Current Advances in the Functional Genes of Edible and Medicinal Fungi: Research Techniques, Functional Analysis, and Prospects
Abstract
1. Introduction
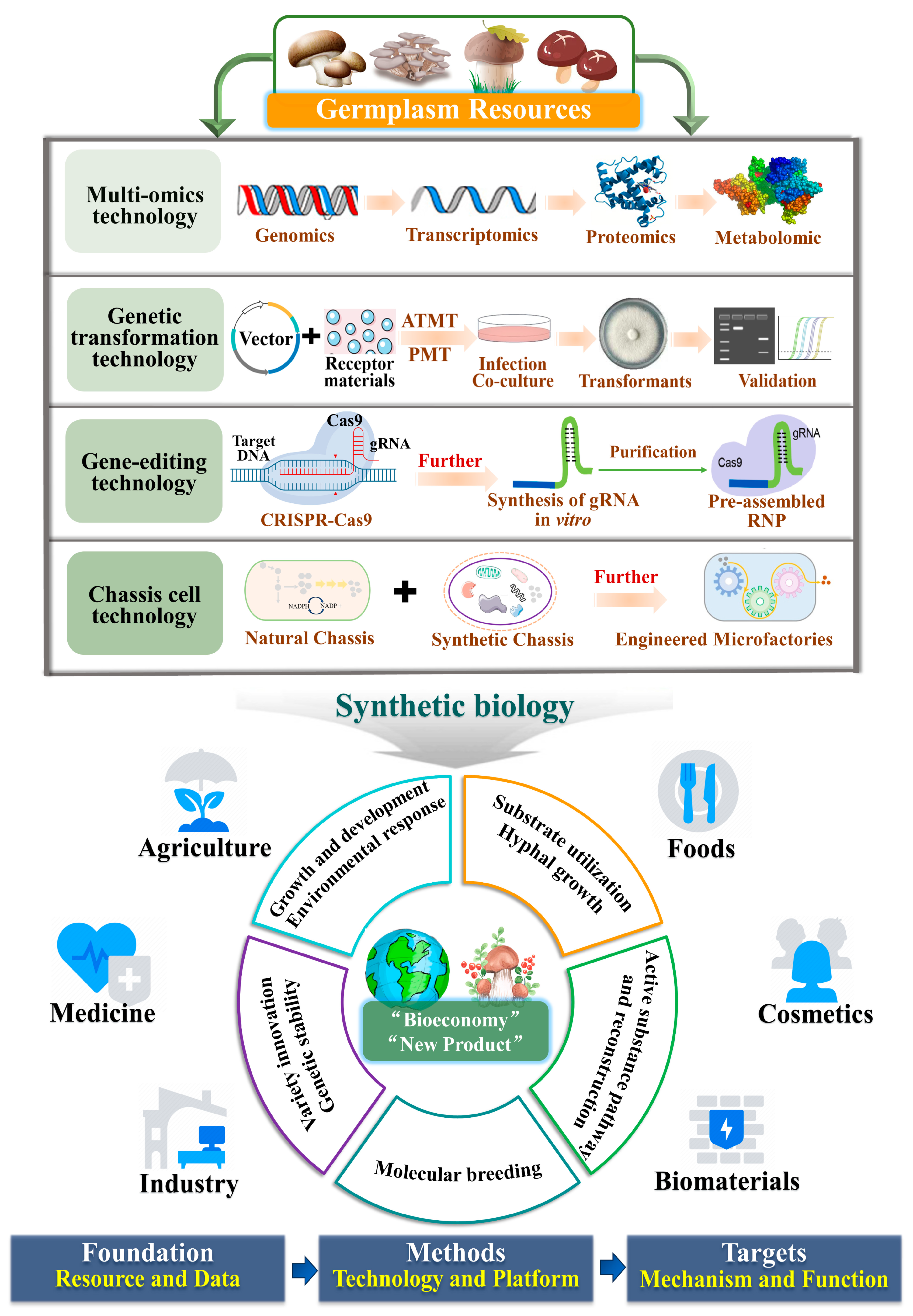
2. Advances in Functional Gene Research Techniques in Edible and Medicinal Fungi
2.1. Multi-Omics Technology
2.2. Molecular Genetic Techniques in Edible and Medicinal Fungi
2.2.1. Genetic Transformation Technology
2.2.2. Gene-Editing Technology
2.3. Chassis Cell Technology
2.4. Synthetic Biology
3. Advances in the Analysis of Important Functional Genes of Edible and Medicinal Fungi
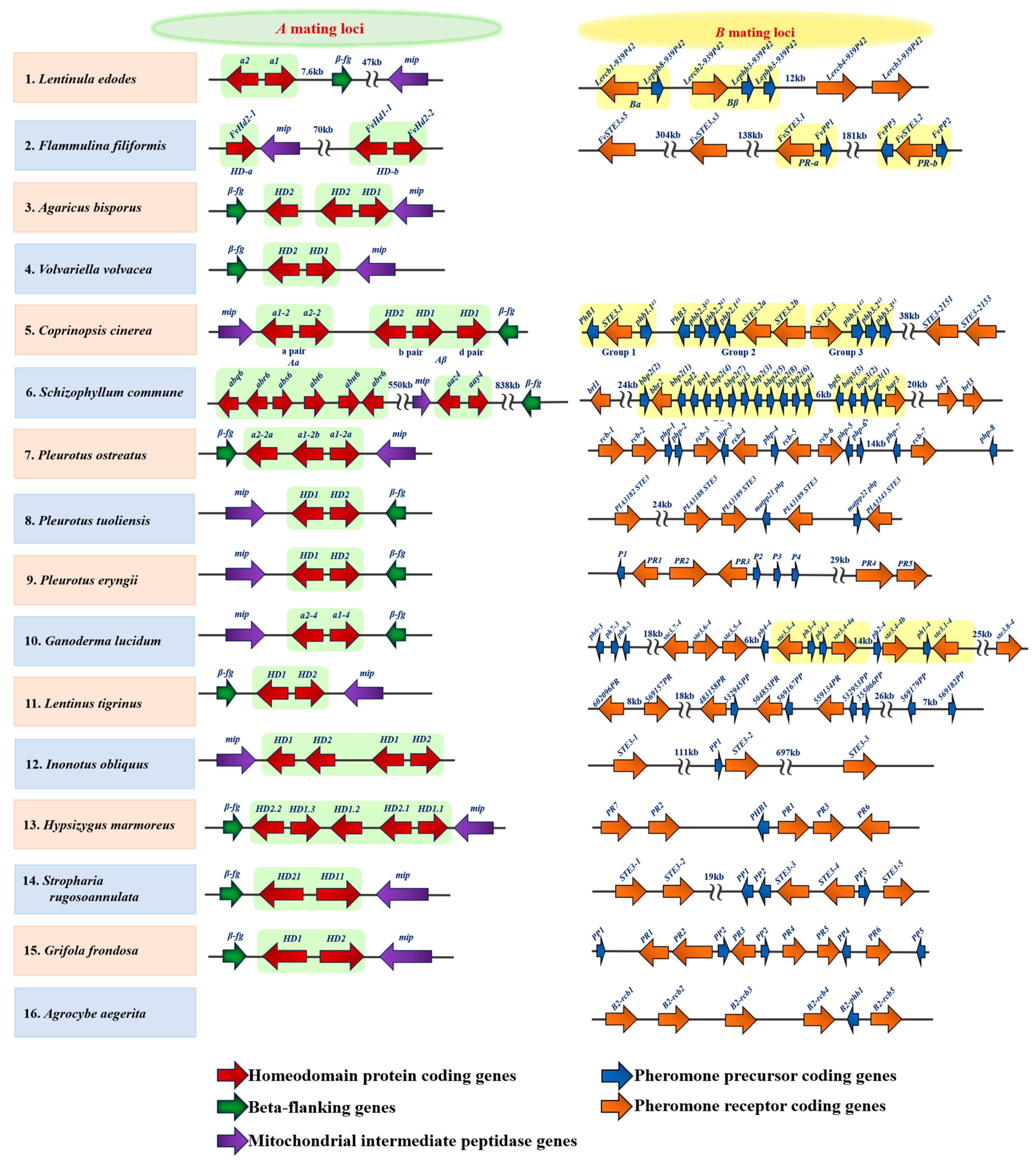
3.1. Mating-Type Genes
3.2. Research Progress on Genes Related to Mycelium and Fruiting Body Development
3.3. Research Progress on Genes Related to Substrate Utilization and Nutrient Transport
3.4. Genes Related to Environmental Response
3.5. Research Progress on Genes Related to the Synthesis and Regulation of Important Active Substances in Edible and Medicinal Fungi
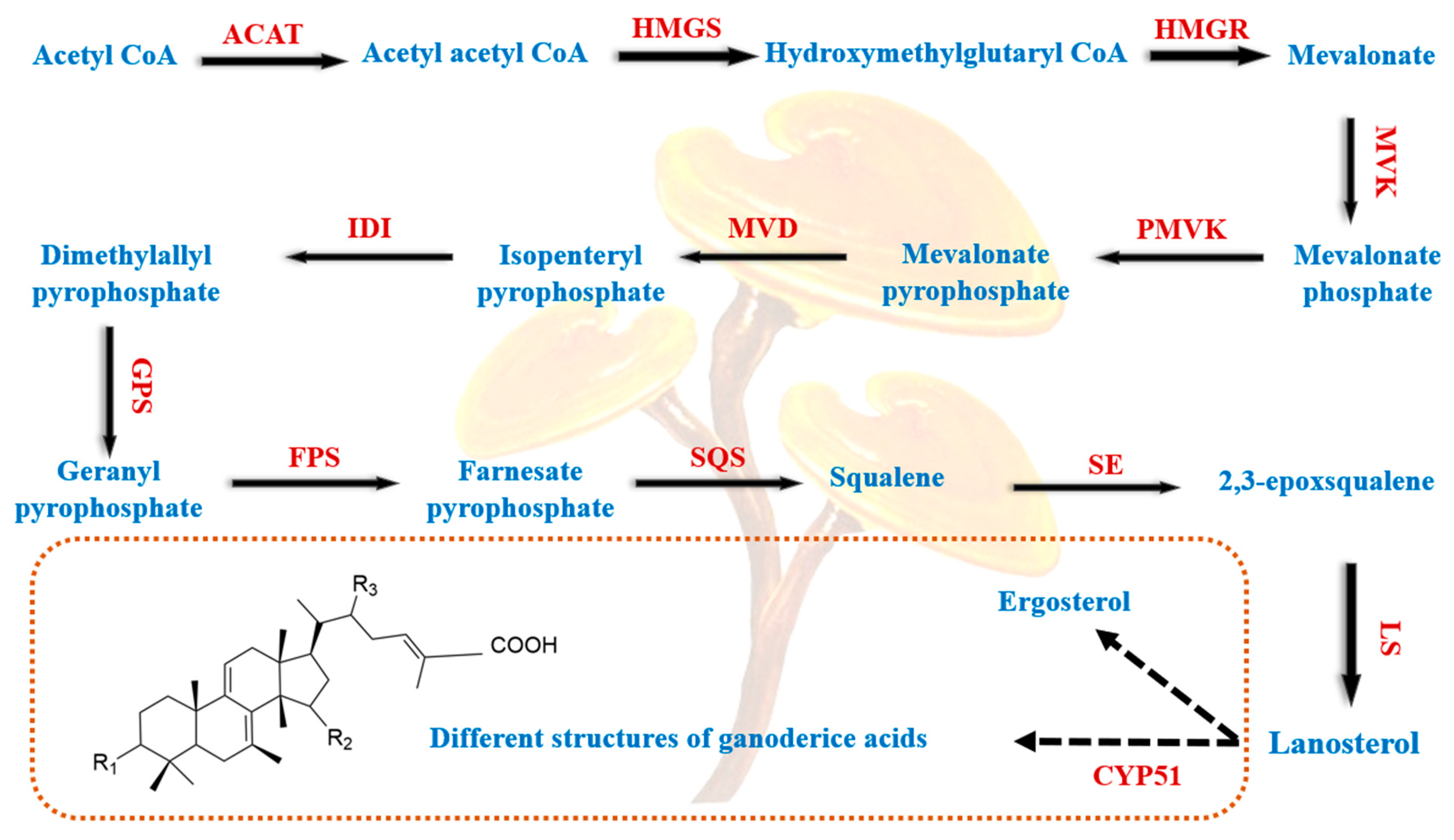
3.5.1. Ganoderma Triterpenoids
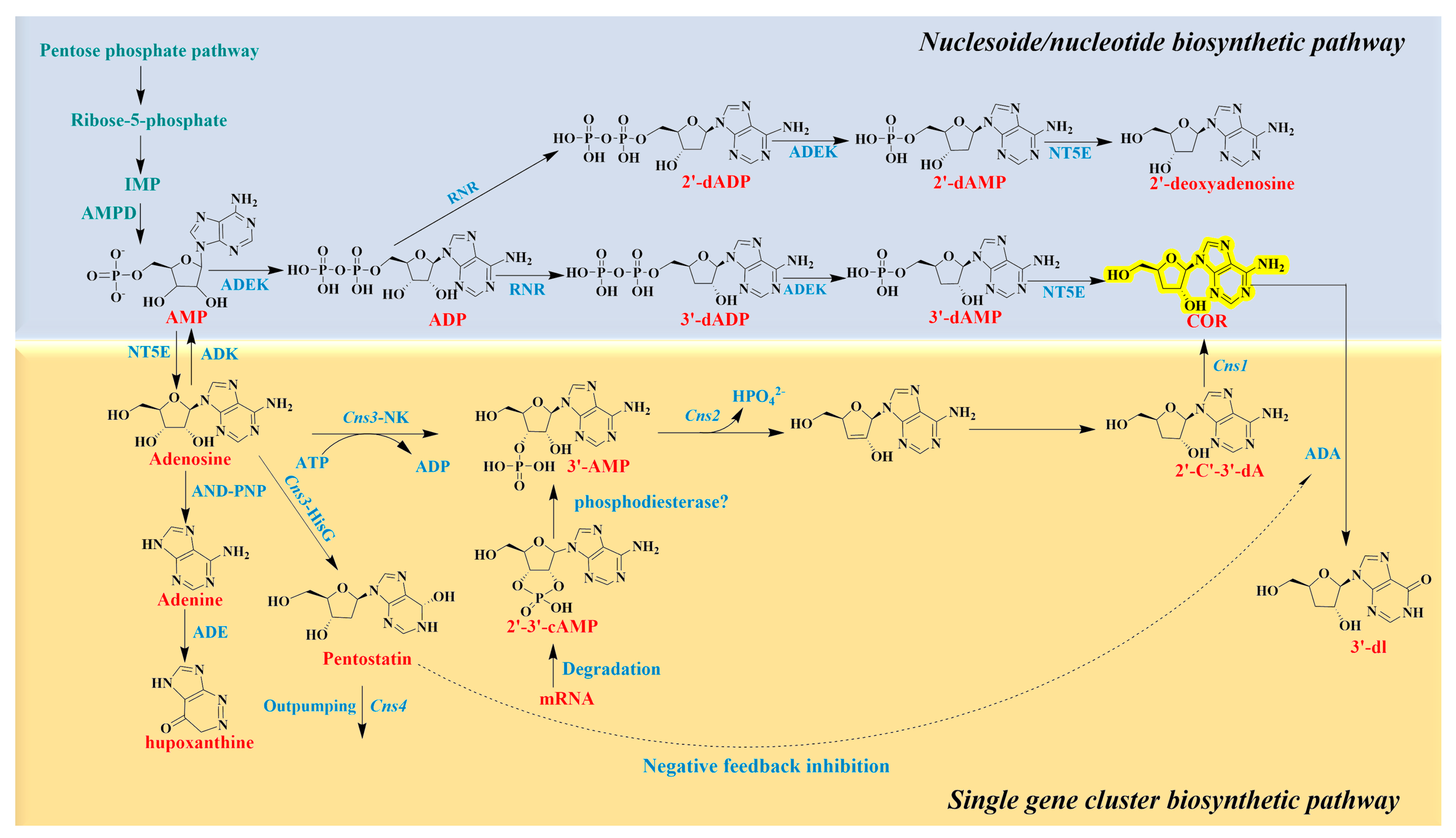
3.5.2. Cordycepin
3.5.3. Polysaccharides
3.5.4. Immunomodulatory Proteins of Edible and Medicinal Fungi
4. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cheung, P.C.K. The nutritional and health benefits of mushrooms. Nutr. Bull. 2010, 35, 292–299. [Google Scholar] [CrossRef]
- Das, S.; Prakash, B. Chapter 11—Edible mushrooms: Nutritional composition and medicinal benefits for improvement in quality life. In Research and Technological Advances in Food Science; Academic Press: Cambridge, MA, USA, 2022; pp. 269–300. [Google Scholar]
- Mingyi, Y.; Belwal, T.; Devkota, H.P.; Li, L.; Luo, Z.S. Trends of utilizing mushroom polysaccharides (MPs) as potent nutraceutical components in food and medicine: A comprehensive review. Trends Food Sci. Technol. 2019, 92, 94–110. [Google Scholar] [CrossRef]
- Wong, J.H.; Ng, T.B.; Cheung, R.C.; Ye, X.J.; Wang, H.X.; Lam, S.K.; Lin, P.; Chan, Y.S.; Fang, E.F.; Ngai, P.H.; et al. Proteins with antifungal properties and other medicinal applications from plants and mushrooms. Appl. Microbiol. Biotechnol. 2010, 87, 1221–1235. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.H.; Sun-Waterhouse, D.; Wang, J.M.; Ma, C.Y.; Waterhouse, G.I.N.; Kang, W.Y. Polysaccharides from edible fungi Pleurotus spp.: Advances and perspectives. J. Future Foods 2021, 1, 128–140. [Google Scholar] [CrossRef]
- Zhang, Y.R.; Wang, D.W.; Chen, Y.T.; Liu, T.T.; Zhang, S.S.; Fan, H.X.; Liu, H.C.; Li, Y. Healthy function and high valued utilization of edible fungi. Food Sci. Hum. Well 2021, 10, 408–420. [Google Scholar] [CrossRef]
- Zhao, J.H.; He, R.J.; Zhong, H.; Liu, S.Z.; Liu, X.F.; Hussain, M.; Sun, P.L. A cold-water extracted polysaccharide-protein complex from Grifola frondosa exhibited anti-tumor activity via TLR4-NF-κB signaling activation and gut microbiota modification in H22 tumor-bearing mice. Int. J. Biol. Macromol. 2023, 239, 124291. [Google Scholar] [CrossRef] [PubMed]
- Wasser, S.P. Current findings, future trends, and unsolved problems in studies of medicinal mushrooms. Appl. Microbiol. Biotechnol. 2011, 89, 1323–1332. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Tian, Y.; Menolli, N., Jr.; Ye, L.; Karunarathna, S.C.; Perez-Moreno, J.; Rahman, M.M.; Rashid, M.H.; Phengsintham, P.; Rizal, L.; et al. Reviewing the world’s edible mushroom species: A new evidence-based classification system. Compr. Rev. Food Sci. Food Saf. 2021, 20, 1982–2014. [Google Scholar] [CrossRef]
- Royse, D.J.; Baars, J.; Tan, Q. Current Overview of Mushroom Production in the World: Technology and Applications. In Edible and Medicinal Mushrooms; John Wiley and Sons Ltd.: Hoboken, NJ, USA, 2017; pp. 5–13. [Google Scholar]
- Grimm, D.; Wösten, H.A.B. Mushroom cultivation in the circular economy. Appl. Microbiol. Biotechnol. 2018, 102, 7795–7803. [Google Scholar] [CrossRef]
- Zou, G.; Nielsen, J.B.; Wei, Y.J. Harnessing synthetic biology for mushroom farming. Trends Biotechnol. 2023, 41, 480–483. [Google Scholar] [CrossRef]
- Ye, D.; Hu, Q.X.; Bai, X.; Zhang, W.J.; Guo, H.W. Increasing the value of Phragmites australis straw in a sustainable integrated agriculture model (SIAM) comprising edible mushroom cultivation and spent mushroom substrate compost. Sci. Total Environ. 2023, 869, 161807. [Google Scholar] [CrossRef]
- Bao, D.P. Development and prospect of genetics of edible mushrooms in China. Mycosystema 2021, 40, 806–821. [Google Scholar]
- Zhao, Z.M.; Fan, D.Y.; Yao, L.; Li, C.T.; Li, Y. Sequencing and Analysis of the Whole Genome of Morchella importuna MT1. J. Fungal Res. 2021, 19, 271–276. [Google Scholar]
- Drott, M.T.; Park, S.C.; Wang, Y.W.; Harrow, L.; Keller, N.P.; Pringle, A. Pangenomics of the death cap mushroom Amanita phalloides, and of Agaricales, reveals dynamic evolution of toxin genes in an invasive range. ISME J. 2023, 17, 1236–1246. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.X.; Liu, Z.H.; Fu, Y.P.; Li, Y.; Dai, Y.T.; Xiao, S.J. Pan-Genomes Provide Insights into the Genetic Basis of Auricularia heimuer Domestication. J. Fungi 2022, 8, 581. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kim, D.Y.; Park, Y.J.; Jang, M.J. Transcriptome analysis of the edible mushroom Lentinula edodes in response to blue light. PLoS ONE 2020, 15, e0230680. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.B.; Zhang, J.J.; Wang, Q.; Huang, J.C.; Juan, J.X.; Kuai, B.K.; Feng, Z.Y.; Chen, H. Transcriptome and Differentially Expressed Gene Profiles in Mycelium, Primordium and Fruiting Body Development in Stropharia rugosoannulata. Genes 2022, 13, 1080. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Zhang, J.C.; Wang, C.C.; Bian, Y.B.; Xiao, Y. Studies on Transcriptome During Fruiting Body Development of Lentinula edodes. Acta Hortic. Sin. 2022, 49, 801–815. [Google Scholar]
- Cao, L.P.; Zhang, Q.; Miao, R.Y.; Lin, J.B.; Feng, R.C.; Ni, Y.Q.; Li, W.S.; Yang, D.L.; Zhao, X. Application of omics technology in the research on edible fungi. Curr. Res. Food Sci. 2023, 6, 100430. [Google Scholar] [CrossRef]
- Jakopovic, B.; Oršolić, N.; Jakopovich, I. Proteomic Research on the Antitumor Properties of Medicinal Mushrooms. Molecules 2021, 26, 6708. [Google Scholar] [CrossRef]
- Lin, T.Y.; Hua, W.J.; Yeh, H.; Tseng, A.J. Functional proteomic analysis reveals that fungal immunomodulatory protein reduced expressions of heat shock proteins correlates to apoptosis in lung cancer cells. Phytomed. Int. J. Phytother. Phytopharm. 2021, 80, 153384. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.P.; Su, Y.; Ma, Z.H.; Guo, L.Z.; Yang, S.; Yu, H. Comparative proteomic analysis reveals differential protein expression of Hypsizygus marmoreus in response to different light qualities. Int. J. Biol. Macromol. 2022, 223, 1320–1334. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.; Zhou, H.B.; Ma, T.; Zheng, Z.H.; Zheng, E.P.; Yang, H.L.; Gao, H.Y. TMT-based quantitative proteomic analysis of postharvest Coprinus comatus fruiting body during storage. Postharvest Biol. Technol. 2022, 185, 111786. [Google Scholar] [CrossRef]
- Jo, C.; Zhang, J.; Tam, J.M.; Church, G.M.; Khalil, A.S.; Segrè, D.; Tang, T.C. Unlocking the magic in mycelium: Using synthetic biology to optimize filamentous fungi for biomanufacturing and sustainability. Mater. Today Bio 2023, 19, 100560. [Google Scholar] [CrossRef] [PubMed]
- Jiang, A.L.; Liu, Y.N.; Liu, R.; Ren, A.; Ma, H.Y.; Shu, L.B.; Shi, L.; Zhu, J.; Zhao, M.W. Integrated Proteomics and Metabolomics Analysis Provides Insights into Ganoderic Acid Biosynthesis in Response to Methyl Jasmonate in Ganoderma Lucidum. Int. J. Mol. Sci. 2019, 20, 6116. [Google Scholar] [CrossRef]
- Liu, Y.C.; Zhang, D.; Ning, Q.y.; Wang, J.H. Growth characteristics and metabonomics analysis of Lactobacillus rhamnosus GG in Ganoderma lucidum aqueous extract medium. Food Biosci. 2023, 53, 102486. [Google Scholar] [CrossRef]
- Zeng, Z.; Wang, J.Q.; Wen, X.F.; Wang, Y.; Li, X.; Liu, D.Y.; Geng, F. Metabolomic analysis provides insights into the mechanism of color and taste changes in Dictyophora indusiata fruiting bodies under different drying processes. Food Res. Int. 2022, 162, 112090. [Google Scholar] [CrossRef]
- Sun, X.Y.; Wu, J.Y.; Zhang, S.H.; Luo, L.; Mo, C.Y.; Sheng, L.; Ma, A.M. Genome and Comparative Transcriptome Dissection Provide Insights into Molecular Mechanisms of Sclerotium Formation in Culinary-Medicinal Mushroom Pleurotus tuber-regium. Front. Microbiol. 2021, 12, 815954. [Google Scholar] [CrossRef]
- Cai, Z.X.; Chen, M.Y.; Lu, Y.P.; Guo, Z.J.; Zeng, Z.H.; Liao, J.H.; Zeng, H. Metabolomics and transcriptomics unravel the mechanism of browning resistance in Agaricus bisporus. PLoS ONE 2022, 17, e0255765. [Google Scholar] [CrossRef]
- Marian, I.M.; Vonk, P.J.; Valdes, I.D.; Barry, K.; Bostock, B.; Carver, A.; Daum, C.; Lerner, H.; Lipzen, A.; Park, H.; et al. The Transcription Factor Roc1 Is a Key Regulator of Cellulose Degradation in the Wood-Decaying Mushroom Schizophyllum commune. mBio 2022, 13, e0062822. [Google Scholar] [CrossRef]
- Liu, C.C.; Kang, L.Q.; Lin, M.; Bi, J.J.; Liu, Z.H.; Yuan, S. Molecular Mechanism by Which the GATA Transcription Factor CcNsdD2 Regulates the Developmental Fate of Coprinopsis cinerea under Dark or Light Conditions. mBio 2021, 13, e0362621. [Google Scholar] [CrossRef]
- Gong, M.; Wang, Y.; Su, E.; Zhang, J.G.; Tang, L.H.; Li, Z.P.; Zhang, L.L.; Zou, G.; Wan, J.N.; Bao, D.P. The promising application of a β-glucosidase inhibitor in the postharvest management of Volvariella volvacea. Postharvest Biol. Technol. 2022, 185, 111784. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Boonyuen, N.; Ranaweera, C.B.; de Zoysa, H.K.S.; Padmathilake, R.E.; Nifla, F.; Dai, D.Q.; Liu, Y.; Suwannarach, N.; Kumla, J.; et al. OMICS and Other Advanced Technologies in Mycological Applications. J. Fungi 2023, 9, 688. [Google Scholar] [CrossRef]
- Shangguan, J.L.; Zhu, J.; Shi, L.; Ren, A.; ZHao, M.W. Research on Edible Fungi Genetic System. J. Fungal Res. 2019, 17, 224–228. [Google Scholar]
- Li, J.G.; Liu, Q.; Liu, D.F.; Wu, M.; Tian, C.G. Advances in metabolic engineering of filamentous fungi. Chin. J. Biotechnol. 2021, 37, 1637–1658. [Google Scholar]
- Bao, D.P.; Huang, Z.H.; Li, Y.; Zhou, C.L.; Li, Y.; Wan, J.N.; Tang, L.H.; Mao, W.J.; Wang, Y.; Gong, M.; et al. Agrobacterium-mediated transformation of arthroconidia obtained from the edible mushroom Hypsizygus marmoreus. J. Microbiol. Method 2020, 171, 105878. [Google Scholar] [CrossRef]
- Liu, L.; Xiao, Z.Y.; Guo, L.Q.; Lin, J.F.; You, L.F.; Liao, J.W. Establishment of genetic transformation system of Hericium erinaceus using PEG mediated method. Mycosystema 2014, 33, 121–128. [Google Scholar]
- Lou, H.W.; Ye, Z.W.; Yu, Y.H.; Lin, J.F.; Guo, L.Q.; Chen, B.X.; Tang, H.B.; Wei, T.; Chen, L.T.; Yun, F. The efficient genetic transformation of Cordyceps militaris by using mononuclear protoplasts. Sci. Hortic. 2019, 243, 307–313. [Google Scholar] [CrossRef]
- Xu, C. The Construction of Liposome-Mediated Transformation Method and the Role of Laccases in the Development of Flammulina velutipes. Master’s Thesis, Nanjing Agricultural University, Nanjing, China, 2016. [Google Scholar]
- Zhang, Y.; Zhang, C.H.; Liu, D.R.; Wang, X.F.; Wang, J.M.; Qiu, L.Y. Liposome-mediated Fruit Body Gill Tissue Transformation of Agaricus bisporus and Function Exploration of 1-aminocyclopropane-1-carboxylic Acid Oxidase Gene (ACO). J. Agric. Biotechnol. 2015, 23, 1112–1120. [Google Scholar]
- Kim, J.K.; Park, Y.J.; Kong, W.S.; Kang, H.W. Highly Efficient Electroporation-mediated Transformation into Edible Mushroom Flammulina velutipes. Mycobiology 2010, 38, 331–335. [Google Scholar] [CrossRef]
- Shi, L.L.; Peer, A.V.; Guo, L.X.; Chen, R.L.; Wang, W.; Yan, J.J.; Deng, Y.J.; Xie, B.G. Agrobacterium-mediated Transformation of an Endogenous HMG-box Transcription Factor fvhom1 in Flammulina velutipes. Genom. Appl. Biol. 2014, 33, 1268–1274. [Google Scholar]
- Lyu, X.M.; Wang, Q.J.; Liu, A.; Liu, F.; Meng, L.; Wang, P.M.; Zhang, Y.; Wang, L.; Li, Z.; Wang, W. The transcription factor Ste12-like increases the mycelial abiotic stress tolerance and regulates the fruiting body development of Flammulina filiformis. Front. Microbiol. 2023, 14, 1139679. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Fan, T.N.; Bai, Y.D.; Guo, Q.Q.; Wang, H.A.; Chen, G.; Gao, Y.X.; Lin, J.W. Native bio-function of FIP-fve towards Flammulina filiformis. J. Huazhong Agric. Univ. 2023, 42, 131–138. [Google Scholar]
- Zhang, X.; Xia, X.; Duan, Z.W.; Guan, S.Y.; Li, H.G.; Chen, L.J.; Zhang, L.; Fan, W.L.; Lin, J.W. Construction of Silencing Vector for FIP-fve Gene and Genetic Transformation of Enoki Mushroom (Flammulina velutipes). Mol. Plant Breed. 2017, 15, 4491–4497. [Google Scholar]
- Meng, L.; Lyu, X.M.; Shi, L.L.; Wang, Q.J.; Wang, L.; Zhu, M.J.; Mukhtar, I.; Xie, B.G.; Wang, W. The transcription factor FvHmg1 negatively regulates fruiting body development in Winter Mushroom Flammulina velutipes. Gene 2021, 785, 145618. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, S.; Yamada, M.; Shibata, K.; Okuhara, T.; Yoshida, M.; Inatomi, S.; Taguchi, G.; Shimosaka, M. Characterization of a gene coding for a putative adenosine deaminase-related growth factor by RNA interference in the Basidiomycete Flammulina velutipes. J. Biosci. Bioeng. 2013, 115, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Du, M.Y.; Xie, Y.B.; Wang, M.; Yang, H.; Hu, B.H.; Mukhtar, I.; Liu, Y.Y.; Tao, Y.X.; Liu, F.; Xie, B.G. FFGA1 Protein Is Essential for Regulating Vegetative Growth, Cell Wall Integrity, and Protection against Stress in Flammunina filiformis. J. Fungi 2022, 8, 401. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.Y.; Ling, Z.L.; Zhao, R.L. Construction of a heat-resistant strain of Lentinus edodes by fungal Hsp20 protein overexpression and genetic transformation. Front. Microbiol. 2022, 13, 1009885. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.B.; Liu, K.F.; Li, X.R.; Zhang, L.; Shen, N.; Bian, Y.B.; Xiao, Y. QTL mapping reveals mating type gene LeHD1 regulating mycelial growth in shiitake mushroom, Lentinula edodes. Sci. Hortic. 2022, 305, 111417. [Google Scholar] [CrossRef]
- Hu, Y.; Cai, Y.L.; Yu, J.J.; Zhou, Y.; Li, J.P.; Bian, Y.B.; Gong, Y.H. Function analysis of HMG-box transcription factor lelcrp1 in Lentinula edodes. Acta Microbiol. Sin. 2018, 58, 2100–2109. [Google Scholar]
- Wang, G.Z.; Zhou, S.S.; Luo, Y.; Ma, C.J.; Gong, Y.H.; Zhou, Y.; Gao, S.S.; Huang, Z.C.; Yan, L.L.; Hu, Y.; et al. The heat shock protein 40 LeDnaJ regulates stress resistance and indole-3-acetic acid biosynthesis in Lentinula edodes. Fungal Genet. Biol. 2018, 118, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.J.; Wang, G.Z.; Zhou, S.S.; Luo, Y.; Gong, Y.H.; Bian, Y.B. Functional analyses of anthranilate synthase gene LetrpE in Lentinula edodes by RNAi mediated gene knockdown. Mycosystema 2018, 37, 576–583. [Google Scholar]
- Zhong, H.Y.; Luo, Y.; Gong, Y.H.; Xu, R.P.; Bian, Y.B. Functional Analysis of Indole-3-pyruvate Monooxygenase Gene YUCCA8 Involved in Heat Resistance of Lentinula edodes by RNAi. Acta Edulis Fungi 2021, 28, 11–18. [Google Scholar]
- Wang, C.; Luo, Y.; Wang, G.Z.; Xu, R.P.; Zhou, Y.; Gong, Y.H.; Bian, Y.B. Functional Analyses of Tryptophan Synthase Gene LetrpB in Lentinula edodes by RNAi Method. Acta Edulis Fungi 2019, 26, 161. [Google Scholar]
- Liu, Y.N.; Zhang, T.J.; Lu, X.X.; Ma, B.L.; Ren, A.; Shi, L.; Jiang, A.L.; Yu, H.S.; Zhao, M.W. Membrane fluidity is involved in the regulation of heat stress induced secondary metabolism in Ganoderma lucidum. Environ. Microbiol. 2017, 19, 1653–1668. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Shi, L.; Ren, A.; Jiang, A.-L.; Wu, F.-L.; Zhao, M.-W. The cloning, characterization and functional analysis of a gene encoding an acetyl-CoA acetyltransferase involved in triterpene biosynthesis in Ganoderma lucidum. Mycoscience 2013, 54, 100–105. [Google Scholar] [CrossRef]
- Xu, J.W.; Xu, Y.N.; Zhong, J.J. Enhancement of ganoderic acid accumulation by overexpression of an N-terminally truncated 3-hydroxy-3-methylglutaryl coenzyme A reductase gene in the basidiomycete Ganoderma lucidum. Appl. Environ. Microbiol. 2012, 78, 7968–7976. [Google Scholar] [CrossRef]
- Lin, Y.L.; Lee, Y.R.; Tsao, N.W.; Wang, S.Y.; Shaw, J.F.; Chu, F.H. Characterization of the 2,3-Oxidosqualene Cyclase Gene from Antrodia cinnamomea and Enhancement of Cytotoxic Triterpenoid Compound Production. J. Nat. Prod. 2015, 78, 1556–1562. [Google Scholar] [CrossRef]
- Qi, Y.C.; Chen, H.J.; Zhang, M.K.; Wen, Q.; Qiu, L.Y.; Shen, J.W. Identification and expression analysis of Pofst3 suggests a role during Pleurotus ostreatus primordia formation. Fungal Biol. 2019, 123, 200–208. [Google Scholar] [CrossRef]
- Lou, H.W.; Lin, J.F.; Guo, L.Q.; Wang, X.W.; Tian, S.Q.; Liu, C.X.; Zhao, Y.; Zhao, R.Y. Advances in research on Cordyceps militaris degeneration. Appl. Microbiol. Biotechnol. 2019, 103, 7835–7841. [Google Scholar] [CrossRef]
- Jiang, N.; Song, C.Y.; Liu, J.Y.; Tan, Q.; Zhang, L.J.; Shang, X.D. Agrobacterium-mediated transformation of the Lentinula edodes mycelia. Mycosystema 2017, 36, 1514–1523. [Google Scholar]
- Lv, S.Y.; Chen, X.; Mou, C.Y.; Dai, S.H.; Bian, Y.B.; Kang, H. Agrobacterium-mediated transformation of the ascomycete mushroom Morchella importuna using polyubiquitin and glyceraldehyde-3-phosphate dehydrogenase promoter-based binary vectors. World J. Microb. Biot. 2018, 34, 148. [Google Scholar] [CrossRef]
- Du, S.S.; Bao, D.P.; Ma, L.J.; Li, X.L.; Shang, J.J. Expression analyses of exogenous genes controlled by different promoters in Hypsizygus marmoreus. Mycosystema 2021, 40, 2065–2073. [Google Scholar]
- Shang, J.J.; Yang, R.H.; Tang, L.H.; Li, Y.; Li, Y.; Mao, W.J.; Gong, M.; Wang, Y.; Honda, Y.C.; Bao, D.P. Differential expression of two gpd genes in the cultivated mushroom Pleurotus eryngii using RNA sequencing analysis. Mycoscience 2019, 60, 351–353. [Google Scholar] [CrossRef]
- Liu, J.Y.; Song, C.Y.; Li, Q.Z.; Xu, Z.; Zhang, D.; Zhang, M.Y.; Tan, Q.; Shang, X.D. A colonized millet grain method for Agrobacterium-mediated transformation of the button mushroom Agaricus bisporus. J. Microbiol. Method 2018, 152, 148–153. [Google Scholar] [CrossRef]
- Liu, J.Y.; Xu, Z.; Zhang, D.; Wang, R.J.; Tan, Q.; Shang, X.D. Efficiency of Flammulina velutipes Transformation by Agrobacterium tumefaciens-mediated Transformation Using Different Receptors. Acta Edulis Fungi 2015, 22, 7–12. [Google Scholar]
- Lei, M.; Wu, X.L.; Zhang, J.X.; Wang, H.X.; Huang, C.Y. Establishment of an efficient transformation system for Pleurotus ostreatus. World J. Microbiol. Biotechnol. 2017, 33, 214. [Google Scholar] [CrossRef]
- Yan, L.L.; Xu, R.P.; Zhou, Y.; Gong, Y.H.; Dai, S.H.; Liu, H.Y.; Bian, Y.B. Effects of Medium Composition and Genetic Background on Agrobacterium-Mediated Transformation Efficiency of Lentinula edodes. Genes 2019, 10, 467. [Google Scholar] [CrossRef]
- Herzog, R.; Solovyeva, I.; Bölker, M.; Lugones, L.G.; Hennicke, F. Exploring molecular tools for transformation and gene expression in the cultivated edible mushroom Agrocybe aegerita. Mol. Genet. Genom. 2019, 294, 663–677. [Google Scholar] [CrossRef]
- Wang, C.C.; Zhang, W.W.; Wong, J.H.; Ng, T.; Ye, X.J. Diversity of potentially exploitable pharmacological activities of the highly prized edible medicinal fungus Antrodia camphorata. Appl. Microbiol. Biotechnol. 2019, 103, 7843–7867. [Google Scholar] [CrossRef]
- Liu, X.F.; Xia, Y.J.; Zhang, Y.; Liang, L.H.; Xiong, Z.Q.; Wang, G.Q.; Song, X.; Ai, L.Z. Enhancement of antroquinonol production via the overexpression of 4-hydroxybenzoate polyprenyltransferase biosynthesis-related genes in Antrodia cinnamomea. Phytochemistry 2021, 184, 112677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhao, L.T.; Shen, M.Y.; Liu, J.Y.; Li, Y.R.; Xu, S.; Chen, L.; Shi, G.Y.; Ding, Z.Y. Establishment of an Efficient Polyethylene Glycol (PEG)-Mediated Transformation System in Pleurotus eryngii var. ferulae Using Comprehensive Optimization and Multiple Endogenous Promoters. J. Fungi 2022, 8, 186. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.J.; Li, Y.; Yang, R.H.; Wang, Y.; Mao, W.J.; Tang, L.H.; Wu, Y.Y.; Nakazawa, T.; Honda, Y.C.; Li, Y.; et al. Efficient transformation of Pleurotus eryngii with a safe selective marker mutated from the Pesdi1 gene. J. Microbiol. Methods 2018, 152, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Zha, L.Y.; Zhou, W.J.; Wang, Y.; Zhang, Z.W.; Wen, H.S.; Mo, M.H. Research Progress on Agrobacterium mediated Transformation of Edible Fungi. Acta Edulis Fungi 2020, 27, 105–118. [Google Scholar]
- Shen, H.M.; Li, C.; Gao, L.; Liu, T.G.; Liu, B.; Chen, W.Q. Research progress in transformation of fungi mediated by protoplasts. Plant Prot. 2017, 43, 25–28. [Google Scholar]
- Wang, P.A.; Xiao, H.; Zhong, J.J. CRISPR-Cas9 assisted functional gene editing in the mushroom Ganoderma lucidum. Appl. Microbiol. Biotechnol. 2020, 104, 1661–1671. [Google Scholar] [CrossRef] [PubMed]
- Waltz, E. Gene-edited CRISPR mushroom escapes US regulation. Nature 2016, 532, 293. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Xiao, H.; Zou, G.; Zhou, Z.; Zhong, J.J. CRISPR-Cas9 assisted gene disruption in the higher fungus Ganoderma species. Process Biochem. 2017, 56, 57–61. [Google Scholar] [CrossRef]
- Liu, K.; Sun, B.; You, H.; Tu, J.L.; Yu, X.Y.; Zhao, P.; Xu, J.W. Dual sgRNA-directed gene deletion in basidiomycete Ganoderma lucidum using the CRISPR/Cas9 system. Microb. Biotechnol. 2020, 13, 386–396. [Google Scholar] [CrossRef]
- Zhang, Z.G.; Zhang, J.S.; Zou, G.; Tang, C.H.; Feng, J.; Bao, D.P.; Chen, J.B.; Tan, Y. Construction of a CRISPR/Cas9-Based Genome Editing System in Ganoderma lucidum “Hunong No.1’Cultivar. Acta Edulis Fungi 2023, 30, 9–18. [Google Scholar]
- Sugano, S.S.; Suzuki, H.; Shimokita, E.; Chiba, H.; Noji, S.; Osakabe, Y.; Osakabe, K. Genome editing in the mushroom-forming basidiomycete Coprinopsis cinerea, optimized by a high-throughput transformation system. Sci. Rep. 2017, 7, 1260. [Google Scholar] [CrossRef] [PubMed]
- Yu, H. Research on the application of CRISPR-Cas9 gene editing system in Coprinus cinereus. Master’s Thesis, Nanjing Normal University, Nanjing, China, 2018. [Google Scholar]
- Chen, B.X.; Wei, T.; Ye, Z.W.; Yun, F.; Kang, L.Z.; Tang, H.B.; Guo, L.i.Q.; Lin, J.F. Efficient CRISPR-Cas9 Gene Disruption System in Edible-Medicinal Mushroom Cordyceps militaris. Front. Microbiol. 2018, 9, 1157. [Google Scholar] [CrossRef] [PubMed]
- Meng, G.L.; Wang, X.P.; Liu, M.Q.; Wang, F.; Liu, Q.Z.; Dong, C.H. Efficient CRISPR/Cas9 system based on autonomously replicating plasmid with an AMA1 sequence and precisely targeted gene deletion in the edible fungus, Cordyceps militaris. Microb. Biotechnol. 2022, 15, 2594–2606. [Google Scholar] [CrossRef]
- Yu, X.; Liang, X.Q.; Xie, H.M.; Kumar, S.; Ravinder, N.; Potter, J.; de Mollerat du Jeu, X.; Chesnut, J.D. Improved delivery of Cas9 protein/gRNA complexes using lipofectamine CRISPRMAX. Biotechnol. Lett. 2016, 38, 919–929. [Google Scholar] [CrossRef]
- Al Abdallah, Q.; Ge, W.; Fortwendel, J.R. A Simple and Universal System for Gene Manipulation in Aspergillus fumigatus: In Vitro-Assembled Cas9-Guide RNA Ribonucleoproteins Coupled with Microhomology Repair Templates. mSphere 2017, 2, e00446-00417. [Google Scholar] [CrossRef] [PubMed]
- Jan Vonk, P.; Escobar, N.; Wösten, H.A.B.; Lugones, L.G.; Ohm, R.A. High-throughput targeted gene deletion in the model mushroom Schizophyllum commune using pre-assembled Cas9 ribonucleoproteins. Sci. Rep. 2019, 9, 7632. [Google Scholar] [CrossRef] [PubMed]
- Zou, G.; Xiao, M.L.; Chai, S.X.; Zhu, Z.H.; Wang, Y.; Zhou, Z.H. Efficient genome editing in filamentous fungi via an improved CRISPR-Cas9 ribonucleoprotein method facilitated by chemical reagents. Microb. Biotechnol. 2021, 14, 2343–2355. [Google Scholar] [CrossRef]
- Yu, L.; Xiao, M.L.; Zhu, Z.H.; Wang, Y.M.; Zhou, Z.H.; Wang, P.P.; Zou, G. Efficient genome editing in Claviceps purpurea using a CRISPR/Cas9 ribonucleoprotein method. Synth. Syst. Biotechnol. 2022, 7, 664–670. [Google Scholar] [CrossRef]
- Liu, J.Y.; Cui, H.Y.; Wang, R.J.; Xu, Z.; Yu, H.L.; Song, C.Y.; Lu, H.; Li, Q.Z.; Xing, D.R.; Tan, Q.; et al. A Simple and Efficient CRISPR/Cas9 System Using a Ribonucleoprotein Method for Flammulina filiformis. J. Fungi 2022, 8, 1000. [Google Scholar] [CrossRef]
- Boontawon, T.; Nakazawa, T.; Choi, Y.J.; Ro, H.S.; Oh, M.J.; Kawauchi, M.; Sakamoto, M.; Honda, Y.C. Double-gene targeting with preassembled Cas9 ribonucleoprotein for safe genome editing in the edible mushroom Pleurotus ostreatus. FEMS Microbiol. Lett. 2023, 370, 370. [Google Scholar] [CrossRef]
- Xu, R.P.; Zhou, Y.; Bian, Y.B. Application of CRISPR/Cas9 Technology in Macrofungi Research. Acta Edulis Fungi 2019, 26, 119–127. [Google Scholar]
- Zhang, Y.; Chen, S.T.; Yang, L.; Zhang, Q. Application progress of CRISPR/Cas9 genome-editing technology in edible fungi. Front. Microbiol. 2023, 14, 1169884. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Ye, C. Microbial cell factories based on filamentous bacteria, yeasts, and fungi. Microb. Cell Fact. 2023, 22, 20. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.F.; Geng, B.N.; Song, H.Y.; Hu, M.M.; He, Q.N.; Chen, S.W.; Bai, F.W.; Yang, S.H. Progress and perspective on development of non-model industrial bacteria as chassis cells for biochemical productionin the synthetic biology era. Chin. J. Biotechnol. 2021, 37, 874–910. [Google Scholar]
- Adams, A.M.; Kaplan, N.A.; Wei, Z.; Brinton, J.D.; Monnier, C.S.; Enacopol, A.L.; Ramelot, T.A.; Jones, J.A. In vivo production of psilocybin in E. coli. Metab. Eng. 2019, 56, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, L.T.; Gu, Z.H.; Li, Y.R.; Shi, G.Y.; Ding, Z.Y. Heterologous expression and characterization of the key enzymes involved in sugar donor synthesis of polysaccharide in Ganoderma lucidum. Microbiol. China 2019, 46, 3233–3247. [Google Scholar]
- Liang, Y.Y.; Zan, X.Y.; Sun, L.; Fu, X.; Cui, F.J.; Tan, M.; Shao, Z.Y.; Sun, W.J. A uridine diphosphate-glycosyltransferase GFUGT88A1 derived from edible mushroom Grifola frondosa extends oligosaccharide chains. Process Biochem. 2022, 112, 80–91. [Google Scholar] [CrossRef]
- Wang, W.F.; Xiao, H.; Zhong, J.J. Biosynthesis of a ganoderic acid in Saccharomyces cerevisiae by expressing a cytochrome P450 gene from Ganoderma lucidum. Biotechnol. Bioeng. 2018, 115, 1842–1854. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Wang, Y.X.; Liu, X.; Zhao, J.; Kang, L.Q.; Liu, Z.H.; Yuan, S. Heterologous expression and characterization of a novel chitin deacetylase, CDA3, from the mushroom Coprinopsis cinerea. Int. J. Biol. Macromol. 2020, 150, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Guo, L.q.; Ye, Z.w.; Zou, Y.; Yu, Y.H.; Lin, J.F. Cloning of the Transglutaminase Gene from Cordyceps militaris and Its Expressionin Pichia pastoris. J. Chin. Inst. Food Sci. Technol. 2022, 22, 107–113. [Google Scholar]
- Lee, J.; Lee, H.; Lee, J.; Chang, P.S. Heterologous expression, purification, and characterization of a recombinant Cordyceps militaris lipase from Candida rugosa-like family in Pichia pastoris. Enzyme Microb. Technol. 2023, 168, 110254. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.R.; Qin, L.N.; Tao, Y.; Huang, J.Z.; Dong, Z.Y. Overexpression and characterization of a laccase gene from Pleurotus ostreatus in Trichoderma reesei. Acta Microbiol. Sin. 2012, 52, 850–856. [Google Scholar]
- Zou, G.; Zhou, Z.H. CRISPR/Cas9-Mediated Genome Editing of Trichoderma reesei. Methods Mol. Biol. 2021, 2234, 87–98. [Google Scholar] [PubMed]
- Lan, X.T.; Yuan, W.; Wang, M.; Xiao, H. Efficient biosynthesis of antitumor ganoderic acid HLDOA using a dual tunable system for optimizing the expression of CYP5150L8 and a Ganoderma P450 reductase. Biotechnol. Bioeng. 2019, 116, 3301–3311. [Google Scholar] [CrossRef]
- Yuan, W.; Jiang, C.J.; Wang, Q.; Fang, Y.B.; Wang, J.; Wang, M.; Xiao, H. Biosynthesis of mushroom-derived type II ganoderic acids by engineered yeast. Nat. Commun. 2022, 13, 7740. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.X.; Xue, L.N.; Wei, T.; Wang, N.; Zhong, J.R.; Ye, Z.W.; Guo, L.Q.; Lin, J.F. Multiplex gene precise editing and large DNA fragment deletion by the CRISPR-Cas9-TRAMA system in edible mushroom Cordyceps militaris. Microb. Biotechnol. 2022, 15, 2982–2991. [Google Scholar] [CrossRef] [PubMed]
- Han, H.Y.; Yu, C.Y.; Qi, J.Z.; Wang, P.C.; Zhao, P.P.; Gong, W.B.; Xie, C.L.; Xia, X.K.; Liu, C.W. High-efficient production of mushroom polyketide compounds in a platform host Aspergillus oryzae. Microb. Cell Fact. 2023, 22, 60. [Google Scholar] [CrossRef] [PubMed]
- Valiante, V. Advances in Synthetic Biology of Fungi and Contributions to the Discovery of New Molecules. Chembiochem. 2023, 24, e202300008. [Google Scholar] [CrossRef]
- Feng, D.L.; Hu, H.P.; Yong, T.Q.; Liu, Y.C.; Xiao, C.; Huang, L.H.; Xie, Y.Z.; WU, Q.P. Induction of sexual fruiting-body formation by pairing the opposite mating-type isolates of Cordyceps militaris. Mycosystema 2023, 42, 344–352. [Google Scholar]
- Guo, Y.X.; Chen, Q.J.; Zhang, G.Q.; Wang, H.X.; Qin, Y. Application of Mating Type Gene Molecular Markers in Morchella Single-spore Cross Breeding. Mol. Plant Breed. 2022, 20, 4067–4074. [Google Scholar]
- Whitehouse, H.L.K. Multiple-Allelomorph Heterothallism in the Fungi. New Phytol. 1949, 48, 212–244. [Google Scholar] [CrossRef]
- Davis, R.H. Genetics of Sexuality in Higher Fungi. BioScience 1967, 17, 200. [Google Scholar] [CrossRef]
- Casselton, L.A. Fungal sex genes-searching for the ancestors. BioEssays News Rev. Mol. Cell. Dev. Biol. 2008, 30, 711–714. [Google Scholar] [CrossRef] [PubMed]
- Raudaskoski, M.; Kothe, E. Basidiomycete mating type genes and pheromone signaling. Eukaryot. Cell 2010, 9, 847–859. [Google Scholar] [CrossRef]
- Casselton, L.A.; Olesnicky, N.S. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol. Mol. Biol. R. 1998, 62, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.J.; Wan, J.N.; Li, Y.; Li, Y.; Shang, J.J.; Bao, D.P. Characterization of clamps and nuclear phases in the monokaryons with transformed mating genes in Pleurotus eryngii. Mycosystema 2020, 39, 1130–1138. [Google Scholar]
- Bao, D.P. Research progress on the mating-typing locus structures of basidiomycete mushrooms. Mycosystema 2019, 38, 2061–2077. [Google Scholar]
- Sun, Y.; Zhang, Y.J.; Cao, X.Y.; Zhou, J.S.; Jiang, J.H. Characteristics related to sexual reproduction and mating type locus of Inonotus obliquus. Mycosystema 2023, 42, 520–529. [Google Scholar]
- Ma, L.J.; Li, X.L.; Bao, D.P.; Shang, J.J.; Zhou, C.L.; Yang, R.H. Genetic Structure and Polymorphism of Mating Type Loci in Different Hypsizygus marmoreus Strains. Acta Edulis Fungi 2022, 29, 23–30. [Google Scholar]
- Shang, J.J.; Hou, D.; Li, Y.; Zhou, C.L.; Guo, T.; Tang, L.H.; Mao, W.J.; Chen, Q.; Bao, D.P.; Yang, R.H. Analyses of mating systems in Stropharia rugosoannulata based on genomic data. Mycosystema 2020, 39, 1152–1161. [Google Scholar]
- Qian, R.; Chen, W.M.; Chai, H.M.; Tao, N.; Zhao, Y.C.; Chen, Y.H. B Mating Locus from Agrocybe aegerita: Structural Analysis. Chin. Agric. Sci. Bull. 2021, 37, 14–19. [Google Scholar]
- Zhang, S.S.; Li, X.; Li, G.J.; Huang, Q.; Tian, J.H.; Wang, J.L.; Li, M.; Li, S.M. Genetic and Molecular Evidence of a Tetrapolar Mating System in the Edible Mushroom Grifola frondosa. J. Fungi 2023, 9, 959. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.J.; Xu, S.J.; Tang, L.H.; Yang, R.H.; Gong, M.; Li, Y.; Wang, Y.; Zou, G.; Wan, J.N.; Bao, D.P. Transformation of Compatible Mating-Type Genes in Monokaryons Triggers Fruiting Body Development by Activating Mating Pathways in Pleurotus eryngii. Microbiol. Spectr. 2023, 11, e0527222. [Google Scholar] [CrossRef] [PubMed]
- Riffiani, R.; Chen, F.C.; Zhang, W.; Wada, T.; Shimomura, N.; Yamaguchi, T.; Aimi, T. Identification, characterization and expression of A-mating type genes in monokaryons and dikaryons of the edible mushroom Mycoleptodonoides aitchisonii (Bunaharitake). Mycoscience 2021, 62, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Han, J.X.; Kawauchi, M.; Schiphof, K.; Terauchi, Y.; Yoshimi, A.; Tanaka, C.; Nakazawa, T.; Honda, Y.C. Features of disruption mutants of genes encoding for hydrophobin Vmh2 and Vmh3 in mycelial formation and resistance to environmental stress in Pleurotus ostreatus. FEMS Microbiol. Lett. 2023, 370, 370. [Google Scholar] [CrossRef] [PubMed]
- Su, W.Y.; Sheng, L.Z.; Dai, Y.T.; Song, B.; Fu, Y.P.; Li, Y. Cloning and function analysis of fruiting body lectin gene from Pleurotus ostreatus. Mycosystema 2017, 36, 1111–1120. [Google Scholar]
- Shen, J.W.; Liang, S.S.; Qiu, L.Y.; Qi, Y.C. Cloning and Expression of a fst3 Gene from Pleurotus ostreatus. Acta Edulis Fungi 2015, 22, 15–20. [Google Scholar]
- Zhao, C.; Wang, X.; Yan, Z.; Hao, J.B.; Wang, F.; Liu, J.Y. Analysis of HMG-box Transcription Factors in Pleurotus ostreatus and Cloning of a Candidate Gene. Acta Edulis Fungi 2023, 30, 1–11. [Google Scholar]
- Nakazawa, T.; Morimoto, R.; Wu, H.L.; Kodera, R.; Sakamoto, M.; Honda, Y.C. Dominant effects of gat1 mutations on the ligninolytic activity of the white-rot fungus Pleurotus ostreatus. Fungal Biol. 2019, 123, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Armas-Tizapantzi, A.; Martínez, Y.P.J.L.; Fernández, F.J.; Mata, G.; Hernández-Cuevas, L.V.; Ortiz Ortiz, E.; García Nieto, E.; Tomasini, A.; Sierra-Palacios, E.; Marcial-Quino, J.; et al. Silencing of the Laccase (lacc2) Gene from Pleurotus ostreatus Causes Important Effects on the Formation of Toxocyst-like Structures and Fruiting Body. Int. J. Mol. Sci. 2023, 24, 8143. [Google Scholar] [CrossRef]
- Hou, L.D.; Liu, Z.Q.; Yan, K.X.; Xu, L.J.; Chang, M.C.; Meng, J.L. Mnsod1 promotes the development of Pleurotus ostreatus and enhances the tolerance of mycelia to heat stress. Microb. Cell Fact. 2022, 21, 155. [Google Scholar] [CrossRef]
- Tao, Y.X.; Chen, R.L.; Yan, J.J.; Long, Y.; Tong, Z.J.; Song, H.B.; Xie, B.G. A hydrophobin gene, Hyd9, plays an important role in the formation of aerial hyphae and primordia in Flammulina filiformis. Gene 2019, 706, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.P.; Chen, R.L.; Long, Y.; Li, X.; Jiang, Y.J.; Xie, B.G. A Jacalin-Related Lectin Regulated the Formation of Aerial Mycelium and Fruiting Body in Flammulina velutipes. Int. J. Mol. Sci. 2016, 17, 1884. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.M.; Jiang, S.Y.; Wang, L.; Chou, T.S.; Wang, Q.J.; Meng, L.; Mukhtar, I.; Xie, B.G.; Wang, W. The Fvclp1 gene regulates mycelial growth and fruiting body development in edible mushroom Flammulina velutipes. Arch. Microbiol. 2021, 203, 5373–5380. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.J.; Hu, C.C.; Xie, B.G.; Zhang, L.; Yan, S.J.; Wang, W.; Tao, Y.X.; Li, S.J. A Single Transcription Factor (PDD1) Determines Development and Yield of Winter Mushroom (Flammulina velutipes). Appl. Environ. Microbiol. 2019, 85, e01735-01719. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.J.; Hu, C.C.; Xie, B.G.; Wei, S.L.; Zhang, L.; Zhu, Z.X.; Zhang, Z.Y.; Li, S.J. A putative transcription factor LFC1 negatively regulates development and yield of winter mushroom. Appl. Microbiol. Biotechnol. 2020, 104, 5827–5844. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wei, Z.Y.; Zhang, Y.L.; Huang, R.M.; Huang, Q.H.; Xie, B.G. Identification and Expression Analysis of Two Oligopeptide Transporter Encoding Genes fvoptl and fvopt2 from Flammulina velutipes. Acta Edulis Fungi 2018, 25, 17–23. [Google Scholar]
- Wu, T.J.; Zhang, Z.Y.; Hu, C.C.; Zhang, L.; Wei, S.L.; Li, S.J. A WD40 Protein Encoding Gene Fvcpc2 Positively Regulates Mushroom Development and Yield in Flammulina velutipes. Front. Microbiol. 2020, 11, 498. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Yan, J.J.; Tong, Z.J.; Liu, Y.Y.; Miao, J.; Tao, Y.X.; Jiang, Y.J.; Xie, B.G. fv-gs6: A glucan synthase gene associated with stipe elongation of Flammulina velutipes. J. Fujian Agric. For. Univ. (Nat. Sci. Ed.) 2018, 47, 698–704. [Google Scholar]
- Yun, Y.H.; Koo, J.S.; Kim, S.H.; Kong, W.S. Cloning and Expression Analysis of Phenylalanine Ammonia-Lyase Gene in the Mycelium and Fruit Body of the Edible Mushroom Flammulina velutipes. Mycobiology 2015, 43, 327–332. [Google Scholar] [CrossRef]
- Liu, A.; Lyu, X.M.; Wang, Q.J.; Wang, X.F.; Li, Z.; ZHang, J.Y.; Wang, W. Differential expression of key genes in pheromone signaling pathway in fruiting body development of Flammulina filiformis. Microbiol. China 2022, 49, 2563–2574. [Google Scholar]
- Wang, R.Q.; Yan, J.J.; Li, Y.N.; Yang, H.; Ma, X.B.; Wang, M.; Tao, Y.X.; Xie, B.G. Cytochrome c peroxidase gene (ffccp) and its differential expression during stipe elongation in Flammulina filiformis. Mycosystema 2020, 39, 993–1005. [Google Scholar]
- Tu, J.L.; Xu, J.W. Subcellular Localization of Ganoderma lucidum CRZl and Transcription Levels of crzl at Different Developmental Stages. Acta Edulis Fungi 2021, 28, 1–9. [Google Scholar]
- Wu, F.L.; Zhang, G.; Ren, A.; Dang, Z.H.; Shi, L.; Jiang, A.L.; Zhao, M.W. The pH-responsive transcription factor PacC regulates mycelial growth, fruiting body development, and ganoderic acid biosynthesis in Ganoderma lucidum. Mycologia 2016, 108, 1104–1113. [Google Scholar]
- Zhang, G.; Ren, A.; Shi, L.; Zhu, J.; Jiang, A.L.; Shi, D.K.; Zhao, M.W. Functional analysis of an APSES transcription factor (GlSwi6) involved in fungal growth, fruiting body development and ganoderic-acid biosynthesis in Ganoderma lucidum. Microbiol. Res. 2018, 207, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Sun, Z.H.; Ren, A.; Shi, L.; Shi, D.K.; Li, X.B.; Zhao, M.W. The mitogen-activated protein kinase GlSlt2 regulates fungal growth, fruiting body development, cell wall integrity, oxidative stress and ganoderic acid biosynthesis in Ganoderma lucidum. Fungal Genet. Biol. 2017, 104, 6–15. [Google Scholar] [CrossRef]
- Li, X.; Liu, M.Q.; Dong, C.H. Hydrophobin Gene Cmhyd4 Negatively Regulates Fruiting Body Development in Edible Fungi Cordyceps militaris. Int. J. Mol. Sci. 2023, 24, 4586. [Google Scholar] [CrossRef] [PubMed]
- Bao, D.P.; Ma, Y.W.; Gong, M.; Li, Y.; Gao, Y.N.; Yang, R.H.; Yang, R.F.; Mao, W.J.; Wang, Y. Sequence analysis and heterologous expression of lectin-like gene CMLec3 from the medicinal fungus Cordyceps militaris. Mycoscience 2018, 59, 119–123. [Google Scholar] [CrossRef]
- Zhang, Z.J.; Yin, Y.Y.; Cui, Y.; Zhang, Y.X.; Liu, B.Y.; Ma, Y.C.; Liu, Y.N.; Liu, G.Q. Chitinase Is Involved in the Fruiting Body Development of Medicinal Fungus Cordyceps militaris. Life 2023, 13, 764. [Google Scholar] [CrossRef]
- Zhang, J.J.; Hao, H.B.; Liu, H.; Wang, Q.; Chen, M.J.; Feng, Z.Y.; Chen, H. Genetic and functional analysis of the Zn(II)(2)Cys(6) transcription factor HADA-1 in Hypsizygus marmoreus. Appl. Microbiol. Biotechnol. 2021, 105, 2815–2829. [Google Scholar] [CrossRef]
- Zhang, J.J.; Chen, H.; Chen, M.J.; Ren, A.; Huang, J.C.; Wang, H.; Zhao, M.W.; Feng, Z.Y. Cloning and functional analysis of a laccase gene during fruiting body formation in Hypsizygus marmoreus. Microbiol. Res. 2015, 179, 54–63. [Google Scholar] [CrossRef]
- Shioya, T.; Nakamura, H.; Ishii, N.; Takahashi, N.; Sakamoto, Y.; Ozaki, N.; Kobayashi, M.; Okano, K.; Kamada, T.; Muraguchi, H. The Coprinopsis cinerea septin Cc.Cdc3 is involved in stipe cell elongation. Fungal Genet. Biol. 2013, 58–59, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Pelkmans, J.F.; Vos, A.M.; Scholtmeijer, K.; Hendrix, E.; Baars, J.J.P.; Gehrmann, T.; Reinders, M.J.T.; Lugones, L.G.; Wösten, H.A.B. The transcriptional regulator c2h2 accelerates mushroom formation in Agaricus bisporus. Appl. Microbiol. Biot. 2016, 100, 7151–7159. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.H.; Zhang, G.; Wen, Y.M.; Li, T.; Gao, Y.Q.; Meng, F.M.; Qiu, L.Y.; Ai, Y.C. Pseudomonas sp. UW4 acdS gene promotes primordium initiation and fruiting body development of Agaricus bisporus. World J. Microb. Biot. 2019, 35, 163. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.H.; Shen, Y.Y.; Cai, W.M.; Jin, Q.L.; Fang, L.J.; Song, T.T.; Feng, W.L.; Zhang, X.H. Cloning and functional prediction of the Ppcsl-1 related to change-temperature fruiting of Pleurotus pulmonarius. Mycosystema 2016, 35, 946–955. [Google Scholar]
- Chen, B.Z.; Li, L.; Chen, T.C.; Meng, Q.M.; Wu, M.W.; Xie, B.G.; Jiang, Y.J. Construction of the overexpression transformant strains with MADS-box transcription factor Vvrin1 gene and analysis on differential growth rate of Volvariella volvacea. Mycosystema 2020, 39, 1049–1055. [Google Scholar]
- Chen, B.Z.; Li, L.; Zhong, Q.G.; Qiu, M.M.; Pan, X.Y.; Xie, B.G.; Jiang, Y.J. Expression analyses of Vvrin1 gene encoding MADS-box transcription factors in different development stages of Volvariella volvacea. Mycosystema 2018, 37, 1628–1634. [Google Scholar]
- Wang, W.; Wang, L.; Chen, B.Z.; Mukhtar, I.; Xie, B.G.; Li, Z.; Meng, L. Characterization and expression pattern of homeobox transcription factors in fruiting body development of straw mushroom Volvariella volvacea. Fungal. Biol. 2019, 123, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.J.; Tong, Z.J.; Liu, Y.Y.; Zhang, L.; Zhang, Y.; Xie, B.G. Sequence Characterization and Differential Expression Analysis of a Aryl Alcohol Oxidase Gene vvaao1 from Volvariella volvacea. Biotechnol. Bull. 2018, 34, 107–114. [Google Scholar]
- Sato, T.; Suzuki, Y.; Naito, M.; Minami, A.; Suzuki, N.; Yaegashi, K.; Hirano, T. Overexpression of the laccase gene, lcc1, in Lentinula edodes using the pChG vector. Mycoscience 2019, 60, 246–249. [Google Scholar] [CrossRef]
- Ma, X.X.; Lu, L.X.; Zhang, Y.M.; Fang, M.; Shao, K.S.; Sun, X.; Yao, F.J.; Wang, P. Velvet Family Members Regulate Pigment Synthesis of the Fruiting Bodies of Auricularia cornea. J. Fungi 2023, 9, 412. [Google Scholar] [CrossRef]
- Zhu, G.; Hayashi, M.; Shimomura, N.; Yamaguchi, T.; Aimi, T. Expression of α-glucosidase during morphological differentiation in the basidiomycetous fungus Pholiota microspora. J. Basic. Microb. 2016, 56, 1036–1045. [Google Scholar] [CrossRef]
- Zan, X.Y.; Zhu, H.A.; Jiang, L.H.; Liang, Y.Y.; Sun, W.J.; Tao, T.L.; Cui, F.J. The role of Rho1 gene in the cell wall integrity and polysaccharides biosynthesis of the edible mushroom Grifola frondosa. Int. J. Biol. Macromol. 2020, 165, 1593–1603. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.Y.; Liu, D.M.; Zhao, X.H. Transcription factors: Switches for regulating growth and development in macrofungi. Appl. Microbiol. Biot. 2023, 107, 6179–6191. [Google Scholar] [CrossRef]
- Li, L.Z.; Qu, M.R.; Liu, C.J.; Xu, L.J.; Pan, K.; Song, X.Z.; OuYang, K.H.; Li, Y.J.; Zhao, X.H. Expression of a Recombinant Lentinula edodes Xylanase by Pichia pastoris and Its Effects on Ruminal Fermentation and Microbial Community in in vitro Incubation of Agricultural Straws. Front. Microbiol. 2018, 9, 2944. [Google Scholar] [CrossRef]
- Li, L.z.; Qu, M.R.; Liu, C.J.; Pan, K.; Xu, L.J.; OuYang, K.H.; Song, X.Z.; Li, Y.J.; Zhao, X.H. Expression of a recombinant Lentinula edodes cellobiohydrolase by Pichia pastoris and its effects on in vitro ruminal fermentation of agricultural straws. Int. J. Biol. Macromol. 2019, 134, 146–155. [Google Scholar] [CrossRef]
- Liu, C.J.; Zhang, W.J.; Li, Y.J.; Pan, K.; OuYang, K.H.; Song, X.Z.; Xiong, X.W.; Zang, Y.T.; Wang, L.; Qu, M.E.; et al. Characterization of yeast cell surface displayed Lentinula edodes xylanase and its effects on the hydrolysis of wheat. Int. J. Biol. Macromol. 2022, 199, 341–347. [Google Scholar] [CrossRef]
- Gong, M.; Wang, Y.; Bao, D.P.; Jiang, S.; Chen, H.Y.; Shang, J.J.; Wang, X.J.; Yu, H.H.; Zou, G. Improving cold-adaptability of mesophilic cellulase complex with a novel mushroom cellobiohydrolase for efficient low-temperature ensiling. Bioresour Technol. 2023, 376, 128888. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Li, T.B.; Guo, J.T.; Wang, G.; Chen, G. Expression and characterization of a novel lytic polysaccharide monooxygenase, PdLPMO9A, from the edible fungus Pleurotus djamor and its synergistic interactions with cellulase in corn straw biomass saccharification. Bioresource Technol. 2022, 348, 126792. [Google Scholar] [CrossRef] [PubMed]
- Zou, G.; Bao, D.P.; Wang, Y.; Zhou, S.C.; Xiao, M.L.; Yang, Z.S.; Wang, Y.M.; Zhou, Z.H. Alleviating product inhibition of Trichoderma reesei cellulase complex with a product-activated mushroom endoglucanase. Bioresour Technol. 2021, 319, 124119. [Google Scholar] [CrossRef]
- Heneghan, M.N.; Burns, C.; Costa, A.M.S.B.; Burton, K.S.; Challen, M.P.; Bailey, A.M.; Foster, G.D. Functional analysis of Agaricus bisporus serine proteinase 1 reveals roles in utilization of humic rich substrates and adaptation to the leaf-litter ecological niche. Environ. Microbiol. 2016, 18, 4687–4696. [Google Scholar] [CrossRef]
- Jiang, N.; Hu, S.; Peng, B.; Li, Z.H.; Yuan, X.H.; Xiao, S.J.; Fu, Y.P. Genome of Ganoderma Species Provides Insights Into the Evolution, Conifers Substrate Utilization, and Terpene Synthesis for Ganoderma tsugae. Front. Microbiol. 2021, 12, 724451. [Google Scholar] [CrossRef] [PubMed]
- Lian, L.D.; Qiao, J.J.; Guo, X.Y.; Xing, Z.Z.; Ren, A.; Zhao, M.W.; Zhu, J. The transcription factor GCN4 contributes to maintaining intracellular amino acid contents under nitrogen-limiting conditions in the mushroom Ganoderma lucidum. Microb. Cell Factories 2023, 22, 205. [Google Scholar] [CrossRef]
- Pareek, M.; Hegedüs, B.; Hou, Z.H.; Csernetics, Á.; Wu, H.L.; Virágh, M.; Sahu, N.; Liu, X.B.; Nagy, L. Preassembled Cas9 Ribonucleoprotein-Mediated Gene Deletion Identifies the Carbon Catabolite Repressor and Its Target Genes in Coprinopsis cinerea. Appl. Environ. Microbiol. 2022, 88, e0094022. [Google Scholar] [CrossRef]
- Yang, H.L.; Chang, T.T.; Zhao, Y.; You, H.F.; Zha, L.; Li, Z.P.; Tong, Z.J.; Chen, M.J. Expression of catalase gene (VvCAT1) from Volvariella volvacea in Escherichia coli and its temperature tolerance. Microbiol. China 2021, 48, 4054–4060. [Google Scholar]
- Yang, H.L.; You, H.F.; Zhao, Y.; Pattana, K.; Chang, T.T.; Yu, C.X.; Cheng, M.J. Heterologous Expression of Manganese Superoxide Dismutase from Volvariella volvacea in E. coli and Characterization of ts Stress Tolerance. Acta Edulis Fungi 2020, 27, 51–58. [Google Scholar]
- Yang, H.L.; Tong, Z.J.; Zhao, Y.; Jiang, J.; Zha, L.; Cheng, M.J. Bioinformatics analysis of manganese peroxidase gene (LeMnP1) in Lentinula edodes and effects of high temperature stress on its expression. Mycosystema 2020, 39, 1056–1064. [Google Scholar]
- Yang, T.; Dong, C.H. Photo morphogenesis and photo response of the blue-light receptor gene Cmwc-1 in different strains of Cordyceps militaris. FEMS Microbiol. Lett. 2014, 352, 190–197. [Google Scholar] [CrossRef][Green Version]
- Pelkmans, J.F.; Patil, M.B.; Gehrmann, T.; Reinders, M.J.T.; Wösten, H.A.B.; Lugones, L.G. Transcription factors of Schizophyllum commune involved in mushroom formation and modulation of vegetative growth. Sci. Rep. 2017, 7, 310. [Google Scholar] [CrossRef]
- Song, L.H.; Shrivastava, N.; Gai, Y.P.; Li, D.; Cai, W.M.; Shen, Y.Y.; Lin, F.C.; Liu, J.Y.; Wang, H.K. Role of the blue light receptor gene Icwc-1 in mycelium growth and fruiting body formation of Isaria cicadae. Front. Microbiol. 2022, 13, 1038034. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, C.; Jing, Z.H.; Li, X.Y.; Li, H.; Chen, Y.Z.; Shao, Y.P.; Cai, J.F.; Wang, B.; Xie, B.G.; et al. Blue light and its receptor white collar complex (FfWCC) regulates mycelial growth and fruiting body development in Flammulina filiformis. Sci. Hortic. 2023, 309, 111623. [Google Scholar] [CrossRef]
- Kurahashi, A.; Shimoda, T.; Sato, M.; Fujimori, F.; Hirama, J.; Nishibori, K. A putative transcription factor Gf.BMR1 in Grifola frondosa, the homolog of BMR1 in Bipolaris oryzae, was strongly induced by near-ultraviolet light and blue light. Mycoscience 2015, 56, 177–182. [Google Scholar] [CrossRef][Green Version]
- Hong, C.P.; Moon, S.; Yoo, S.I.; Noh, J.H.; Ko, H.G.; Kim, H.A.; Ro, H.S.; Cho, H.; Chung, J.W.; Lee, H.Y.; et al. Functional Analysis of a Novel ABL (Abnormal Browning Related to Light) Gene in Mycelial Brown Film Formation of Lentinula edodes. J. Fungi 2020, 6, 272. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Lu, Z.L.; Shen, Y.M.; Tao, Y.; Song, S.Y. Para-aminobenzoic acid synthase from mushroom Agaricus bisporus enhances UV-C tolerance in Arabidopsis by reducing oxidative DNA damage. Acta Physiol. Plant. 2019, 41, 160. [Google Scholar] [CrossRef]
- Chen, X.; Lv, S.Y.; Mou, C.Y.; Bian, Y.B.; Kang, H. Functions of gene ATX1 under cadmium stress in Morchella importuna. Mycosystema 2020, 39, 827–838. [Google Scholar]
- Shen, N.; Xu, C.J.; Zhang, J.C.; Liu, K.F.; Liu, G.L.; He, S.F.; Wang, L.; Bian, Y.B.; Xiao, Y. Molecular mechanism underlying cadmium tolerance differentiation in Lentinula edodes as revealed by mRNA and milRNA analyses. J. Hazard. Mater. 2022, 440, 129841. [Google Scholar] [CrossRef]
- Elsayed, E.A.; El Enshasy, H.; Wadaan, M.A.; Aziz, R. Mushrooms: A potential natural source of anti-inflammatory compounds for medical applications. Mediat. Inflamm. 2014, 2014, 805841. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Pu, J.F.; Zhang, Z.X.; Feng, Z.; Han, J.; Su, X.J.; Shi, L.S. Triterpenoids of Ganoderma lucidum inhibited S180 sarcoma and H22 hepatoma in mice by regulating gut microbiota. Heliyon 2023, 9, e16682. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.F.; Ahmad, F.A.; Khan, M.I.; Alsayegh, A.A.; Wahab, S.; Alam, M.I.; Ahmed, F. Ganoderma lucidum: A potential source to surmount viral infections through β-glucans immunomodulatory and triterpenoids antiviral properties. Int. J. Biol. Macromol. 2021, 187, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.Z.; Ji, B.Y.; Wang, Z.Z.; Si, Y.Y.; Sun, Y.J.; Chen, H.; Feng, W.S.; Zheng, X.K.; Liu, J.K. Lanostane triterpenoids with anti-proliferative and anti-inflammatory activities from medicinal mushroom Ganoderma lingzhi. Phytochemistry 2023, 213, 113791. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Q.; Gao, X.X.; Long, G.Q.; Yang, Y.C.; Chen, G.; Hou, G.L.; Huo, X.T.; Jia, J.M.; Wang, A.H.; Hu, G.S. Lanostane-type triterpenoids from the mycelial mat of Ganoderma lucidum and their hepatoprotective activities. Phytochemistry 2022, 198, 113131. [Google Scholar] [CrossRef]
- Mi, X.; Zeng, G.R.; Liu, J.Q.; Luo, Z.S.; Zhang, L.; Dai, X.M.; Fang, W.T.; Zhang, J.; Chen, X.C. Ganoderma Lucidum Triterpenoids Improve Maternal Separation-Induced Anxiety- and Depression-like Behaviors in Mice by Mitigating Inflammation in the Periphery and Brain. Nutrients 2022, 14, 2268. [Google Scholar] [CrossRef]
- Tan, Y.; Tang, C.H.; Feng, J.; Feng, N.; Wu, Y.Y.; Bao, D.P.; Yang, Y.; Zhang, J.S. Current Progress in the Study on Biosynthesisand Regulation of Ganoderma Triterpenoids. Acta Edulis Fungi 2019, 26, 125–140. [Google Scholar]
- Gu, L.; Zhong, X.; Lian, D.H.; Zheng, Y.M.; Wang, H.Z.; Liu, X. Triterpenoid biosynthesis and the transcriptional response elicited by nitric oxide in submerged fermenting Ganoderma lucidum. Process Biochem. 2017, 60, 19–26. [Google Scholar] [CrossRef]
- He, X.Y.; Chen, Y.W.; Li, Z.H.; Fang, L.; Chen, H.M.; Liang, Z.S.; Abozeid, A.; Yang, Z.Q.; Yang, D.F. Germplasm resources and secondary metabolism regulation in Reishi mushroom (Ganoderma lucidum). Chin. Herb. Med. 2023, 15, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, Y.Y.; Zhang, Y.; Xiong, K.H.; Yan, X.Y.; Ruan, S.Y.; Wu, X.Q. Identification of a Novel Metabolic Target for Bioactive Triterpenoids Biosynthesis in Ganoderma lucidum. Front. Microbiol. 2022, 13, 878110. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.L.; Shi, L.; Yao, J.; Ren, A.; Zhou, C.; Mu, D.S.; ZHao, M.W. The cloning, characterization, and functional analysis of a gene encoding an isopentenyl diphosphate isomerase involved in triterpene biosynthesis in the Lingzhi or reishi medicinal mushroom Ganoderma lucidum (higher Basidiomycetes). Int. J. Med. Mushrooms 2013, 15, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Ren, A.; Ouyang, X.; Shi, L.; Jiang, A.L.; Mu, D.S.; Li, M.J.; Han, Q.; Zhao, M.W. Molecular characterization and expression analysis of GlHMGS, a gene encoding hydroxymethylglutaryl-CoA synthase from Ganoderma lucidum (Ling-zhi) in ganoderic acid biosynthesis pathway. World J. Microb. Biot. 2013, 29, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Fei, Y.; Li, N.; Zhang, D.H.; Xu, J.W. Increased production of ganoderic acids by overexpression of homologous farnesyl diphosphate synthase and kinetic modeling of ganoderic acid production in Ganoderma lucidum. Microb. Cell Fact. 2019, 18, 115. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Qin, L.; Xu, Y.J.; Ren, A.; Fang, X.; Mu, D.S.; Tan, Q.; ZHao, M.W. Molecular cloning, characterization, and function analysis of a mevalonate pyrophosphate decarboxylase gene from Ganoderma lucidum. Mol. Biol. Rep. 2012, 39, 6149–6159. [Google Scholar] [CrossRef]
- Jiang, L.X.; Wang, J.J.; Li, N.; Xu, J.W. Identification and Analysis of Candidate Cytochrome P450 Genes Involved in the Biosynthesis of Ganoderic Acid in Ganoderma lingzhi. Acta Edulis Fungi 2018, 25, 35–41. [Google Scholar]
- Shi, D.K.; Zhu, J.; Sun, Z.H.; Zhang, G.; Liu, R.; Zhang, T.J.; Wang, S.L.; Ren, A.; ZHao, M.W. Alternative oxidase impacts ganoderic acid biosynthesis by regulating intracellular ROS levels in Ganoderma lucidum. Microbiology 2017, 163, 1466–1476. [Google Scholar] [CrossRef]
- Lian, L.D.; Wang, L.S.; Song, S.Q.; Zhu, J.; Liu, R.; Shi, L.; Ren, A.; ZHao, M.W. GCN4 Regulates Secondary Metabolism through Activation of Antioxidant Gene Expression under Nitrogen Limitation Conditions in Ganoderma lucidum. Appl. Environ. Microbiol. 2021, 87, e0015621. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.R.; Xu, W.Z.; Hu, S.S.; Lian, L.D.; Zhu, J.; Shi, L.; Ren, A.; ZHao, M.W. In Ganoderma lucidum, Glsnf1 regulates cellulose degradation by inhibiting GlCreA during the utilization of cellulose. Environ. Microbiol. 2020, 22, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, C.H.; Leng, D.D.; Yan, P.; Wang, Z.H.; Zhang, M.X.; Wu, Z.W. The non-canonical functions of telomerase reverse transcriptase gene GlTert on regulating fungal growth, oxidative stress, and ganoderic acid biosynthesis in Ganoderma lucidum. Appl. Microbiol. Biotechnol. 2021, 105, 7353–7365. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ren, A.; Li, M.J.; Cao, P.F.; Chen, T.X.; Zhang, G.; Shi, L.; Jiang, A.L.; ZHao, M.W. Heat Stress Modulates Mycelium Growth, Heat Shock Protein Expression, Ganoderic Acid Biosynthesis, and Hyphal Branching of Ganoderma lucidum via Cytosolic Ca2. Appl. Environ. Microb. 2016, 82, 4112–4125. [Google Scholar] [CrossRef]
- Shi, L.; Yue, S.N.; Gao, T.; Zhu, J.; Ren, A.; Yu, H.S.; Wang, H.; ZHao, M.W. Nitrate reductase-dependent nitric oxide plays a key role on MeJA-induced ganoderic acid biosynthesis in Ganoderma lucidum. Appl. Microbiol. Biotechnol. 2020, 104, 10737–10753. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Song, S.Q.; Sun, Z.H.; Lian, L.D.; Shi, L.; Ren, A.; ZHao, M.W. Regulation of glutamine synthetase activity by transcriptional and posttranslational modifications negatively influences ganoderic acid biosynthesis in Ganoderma lucidum. Environ. Microbiol. 2021, 23, 1286–1297. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.F.; Xiao, H.; Zhong, J.J. Biosynthesis of a novel ganoderic acid by expressing CYP genes from Ganoderma lucidum in Saccharomyces cerevisiae. Appl. Microbiol. Biot. 2022, 106, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.J.; Cheng, S.; Bian, G.K.; Yan, P.; Ma, Z.N.; Dai, W.; Chen, R.; Fu, S.; Huang, H.W.; Chi, H.M.; et al. Efficient exploration of terpenoid biosynthetic gene clusters in filamentous fungi. Nat. Catal. 2022, 5, 277–287. [Google Scholar] [CrossRef]
- Cunningham, K.G.; Manson, W.; Spring, F.S.; Hutchinson, S.A. Cordycepin, a metabolic product isolated from cultures of Cordyceps militaris (Linn.) Link. Nature 1950, 166, 949. [Google Scholar] [CrossRef]
- Deng, Q.F.; Li, X.R.; Fang, C.Q.; Li, X.; Zhang, J.; Xi, Q.; Li, Y.; Zhang, R.X. Cordycepin enhances anti-tumor immunity in colon cancer by inhibiting phagocytosis immune checkpoint CD47 expression. Int. Immunopharmacol. 2022, 107, 108695. [Google Scholar] [CrossRef]
- Wei, P.J.; Wang, K.; Luo, C.; Huang, Y.C.; Misilimu, D.; Wen, H.M.; Jin, P.; Li, C.H.; Gong, Y.; Gao, Y.Q. Cordycepin confers long-term neuroprotection via inhibiting neutrophil infiltration and neuroinflammation after traumatic brain injury. J. Neuroinflamm. 2021, 18, 137. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.H.; Zha, Z.B.; Zheng, Z.; Pan, Y.M. Cordycepin suppresses vascular inflammation, apoptosis and oxidative stress of arterial smooth muscle cell in thoracic aortic aneurysm with VEGF inhibition. Int. Immunopharmacol. 2023, 116, 109759. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Madaan, K.; Kaur, R. Cordycepin as a Metabolite with Pharmacological Potential: A Review. Int. J. Med. Mushrooms 2022, 24, 1–20. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Li, Q.; Liu, W.J.; Guan, H.Q.; Li, C.; Wang, J.H.; Wang, L. Advances in biosynthesis of cordycepin from Cordyceps militaris. Chin. J. Biotechnol. 2020, 36, 1293–1304. [Google Scholar]
- Duan, X.Y.; Yang, H.; Wang, C.; Liu, H.H.; Lu, X.Y.; Tian, Y. Microbial synthesis of cordycepin, current systems and future perspectives. Trends Food Sci. Technol. 2023, 132, 162–170. [Google Scholar] [CrossRef]
- Tan, H.P.; Wang, L.; Wang, H.G.; Cheng, Y.H.; Li, X.; Wan, H.H.; Liu, C.G.; Liu, T.; Li, Q. Engineering Komagataella phaffii to biosynthesize cordycepin from methanol which drives global metabolic alterations at the transcription level. Synth. Syst. Biotechnol. 2023, 8, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.L.; Luo, F.F.; Shang, Y.F.; Chen, P.L.; Lu, Y.Z.; Wang, C.S. Fungal Cordycepin Biosynthesis Is Coupled with the Production of the Safeguard Molecule Pentostatin. Cell Chem. Biol. 2017, 24, 1479–1489.e1474. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Yan, P.F.; Lu, X.J.; Qiu, Y.L.; Liang, S.; Liu, G.; Li, S.F.; Mou, L.; Xie, N. Involvement of PaSNF1 in Fungal Development, Sterigmatocystin Biosynthesis, and Lignocellulosic Degradation in the Filamentous Fungus Podospora anserina. Front. Microbiol. 2020, 11, 1038. [Google Scholar] [CrossRef] [PubMed]
- Wāng, Y.; Wang, R.; Wáng, Y.; Li, Y.; Yang, R.H.; Gong, M.; Shang, J.J.; Zhang, J.S.; Mao, W.J.; Zou, G.; et al. Diverse function and regulation of CmSnf1 in entomopathogenic fungus Cordyceps militaris. Fungal Genet. Biol. 2020, 142, 103415. [Google Scholar] [CrossRef]
- Chen, B.X.; Wei, T.; Xue, L.N.; Zheng, Q.W.; Ye, Z.W.; Zou, Y.; Yang, Y.; Yun, F.; Guo, L.Q.; Lin, J.F. Transcriptome Analysis Reveals the Flexibility of Cordycepin Network in Cordyceps militaris Activated by L-Alanine Addition. Front. Microbiol. 2020, 11, 577. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Z.S.; Bao, D.P.; Li, B.; Yin, X.; Wu, Y.Y.; Chen, H.Y.; Tang, G.R.; Li, N.Y.; Zou, G. Improving Hypoxia Adaption Causes Distinct Effects on Growth and Bioactive Compounds Synthesis in an Entomopathogenic Fungus Cordyceps militaris. Front. Microbiol. 2021, 12, 698436. [Google Scholar] [CrossRef]
- Zou, G.; Li, B.; Wang, Y.; Yin, X.; Gong, M.; Shang, J.; Wei, Y.; Li, X.; Bao, D. Efficient conversion of spent mushroom substrate into a high value-added anticancer drug pentostatin with engineered Cordyceps militaris. Green Chem. 2021, 23, 10030–10038. [Google Scholar] [CrossRef]
- Hu, T.; Lin, Q.L.; Guo, T.; Yang, T.; Zhou, W.H.; Deng, X.F.; Yan, J.K.; Luo, Y.; Ju, M.M.; Luo, F.J. Polysaccharide isolated from Phellinus linteus mycelia exerts anti-inflammatory effects via MAPK and PPAR signaling pathways. Carbohydr. Polym. 2018, 200, 487–497. [Google Scholar] [CrossRef]
- Rao, Z.L.; Dong, Y.T.; Zheng, X.J.; Tang, K.Y.; Liu, J. Extraction, purification, bioactivities and prospect of lentinan: A review. Biocatal. Agric. BioTechnol. 2021, 37, 102163. [Google Scholar] [CrossRef]
- Yang, Y.X.; Chen, J.L.; Lei, L.; Li, F.H.; Tang, Y.; Yuan, Y.; Zhang, Y.Q.; Wu, S.R.; Yin, R.; Ming, J. Acetylation of polysaccharide from Morchella angusticeps peck enhances its immune activation and anti-inflammatory activities in macrophage RAW264.7 cells. Food Chem. Toxicol. 2019, 125, 38–45. [Google Scholar] [CrossRef]
- Bisen, P.S.; Baghel, R.K.; Sanodiya, B.S.; Thakur, G.S.; Prasad, G.B. Lentinus edodes: A macrofungus with pharmacological activities. Curr. Med. Chem. 2010, 17, 2419–2430. [Google Scholar] [CrossRef]
- Zhou, J.S.; Bai, Y.; Dai, R.J.; Guo, X.L.; Liu, Z.H.; Yuan, S. Improved Polysaccharide Production by Homologous Co-overexpression of Phosphoglucomutase and UDP Glucose Pyrophosphorylase Genes in the Mushroom Coprinopsis cinerea. J. Agric. Food Chem. 2018, 66, 4702–4709. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.F.; Yang, X.; Chen, P.; Yang, S.L.; Zhang, H. Homologous overexpression of genes in Cordyceps militaris improves the production of polysaccharides. Food Res. Int. 2021, 147, 110452. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.J.; Wu, X.H.; Tao, T.L.; Zan, X.Y.; Sun, W.J.; Mu, D.S.; Yang, Y.; Wu, D. Functions of a Glucan Synthase Gene GFGLS in Mycelial Growth and Polysaccharide Production of Grifola frondosa. J. Agric. Food Chem. 2019, 67, 8875–8883. [Google Scholar] [CrossRef]
- Jiang, L.H.; Li, X.F.; Zan, X.Y.; Fu, X.; Cui, F.J.; Zhu, H.A.; Sun, W.J.; Tao, T.L. The β-1,3-glucan synthase gene GFGLS2 plays major roles in mycelial growth and polysaccharide synthesis in Grifola frondosa. Appl. Microbiol. Biot. 2022, 106, 563–578. [Google Scholar] [CrossRef]
- Zan, X.Y.; Wu, X.H.; Cui, F.J.; Zhu, H.A.; Sun, W.J.; Jiang, L.H.; Tao, T.L.; Zhao, X. UDP-glucose pyrophosphorylase gene affects mycelia growth and polysaccharide synthesis of Grifola frondosa. Int. J. Biol. Macromol. 2020, 161, 1161–1170. [Google Scholar] [CrossRef]
- Xu, J.W.; Ji, S.L.; Li, H.J.; Zhou, J.S.; Duan, Y.Q.; Dang, L.Z.; Mo, M.H. Increased polysaccharide production and biosynthetic gene expressions in a submerged culture of Ganoderma lucidum by the overexpression of the homologous α-phosphoglucomutase gene. Bioproc. Biosyst. Eng. 2015, 38, 399–405. [Google Scholar] [CrossRef]
- Li, H.J.; Zhang, D.H.; Yue, T.H.; Jiang, L.X.; Yu, X.; Zhao, P.; Li, T.; Xu, J.W. Improved polysaccharide production in a submerged culture of Ganoderma lucidum by the heterologous expression of Vitreoscilla hemoglobin gene. J. Biotechnol. 2016, 217, 132–137. [Google Scholar] [CrossRef]
- Xu, Y.L.; Yuan, H.; Li, N.; Xiao, J.H.; Xu, J.W. Increased production and anti-senescence activity of exopolysaccharides in Ganoderma lingzhi by co-overexpression of β-1,3-glucan synthase and UDP-glucose pyrophosphorylase. Int. J. Biol. Macromol. 2023, 253, 126778. [Google Scholar] [CrossRef]
- Zou, G.; Ren, J.B.; Wu, D.; Zhang, H.N.; Gong, M.; Li, W.; Zhang, J.S.; Yang, Y. Characterization and Heterologous Expression of UDP-Glucose 4-Epimerase From a Hericium erinaceus Mutant with High Polysaccharide Production. Front. Bioeng. BioTechnol. 2021, 9, 796278. [Google Scholar] [CrossRef]
- Wu, J.Y.; Siu, K.C.; Geng, P. Bioactive Ingredients and Medicinal Values of Grifola frondosa (Maitake). Foods 2021, 10, 95. [Google Scholar] [CrossRef]
- Jędrzejewski, T.; Pawlikowska, M.; Sobocińska, J.; Wrotek, S. COVID-19 and Cancer Diseases-The Potential of Coriolus versicolor Mushroom to Combat Global Health Challenges. Int. J. Mol. Sci. 2023, 24, 4864. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Kong, H.L.; Fang, Y.P.; Nishinari, K.; Phillips, G.O. Schizophyllan: A review on its structure, properties, bioactivities and recent developments. Bioact. Carbohydr. Diet. Fibre 2013, 1, 53–71. [Google Scholar] [CrossRef]
- Sivanesan, I.; Muthu, M.; Gopal, J.; Oh, J.W. Mushroom Polysaccharide-Assisted Anticarcinogenic Mycotherapy: Reviewing Its Clinical Trials. Molecules 2022, 27, 4090. [Google Scholar] [CrossRef]
- Xu, J.; Xu, D.Z.; Hu, Q.H.; Ma, N.; Pei, F.; Su, A.X.; Ma, G.X. Immune regulatory functions of biologically active proteins from edible fungi. Front. Immunol. 2022, 13, 1034545. [Google Scholar] [CrossRef]
- Lin, Z.H.; Yeh, H.; Lo, H.C.; Hua, W.J.; Ni, M.Y.; Wang, L.K.; Chang, T.T.; Yang, M.H.; Lin, T.Y. GMI, a fungal immunomodulatory protein, ameliorates SARS-CoV-2 envelope protein-induced inflammation in macrophages via inhibition of MAPK pathway. Int. J. Biol. Macromol. 2023, 241, 124648. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.Z.; Zheng, Y.Z.; Zhou, X.W. Fungal immunomodulatory proteins: Characteristic, potential antitumor activities and their molecular mechanisms. Drug Discov. Today 2019, 24, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Rezvani, V.; Pourianfar, H.R.; Mohammadnejad, S.; Madjid Ansari, A.; Farahmand, L. Anticancer potentiality and mode of action of low-carbohydrate proteins and peptides from mushrooms. Appl. Microbiol. Biot. 2020, 104, 6855–6871. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.W.; Jia, J.; Shen, Y.H.; Zhong, M.; Chen, L.J.; Li, H.G.; Ma, H.; Guo, Z.F.; Qi, M.F.; Liu, L.X.; et al. Functional expression of FIP-fve, a fungal immunomodulatory protein from the edible mushroom Flammulina velutipes in Pichia pastoris GS115. J. Biotechnol. 2013, 168, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Li, C.C.; Zhu, C.; Yang, G.D.; Wu, J.F.; Wang, J.J.; Qiao, Y.; Fu, M.R.; Cui, X.X.; Dong, B.X. Overexpression and activity analysis of Antrodia camphorata immunomodulatory protein. Acta Microbiol. Sin. 2021, 61, 2397–2412. [Google Scholar]
- Wu, G.G.; Sun, Y.; Deng, T.S.; Song, L.L.; Li, P.; Zeng, H.J.; Tang, X.M. Identification and Functional Characterization of a Novel Immunomodulatory Protein From Morchella conica SH. Front. Immunol. 2020, 11, 559770. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.Y.; Guan, S.X.; Duan, Z.W.; Han, X.; Zhang, X.; Fan, W.L.; Li, H.G.; Chen, L.J.; Ma, H.; Liu, H.M.; et al. Molecular cloning, codon-optimized gene expression, and bioactivity assessment of two novel fungal immunomodulatory proteins from Ganoderma applanatum in Pichia. Appl. Microbiol. Biotechnol. 2018, 102, 5483–5494. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Gao, Y.N.; Li, Y.; Wan, J.N.; Yang, R.H.; Mao, W.J.; Zhou, C.L.; Tang, L.H.; Gong, M.; et al. Discovery and Characterization of the Highly Active Fungal Immunomodulatory Protein Fip-vvo82. J. Chem. Inf. Model. 2016, 56, 2103–2114. [Google Scholar] [CrossRef]
- Bao, D.P.; Bai, R.; Gao, Y.N.; Li, Y.; Wang, Y. Comparative analysis of two fungal immunomodulatory proteins LZ-8 and LZ-9 of Ganoderma lucidum. Acta Agric. Shanghai 2017, 33, 1–5. [Google Scholar]
- Bao, D.P.; Bai, R.; Gao, Y.N.; Wu, Y.Y.; Wang, Y. Computational Insights into the Molecular Mechanism of the High Immunomodulatory Activity of LZ-8 Protein Isolated from the Lingzhi or Reishi Medicinal Mushroom Ganoderma lucidum (Agaricomycetes). Int. J. Med. Mushrooms 2018, 20, 537–548. [Google Scholar] [CrossRef]
- Gao, Y.N.; Wáng, Y.; Wāng, Y.; Wu, Y.Y.; Chen, H.Y.; Yang, R.H.; Bao, D.P. Protective Function of Novel Fungal Immunomodulatory Proteins Fip-lti1 and Fip-lti2 from Lentinus tigrinus in Concanavalin A-Induced Liver Oxidative Injury. Oxid. Med. Cell. Longev. 2019, 2019, 3139689. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.H.; Wang, Y.; Wu, Y.Y.; Bao, D.P.; Bing, W.; Li, Y.; Chen, H.Y. Characterization, Recombinant Production, and Bioactivity of a Novel Immunomodulatory Protein from Hypsizygus marmoreus. Molecules 2023, 28, 4796. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Wei, X.Y.; Shi, T.Q.; Sun, X.M.; Xu, N.; Gao, C.; Zou, W. Genome-scale metabolic network models: From first-generation to next-generation. Appl. Microbiol. Biotechnol. 2022, 106, 4907–4920. [Google Scholar] [CrossRef] [PubMed]
| Method | Advantage | Disadvantage | Application Case | Refs. |
|---|---|---|---|---|
| ATMT | Efficient; copy number of DNA is inserted low; high efficiency of homologous recombination | Time-consuming; partially sensitive to acetosyringone | Construction of ATMT in H. marmoreus, with CaMV35s as a promoter to efficiently drive the expression of GUS genes | [37,38] |
| PMT | Efficient; high copy number of inserted DNA; suitable for most edible and medicinal fungi | Requires special lytic enzyme; difficult to prepare and not easy to store; low regeneration efficiency | Successful PMT of linear DNA fragments containing the bar gene into C. militaris protoplasts yielded 87 positive mutants containing the bar gene | [39,40] |
| LMT | Easy operation; transient transfection; wide range of cell types | Combining other methods for cell wall treatment; only a single copy of the gene is integrated into the chromosome | Through LMT of A. bisporus, the ACO gene was verified, and the ethylene biosynthesis pathway was explored | [41,42] |
| EP | Easy to operate; high copy number of inserted DNA; suitable for a variety of cell types | The cells are highly damaged and inefficient; requires special equipment | The pFTXHg plasmid was constructed from the ftx gene of F. filiformis and was transformed by EP, and high conversion efficiency was obtained | [37,43] |
| Species | Gene | Function | Method | Vector | Refs. | |
|---|---|---|---|---|---|---|
| Plasmid | Promoter | |||||
| Flammulina filiformis | HMG-box transcription factor (fvhom1) | G | OE | pBHG-fvhom1 | A. bisporus gpd | [44] |
| Transcription factor (Ste12-like) | E | OE | Ste12-likeOE | Pgpd | [45] | |
| Fungal immunomodulatory proteins (FIP-fve) | G | OE | pCAMBIA1301-pGPD-FIP-fve | pGPD | [46] | |
| Fungal immunomodulatory proteins (FIP) | G | RNAi | pTCK303-fve (F)-fve (R) | Ubi-1 | [47] | |
| Transcription factor (FvHmg1) | G | RNAi | Fvhmg1-RNAi | Pgpd | [48] | |
| Adenosine deaminase (Fv-ada) | G | RNAi | pFungiway-Fv-ada | Pgpd | [49] | |
| Heterotrimeric G protein α subunits (FfGa1) | E | OE RNAi | FfGa1-OE FfGa1-RNAi | Pgpd | [50] | |
| Lentinus edodes | Fungal Hsp20 protein (hsp20) | E | OE | pEHg-gdp-hsp20 | L. edodes gpd | [51] |
| Mating-type gene (LeHD1) | G | OE RNAi | LeHD1-OE LeHD1-RNAi | L. edodes gpd | [52] | |
| HMG-box transcription factor (Lelcrp1) | B | RNAi | pCAMBIA1300-g-lelcrpl | L. edodes gpd | [53] | |
| Heat shock protein 40 (LeDnaJ) | E | RNAi | pCAMBIA1300-g-dual | L. edodes gpd | [54] | |
| Anthranilate synthase (TrpE) | E | RNAi | pCAMBIA1300-g-trpE | L. edodes gpd | [55] | |
| Flavin-containing monooxygenases (YUCCA) | E | RNAi | pCAMBIA1300-g-YUCCA8 | L. edodes gpd | [56] | |
| Tryptophan synthase (LetrpB) | E | RNAi | pCAMBIA1300-g-LetrpB | L. edodes gpd | [57] | |
| Ganoderma lucidum | Delta 9 fatty acid desaturase (D9desA) | A | OE | OE::D9desA | G. lucidum gpd | [58] |
| Acelyl-CoA acelyltransferase (Gl-aact) | A | OE | pGl-aact | G. lucidum gpd | [59] | |
| 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) | A | OE | pJW-tHMGR | Pgpd | [60] | |
| Antrodia cinnamomea | 2,3-Oxidosqualene Cyclase (OSC) | A | OE | pCAM-AcOSC | CaMV35s | [61] |
| Pleurotus ostreatus | Transcription factor (Pofst3) | G | OE RNAi | pPo-GPD-Pofst3+ pPo-GPD-Pofst3− | P. ostreatus gpd | [62] |
| Cordyceps militaris | Glutathione peroxidase (gpxA) | E | OE | pDHt-gpd-bar | A. nidulans gpdA | [63] |
| Species | Name | Category | Function | Refs. |
|---|---|---|---|---|
| Pleurotus ostreatus | Vmh2, Vmh3 | Hydrophobin | Regulate the fungus to form aerial hyphae | [129] |
| polectin2 | Lectin | Promote low-temperature resistance of fruiting body | [130] | |
| fst3 | Transcription factor | Regulation of primordium and fruiting body development | [131] | |
| PoHMG11 | Transcription factor | Regulation of fruiting body maturation | [132] | |
| PoGat1 | Transcription factor | Promote lignin decomposition and fruiting body formation | [133] | |
| Pofst3 | Transcription factor | Promote the growth and development of fruiting bodies | [62] | |
| Lacc2 | Laccase | Promote the growth of aerial hyphae, primordia, and fruiting bodies | [134] | |
| Mnsod1 | Manganese superoxide dismutase | Promote the development of fruiting bodies and enhance mycelia tolerance to heat stress | [135] | |
| Flammulina filiformis | Hyd9 | Hydrophobin | Promote mycelia and fruiting body development | [136] |
| Fv-JRL1 | Lectin | Increase aerial hyphae and promote fruiting body development | [137] | |
| Fvclp1 | Transcription factor | Involved in sexual reproduction and fruiting body development | [138] | |
| ste12-like | Transcription factor | Increase aerial hyphae and promote fruiting body development | [45] | |
| pdd1 | Transcription factor | Promote fruiting body development and increase yield | [139] | |
| FvHmg1 | Negative transcription factor | Negatively regulate the fruiting body development | [48] | |
| LFC1 | Negative transcription factor | Negatively regulates fruiting body development and yield | [140] | |
| fvopt1, fvopt2 | Oligopeptide transporter | Regulation of primordium and fruiting body development | [141] | |
| Fvcpc2 | WD40 Protein | Positively regulates the development and yield of fruiting bodies | [142] | |
| fv-gs6 | β-1,6-glucan synthase | Promote cell wall synthesis and stalk elongation process | [143] | |
| Fvpal | Phenylalanine Ammonia-Lyase | Involved in fruiting body growth and development | [144] | |
| FfPkac | Adenylate-dependent protein kinase A pathway | Promote mycelia and fruiting body development | [145] | |
| ffccp | Cytochrome C peroxidase | Promote elongation of fruiting body stalk | [146] | |
| Ganoderma lucidum | crzl | Transcription factor | Involved in fruiting body growth and development | [147] |
| GlPacC | Transcription factor | Involved in mycelium growth and fruiting body development | [148] | |
| GlSwi6 | Transcription factor | Promote mycelium growth and fruiting body development | [149] | |
| GlSlt2 | Mitogen-activated protein kinases | Regulates mycelium growth, fruiting body development, and cell wall integrity | [150] | |
| Cordyceps militaris | Cmhyd4 | Hydrophobin | Negative regulation of fruiting body growth and development | [151] |
| CMLec3 | Lectin | Involved in fruiting body growth and development | [152] | |
| Chi1, Chi4 | Chitinase | Promote fruiting body growth and development | [153] | |
| Hypsizygus marmoreus | HADA-1 | Transcription factor | Promote mycelium growth and fruiting body development | [154] |
| lcc1 | Laccase | Promote fruiting body growth and development | [155] | |
| Coprinopsis cinerea | CcNsdD2 | Transcription factor | Promote secondary mycelia and fruiting body growth and development | [33] |
| Cc.Cdc3 | a homolog of S. cerevisiae CDC3 septin | Promote the elongation of fruiting body stalk cells | [156] | |
| Agaricus bisporus | c2h2 | Transcription factor | Shorten fruiting body development period | [157] |
| acdS | 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase (AcdS) | Promote primordium initiation and fruiting body development | [158] | |
| Pleurotus pulmonarius | Ppcsl-1 | Transcription factor | Regulates fruiting body development | [159] |
| Volvariella volvacea | Vvrin1 | Transcription factor | Promote mycelium growth and pigment accumulation, elongation of fruiting stalk, and opening of the lid | [160,161] |
| VvHox1-VvHox8 | Transcription factor | Affect the elongation of fruiting bodies and the formation of primordia | [162] | |
| vvaao1 | Aryl Alcohol Oxidase | Promote the formation and development of fruiting bodies | [163] | |
| Lentinula edodes | lcc1 | Laccase | Involved in fruiting body growth and development | [164] |
| Auricularia cornea | AcveA | Velvet factor family protein | Regulation of fruiting body pigment synthesis | [165] |
| Pholiota microspora | PnGcs | α-glucosidase | Involved in fruiting body development | [166] |
| Griflola frondosa | Rho1 | β-1,3-glucan synthase | Promote mycelium growth | [167] |
| Type | Species | Name | Category | Function | Refs. |
|---|---|---|---|---|---|
| Temperature | V. volvacea | VvCAT1 | Catalase | Low-temperature stress defense response | [179] |
| V. volvacea | Mn-SOD | Mn-superoxide dismutase | Heat, cold, and salt stress tolerance | [180] | |
| L. edodes | LeMnP1 | Manganese peroxidase | Rapid growth recovery under high-temperature stress | [181] | |
| L. edodes | LeDnaJ | Heat shock protein 40 | Enhance the heat resistance of mycelium | [54] | |
| L. edodes | YUCCA8 | Flavin-containing monooxygenases | Enhance the heat resistance of mycelium | [56] | |
| L. edodes | TrpB | Trytophan synthase | Defense response to heat stress | [57] | |
| L. edodes | LetrpE | Anthranilate synthase | Enhance the heat resistance of mycelium | [55] | |
| F. filiformis | FfGa1 | Heterotrimeric G protein α subunits | Strong tolerance to heat stress and maintenance of cell wall integrity | [50] | |
| Light | C. militaris | Cmwc-1 | Blue-light receptor | Colony pigmentation and promote conidia production | [182] |
| S. commune | Wc-1 Wc-2 | Blue light complex transcription factor | Inhibit vegetative growth Phototoxic protection | [183] | |
| Isaria cicadae | Icwc-1 | Blue light receptor | Regulation of fruiting body, pigment, and enzyme synthesis | [184] | |
| F. filiformis | FfWc1 FfWc2 | Blue light and its receptor white collar complex | Regulatory morphogenesis | [185] | |
| G. frondosa | Gf.BMR1 | Transcription factor | Fruiting body development and pigment accumulation | [186] | |
| L. edodes | abl-D | Abnormal browning related to light | Brown film formation of mycelial tissue | [187] | |
| A. bisporus | Pabs | Para-aminobenzoic acid synthase | Enhance UV tolerance | [188] | |
| Metal stress | M. esculenta | ATX1 | Copper chaperones | Regulation of cadmium metabolism | [189] |
| L. edodes | LeAmy | α amylase | Enhance cadmium tolerance | [190] |
| Gene | Mode of Action and Change of GA Content | Refs. |
|---|---|---|
| Alternative oxidase (AOX) | Silent AOX, ROS content ↑, GA content ↑ | [206] |
| Transcription factor (GCN4) | Silent GCN4, ROS content ↑, GA content ↑ | [207] |
| AMP-activated protein kinase homolog (GlSnf1) | Silent GlSnf1, ROS content ↑, GA content ↑ | [208] |
| Telomerase reverse transcriptase (GlTert) | Silent GlTert, ROS content ↑, GA content ↓ | [209] |
| Calcium-permeable channel (Cch) | Silent Cch, Ca2+ content ↓, GA content ↓ | [210] |
| Nitrate reductase (NR) | Silent NR, GA content ↑ | [211] |
| Glutamine synthetase (GS) | Silent GS, GA content ↓ | [212] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Zou, G.; Bao, D.; Wu, Y. Current Advances in the Functional Genes of Edible and Medicinal Fungi: Research Techniques, Functional Analysis, and Prospects. J. Fungi 2024, 10, 311. https://doi.org/10.3390/jof10050311
Li W, Zou G, Bao D, Wu Y. Current Advances in the Functional Genes of Edible and Medicinal Fungi: Research Techniques, Functional Analysis, and Prospects. Journal of Fungi. 2024; 10(5):311. https://doi.org/10.3390/jof10050311
Chicago/Turabian StyleLi, Wenyun, Gen Zou, Dapeng Bao, and Yingying Wu. 2024. "Current Advances in the Functional Genes of Edible and Medicinal Fungi: Research Techniques, Functional Analysis, and Prospects" Journal of Fungi 10, no. 5: 311. https://doi.org/10.3390/jof10050311
APA StyleLi, W., Zou, G., Bao, D., & Wu, Y. (2024). Current Advances in the Functional Genes of Edible and Medicinal Fungi: Research Techniques, Functional Analysis, and Prospects. Journal of Fungi, 10(5), 311. https://doi.org/10.3390/jof10050311






