takusagawa, S.; Satoh, Y.; Ohtsu, I.; Dairi, T. Ergothioneine production with Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2019, 83, 181–184. [Google Scholar] [CrossRef]
- Bok, J.W.; Keller, N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 2004, 3, 527–535. [Google Scholar] [CrossRef]
- Bok, J.W.; Balajee, S.A.; Marr, K.A.; Andes, D.; Nielsen, K.F.; Frisvad, J.C.; Keller, N.P. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell 2005, 4, 1574–1582. [Google Scholar] [CrossRef]
- Jain, S.; Keller, N. Insights to fungal biology through LaeA sleuthing. Fungal Biol. Rev. 2013, 27, 51–59. [Google Scholar] [CrossRef]
- Bayram, O.; Braus, G.H. Coordination of secondary metabolism and development in fungi: The velvet family of regulatory proteins. FEMS Microbiol. Rev. 2012, 36, 1–24. [Google Scholar] [CrossRef]
- Jiang, T.; Wang, M.H.; Li, L.; Si, J.G.; Song, B.; Zhou, C.; Yu, M.; Wang, X.W.; Zhang, Y.G.; Ding, G.; et al. Overexpression of the Global Regulator LaeA in Chaetomium globosum Leads to the Biosynthesis of Chaetoglobosin Z. J. Nat. Prod. 2016, 79, 2487–2494. [Google Scholar] [CrossRef]
- Kawauchi, M.; Nishiura, M.; Iwashita, K. Fungus-Specific Sirtuin HstD Coordinates Secondary Metabolism and Development through Control of LaeA. Eukaryot. Cell 2013, 12, 1087–1096. [Google Scholar] [CrossRef]
- Feng, W.M.; Liang, J.R.; Wang, B.B.; Chen, J.H. Improvement of kojic acid production in Aspergillus oryzae AR-47 mutant strain by combined mutagenesis. Bioprocess Biosyst. Eng. 2019, 42, 753–761. [Google Scholar] [CrossRef]
- Terabayashi, Y.; Sano, M.; Yamane, N.; Marui, J.; Tamano, K.; Sagara, J.; Dohmoto, M.; Oda, K.; Ohshima, E.; Tachibana, K.; et al. Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal Genet. Biol. 2010, 47, 953–961. [Google Scholar] [CrossRef]
- Marui, J.; Yamane, N.; Ohashi-Kunihiro, S.; Ando, T.; Terabayashi, Y.; Sano, M.; Ohashi, S.; Ohshima, E.; Tachibana, K.; Higa, Y. Kojic acid biosynthesis in Aspergillus oryzae is regulated by a Zn(II)(2)Cys(6) transcriptional activator and induced by kojic acid at the transcriptional level. J. Biosci. Bioeng. 2011, 112, 40–43. [Google Scholar] [CrossRef]
- Calvo, A.M. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet. Biol. 2008, 45, 1053–1061. [Google Scholar] [CrossRef]
- Shin, K.S.; Kim, Y.H.; Yu, J.H. Proteomic analyses reveal the key roles of BrlA and AbaA in biogenesis of gliotoxin in Aspergillus fumigatus. Biochem. Biophys. Res. Commun. 2015, 463, 428–433. [Google Scholar] [CrossRef]
- Jia, L.L.; Yu, J.H.; Chen, F.S.; Chen, W.P. Characterization of the asexual developmental genes brlA and wetA in Monascus ruber M7. Fungal Genet. Biol. 2021, 151, 103564. [Google Scholar] [CrossRef]
- Arakawa, G.Y.; Kudo, H.; Yanase, A.; Eguchi, Y.; Kodama, H.; Ogawa, M.; Koyama, Y.; Shindo, H.; Hosaka, M.; Tokuoka, M. A unique Zn(II)(2)-Cys(6)-type protein, KpeA, is involved in secondary metabolism and conidiation in Aspergillus oryzae. Fungal Genet. Biol. 2019, 127, 35–44. [Google Scholar] [CrossRef]
- Watanabe, M.; Watanabe, D.; Akao, T.; Shimoi, H. Overexpression of MSN2 in a sake yeast strain promotes ethanol tolerance and increases ethanol production in sake brewing. J. Biosci. Bioeng. 2009, 107, 516–518. [Google Scholar] [CrossRef]
- Chang, P.-K.; Scharfenstein, L.L.; Luo, M.; Mahoney, N.; Molyneux, R.J.; Yu, J.; Brown, R.L.; Campbell, B.C. Loss of msnA, a Putative Stress Regulatory Gene, in Aspergillus parasiticus and Aspergillus flavus Increased Production of Conidia, Aflatoxins and Kojic Acid. Toxins 2011, 3, 82–104. [Google Scholar] [CrossRef]
- Kim, N.Y.; Lee, J.H.; Lee, I.; Ji, G.E. An Evaluation of Aflatoxin and Cyclopiazonic Acid Production in Aspergillus oryzae. J. Food Prot. 2014, 77, 1010–1016. [Google Scholar] [CrossRef]
- Saha, P.; Ghosh, S.; Roy-Barman, S. MoLAEA Regulates Secondary Metabolism in Magnaporthe oryzae. mSphere 2020, 5, e00936-19. [Google Scholar] [CrossRef]
- Wang, B.; Guo, G.W.; Wang, C.; Lin, Y.; Wang, X.N.; Zhao, M.M.; Guo, Y.; He, M.H.; Zhang, Y.; Pan, L. Survey of the transcriptome of Aspergillus oryzae via massively parallel mRNA sequencing. Nucleic Acids Res. 2010, 38, 5075–5087. [Google Scholar] [CrossRef]
- Shimizu, T.; Kinoshita, H.; Nihira, T. Identification and in vivo functional analysis by gene disruption of ctnA, an activator gene involved in citrinin biosynthesis in Monascus purpureus. Appl. Environ. Microbiol. 2007, 73, 5097–5103. [Google Scholar] [CrossRef]
- Sakai, K.; Kinoshita, H.; Shimizu, T.; Nihira, T. Construction of a Citrinin Gene Cluster Expression System in Heterologous Aspergillus oryzae. J. Biosci. Bioeng. 2008, 106, 466–472. [Google Scholar] [CrossRef]
- Aghcheh, R.K.; Kubicek, C.P. Epigenetics as an emerging tool for improvement of fungal strains used in biotechnology. Appl. Microbiol. Biotechnol. 2015, 99, 6167–6181. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Schroeckh, V. Fungal secondary metabolites—Strategies to activate silent gene clusters. Fungal Genet. Biol. 2011, 48, 15–22. [Google Scholar] [CrossRef]
- Xue, M.Y.; Hou, X.W.; Fu, J.J.; Zhang, J.Y.; Wang, J.C.; Zhao, Z.T.; Xu, D.; Lai, D.W.; Zhou, L.G. Recent Advances in Search of Bioactive Secondary Metabolites from Fungi Triggered by Chemical Epigenetic Modifiers. J. Fungi 2023, 9, 172. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, Y.; Kawatani, M.; Futamura, Y.; Osada, H.; Koyama, Y. An overproduction of astellolides induced by genetic disruption of chromatin-remodeling factors in Aspergillus oryzae. J. Antibiot. 2016, 69, 4–8. [Google Scholar] [CrossRef]
- Asai, T.; Yamamoto, T.; Shirata, N.; Taniguchi, T.; Monde, K.; Fujii, I.; Gomi, K.; Oshima, Y. Structurally Diverse Chaetophenol Productions Induced by Chemically Mediated Epigenetic Manipulation of Fungal Gene Expression. Org. Lett. 2013, 15, 3346–3349. [Google Scholar] [CrossRef] [PubMed]
- Kawauchi, M.; Iwashita, K. Functional analysis of histone deacetylase and its role in stress response, drug resistance and solid-state cultivation in Aspergillus oryzae. J. Biosci. Bioeng. 2014, 118, 172–176. [Google Scholar] [CrossRef]
- Gorog, S. Advances in the analysis of steroid hormone drugs in pharmaceuticals and environmental samples (2004–2010). J. Pharm. Biomed. Anal. 2011, 55, 728–743. [Google Scholar] [CrossRef]
- Hu, Z.H.; He, B.; Ma, L.; Sun, Y.L.; Niu, Y.L.; Zeng, B. Recent Advances in Ergosterol Biosynthesis and Regulation Mechanisms in Saccharomyces cerevisiae. Indian J. Microbiol. 2017, 57, 270–277. [Google Scholar] [CrossRef]
- Kristan, K.; Rizner, T.L. Steroid-transforming enzymes in fungi. J. Steroid Biochem. Mol. Biol. 2012, 129, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Mercer, E.I. Inhibitors of sterol biosynthesis and their applications. Prog. Lipid Res. 1993, 32, 357–416. [Google Scholar] [CrossRef] [PubMed]
- Ryder, N.S. Terbinafine: Mode of action and properties of the squalene epoxidase inhibition. Br. J. Dermatol. 1992, 126 (Suppl. 39), 2–7. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.H.; Li, G.H.; Sun, Y.L.; Niu, Y.L.; Ma, L.; He, B.; Ai, M.Q.; Han, J.Z.; Zeng, B. Gene transcription profiling of Aspergillus oryzae 3.042 treated with ergosterol biosynthesis inhibitors. Braz. J. Microbiol. 2019, 50, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Kim, J.; Liew, M.; Yan, J.K.; Herrera, O.; Bok, J.W.; Kelleher, N.L.; Keller, N.P.; Wang, Y. Redox Metabolites Signal Polymicrobial Biofilm Development via the NapA Oxidative Stress Cascade in Aspergillus. Curr. Biol. 2015, 25, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Caceres, I.; Al Khoury, A.; El Khoury, R.; Lorber, S.; Oswald, I.P.; El Khoury, A.; Atoui, A.; Puel, O.; Bailly, J.D. Aflatoxin Biosynthesis and Genetic Regulation: A Review. Toxins 2020, 12, 150. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jiang, Y.R.; Ma, L.X.; Ma, X.Y.; Liu, Y.; Shan, J.H.; Ma, K.; Xing, F.G. Comprehensive Transcriptome and Proteome Analyses Reveal the Modulation of Aflatoxin Production byAspergillus flavuson Different Crop Substrates. Front. Microbiol. 2020, 11, 1497. [Google Scholar]
- Si, P.D.; Wang, G.; Wu, W.Q.; Hussain, S.; Guo, L.; Wu, W.; Yang, Q.L.; Xing, F.G. SakA Regulates Morphological Development, Ochratoxin A Biosynthesis and Pathogenicity of Aspergillus westerdijkiae and the Response to Different Environmental Stresses. Toxins 2023, 15, 292. [Google Scholar] [CrossRef]
- Daba, G.M.; Mostafa, F.A.; Elkhateeb, W.A. The ancient koji mold (Aspergillus oryzae) as a modern biotechnological tool. Bioresour. Bioprocess. 2021, 8, 52. [Google Scholar] [CrossRef]
- de Vries, R.P.; Riley, R.; Wiebenga, A.; Aguilar-Osorio, G.; Amillis, S.; Uchima, C.A.; Anderluh, G.; Asadollahi, M.; Askin, M.; Barry, K.; et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 28. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.M.; Chen, C.C.; Giridhar, R.; Chang, T.S.; Wu, W.T. Repeated-batch production of kojic acid in a cell-retention fermenter using Aspergillus oryae M3B9. J. Ind. Microbiol. Biotechnol. 2005, 32, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Rosfarizan, M.; Ariff, A.B. Kinetics of kojic acid fermentation by Aspergillus flavus using different types and concentrations of carbon and nitrogen sources. J. Ind. Microbiol. Biotechnol. 2000, 25, 20–24. [Google Scholar] [CrossRef]
- Ochsenreither, K.; Fischer, C.; Neumann, A.; Syldatk, C. Process characterization and influence of alternative carbon sources and carbon-to-nitrogen ratio on organic acid production by Aspergillus oryzae DSM1863. Appl. Microbiol. Biotechnol. 2014, 98, 5449–5460. [Google Scholar] [CrossRef] [PubMed]
- Vorapreeda, T.; Khongto, B.; Thammarongtham, C.; Srisuk, T.; Laoteng, K. Metabolic Regulation of Sugar Assimilation for Lipid Production in Aspergillus oryzae BCC7051 through Comparative Transcriptome Perspective. Biology 2021, 10, 885. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.P.; Zhao, J.; Duan, W.L.; Tian, S.; Wang, X.D.; Zhuang, H.; Fu, J.; Kang, Z.S. TaAMT2;3a, a wheat AMT2-type ammonium transporter, facilitates the infection of stripe rust fungus on wheat. BMC Plant Biol. 2019, 19, 239. [Google Scholar] [CrossRef] [PubMed]
- Chutrakul, C.; Panchanawaporn, S.; Vorapreeda, T.; Jeennor, S.; Anantayanon, J.; Laoteng, K. The Exploring Functional Role of Ammonium Transporters of Aspergillus oryzae in Nitrogen Metabolism: Challenges towards Cell Biomass Production. Int. J. Mol. Sci. 2022, 23, 7567. [Google Scholar] [CrossRef] [PubMed]
- Christensen, T.; Hynes, M.J.; Davis, M.A. Role of the regulatory gene areA of Aspergillus oryzae in nitrogen metabolism. Appl. Environ. Microbiol. 1998, 64, 3232–3237. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.A.; Arst, H.N., Jr. Mutational analysis of AREA, a transcriptional activator mediating nitrogen metabolite repression in Aspergillus nidulans and a member of the “streetwise” GATA family of transcription factors. Microbiol. Mol. Biol. Rev. 1998, 62, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Chutrakul, C.; Panchanawaporn, S.; Jeennor, S.; Anantayanon, J.; Laoteng, K. Promoter exchange of the cryptic nonribosomal peptide synthetase gene for oligopeptide production in Aspergillus oryzae. J. Microbiol. 2022, 60, 47–56. [Google Scholar] [CrossRef]
- Dalirfardouei, R.; Karimi, G.; Jamialahmadi, K. Molecular mechanisms and biomedical applications of glucosamine as a potential multifunctional therapeutic agent. Life Sci. 2016, 152, 21–29. [Google Scholar] [CrossRef]
- Li, J.S.; Chew, Y.M.; Lin, M.C.; Lau, Y.Q.; Chen, C.S. Enhanced glucosamine production from Aspergillus oryzae NCH-42 via acidic stress under submerged fermentation. Cyta-J. Food 2021, 19, 614–624. [Google Scholar] [CrossRef]
- Wattanachaisaereekul, S.; Tachaleat, A.; Punya, J.; Haritakun, R.; Boonlarppradab, C.; Cheevadhanarak, S. Assessing medium constituents for optimal heterologous production of anhydromevalonolactone in recombinant Aspergillus oryzae. Amb Express 2014, 4, 52. [Google Scholar] [CrossRef]
- Sano, M. Aspergillus oryzae nrtA affects kojic acid production. Biosci. Biotechnol. Biochem. 2016, 80, 1776–1780. [Google Scholar] [CrossRef]
- Jiang, C.M.; Ge, J.X.; He, B.; Zhang, Z.; Hu, Z.H.; Li, Y.K.; Zeng, B. Transcriptomic analysis reveals Aspergillus oryzae responds to temperature stress by regulating sugar metabolism and lipid metabolism. PLoS ONE 2022, 17, e0274394. [Google Scholar] [CrossRef]
- Machida, M.; Yamada, O.; Gomi, K. Genomics of Aspergillus oryzae: Learning from the History of Koji Mold and Exploration of Its Future. DNA Res. 2008, 15, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Umemura, M.; Koike, H.; Yamane, N.; Koyama, Y.; Satou, Y.; Kikuzato, I.; Teruya, M.; Tsukahara, M.; Imada, Y.; Wachi, Y.; et al. Comparative Genome Analysis Between Aspergillus oryzae Strains Reveals Close Relationship Between Sites of Mutation Localization and Regions of Highly Divergent Genes among Aspergillus Species. DNA Res. 2012, 19, 375–382. [Google Scholar] [CrossRef]
- Sakurai, Y.; Misawa, S.; Shiota, H. Growth and respiratory activity of aspergillus-oryzae grown on solid-state medium. Agric. Biol. Chem. 1985, 49, 745–750. [Google Scholar] [CrossRef]
- Chen, T.T.; Xiong, S.Q.; Jiang, S.Y.; Wang, M.J.; Wu, Q.L.; Wei, H. Molecular identification of microbial community in Chinese douchi during post-fermentation process. Food Sci. Biotechnol. 2011, 20, 1633–1638. [Google Scholar] [CrossRef]
- Ge, J.X.; Zhang, Z.; Li, Y.; Hu, Z.H.; He, B.; Li, Y.K.; Zeng, B.; Jiang, C.M. Inhibition of AoAur1 increases mycelial growth, hyphal fusion and improves physiological adaptation to high-temperature stress in Aspergillus oryzae. Arch. Microbiol. 2022, 204, 447. [Google Scholar] [CrossRef]
- Hayakawa, H.; Sobue, F.; Motoyama, K.; Yoshimura, T.; Hemmi, H. Identification of enzymes involved in the mevalonate pathway of Flavobacterium johnsoniae. Biochem. Biophys. Res. Commun. 2017, 487, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Krepkiy, D.; Miziorko, H.M. Identification of active site residues in mevalonate diphosphate decarboxylase: Implications for a family of phosphotransferases. Protein Sci. 2004, 13, 1875–1881. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.H.; Huang, H.; Sun, Y.L.; Niu, Y.L.; Xu, W.Z.S.; Liu, Q.C.; Zhang, Z.; Jiang, C.M.; Li, Y.K.; Zeng, B. Effects on Gene Transcription Profile and Fatty Acid Composition by Genetic Modification of Mevalonate Diphosphate Decarboxylase MVD/Erg19 in Aspergillus oryzae. Microorganisms 2019, 7, 342. [Google Scholar] [CrossRef] [PubMed]
- Cordier, H.; Lacombe, C.; Karst, F.; Berges, T. The Saccharomyces cerevisiae mevalonate diphosphate decarboxylase (Erg19p) forms homodimers in vivo, and a single substitution in a structurally conserved region impairs dimerization. Curr. Microbiol. 1999, 38, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.L.; Niu, Y.L.; Huang, H.; He, B.; Ma, L.; Tu, Y.Y.; Tran, V.T.; Zeng, B.; Hu, Z.H. Mevalonate Diphosphate Decarboxylase MVD/Erg19 Is Required for Ergosterol Biosynthesis, Growth, Sporulation and Stress Tolerance in Aspergillus oryzae. Front. Microbiol. 2019, 10, 1074. [Google Scholar] [CrossRef]
- Alvarado-Obando, M.; Contreras, N.; Leon, D.; Botero, L.; Beltran, L.; Diaz, D.; Rodriguez-Lopez, A.; Reyes, L.H.; Almeciga-Diaz, C.J.; Sanchez, O.F. Engineering a heterologously expressed fructosyltransferase from Aspergillus oryzae N74 in Komagataella phaffii (Pichia pastoris) for kestose production. New Biotechnol. 2022, 69, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Landraud, P.; Chuzeville, S.; Billon-Grande, G.; Poussereau, N.; Bruel, C. Adaptation to pH and Role of PacC in the Rice Blast Fungus Magnaporthe oryzae. PLoS ONE 2013, 8, e69236. [Google Scholar] [CrossRef]
- Barda, O.; Maor, U.; Sadhasivam, S.; Bi, Y.; Zakin, V.; Prusky, D.; Sionov, E. The pH-Responsive Transcription Factor PacC Governs Pathogenicity and Ochratoxin A Biosynthesis in Aspergillus carbonarius. Front. Microbiol. 2020, 11, 210. [Google Scholar] [CrossRef]
- McKelvey, S.M.; Murphy, R.A. Analysis of wide-domain transcriptional regulation in solid-state cultures of Aspergillus oryzae. J. Ind. Microbiol. Biotechnol. 2010, 37, 455–469. [Google Scholar] [CrossRef]
- Shao, H.H.; Tu, Y.Y.; Wang, Y.J.; Jiang, C.M.; Ma, L.; Hu, Z.H.; Wang, J.F.; Zeng, B. Oxidative Stress Response of Aspergillus oryzae Induced by Hydrogen Peroxide and Menadione Sodium Bisulfite. Microorganisms 2019, 7, 225. [Google Scholar] [CrossRef]
- Zhao, G.Z.; Hou, L.H.; Yao, Y.P.; Wang, C.L.; Cao, X.H. Comparative proteome analysis of Aspergillus oryzae 3.042 and A. oryzae 100-8 strains: Towards the production of different soy sauce flavors. J. Proteom. 2012, 75, 3914–3924. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.Z.; Yao, Y.P.; Hao, G.F.; Fang, D.S.; Yin, B.X.; Cao, X.H.; Chen, W. Gene regulation in Aspergillus oryzae promotes hyphal growth and flavor formation in soy sauce koji. Rsc Adv. 2015, 5, 24224–24230. [Google Scholar] [CrossRef]
- Park, M.K.; Seo, J.A.; Kim, Y.S. Comparative study on metabolic changes of Aspergillus oryzae isolated from fermented foods according to culture conditions. Int. J. Food Microbiol. 2019, 307, 108270. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Nakamura, H.; Zhang, Y.; Pascal, A.; Fujii, W.; Maruyama, J. Forced Recycling of an AMA1-Based Genome-Editing Plasmid Allows for Efficient Multiple Gene Deletion/Integration in the Industrial Filamentous Fungus Aspergillus oryzae. Appl. Environ. Microbiol. 2019, 85, e01896-18. [Google Scholar] [CrossRef]
- Katayama, T.; Tanaka, Y.; Okabe, T.; Nakamura, H.; Fujii, W.; Kitamoto, K.; Maruyama, J. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol. Lett. 2016, 38, 637–642. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, X.; Song, J.; Wu, Y.; Feng, S.; Sun, Z.; Hu, Y.; Yu, M.; Han, R.; Zeng, B. Strategies for the Enhancement of Secondary Metabolite Production via Biosynthesis Gene Cluster Regulation in Aspergillus oryzae. J. Fungi 2024, 10, 312. https://doi.org/10.3390/jof10050312
Jia X, Song J, Wu Y, Feng S, Sun Z, Hu Y, Yu M, Han R, Zeng B. Strategies for the Enhancement of Secondary Metabolite Production via Biosynthesis Gene Cluster Regulation in Aspergillus oryzae. Journal of Fungi. 2024; 10(5):312. https://doi.org/10.3390/jof10050312
Chicago/Turabian StyleJia, Xiao, Jiayi Song, Yijian Wu, Sai Feng, Zeao Sun, Yan Hu, Mengxue Yu, Rui Han, and Bin Zeng. 2024. "Strategies for the Enhancement of Secondary Metabolite Production via Biosynthesis Gene Cluster Regulation in Aspergillus oryzae" Journal of Fungi 10, no. 5: 312. https://doi.org/10.3390/jof10050312
APA StyleJia, X., Song, J., Wu, Y., Feng, S., Sun, Z., Hu, Y., Yu, M., Han, R., & Zeng, B. (2024). Strategies for the Enhancement of Secondary Metabolite Production via Biosynthesis Gene Cluster Regulation in Aspergillus oryzae. Journal of Fungi, 10(5), 312. https://doi.org/10.3390/jof10050312









