Discovery of a Unique Set of Dog-Seroreactive Coccidioides Proteins Using Nucleic Acid Programmable Protein Array
Abstract
1. Introduction
2. Materials and Methods
2.1. NAPPA
2.2. Serum Specimens
2.3. Data Analysis
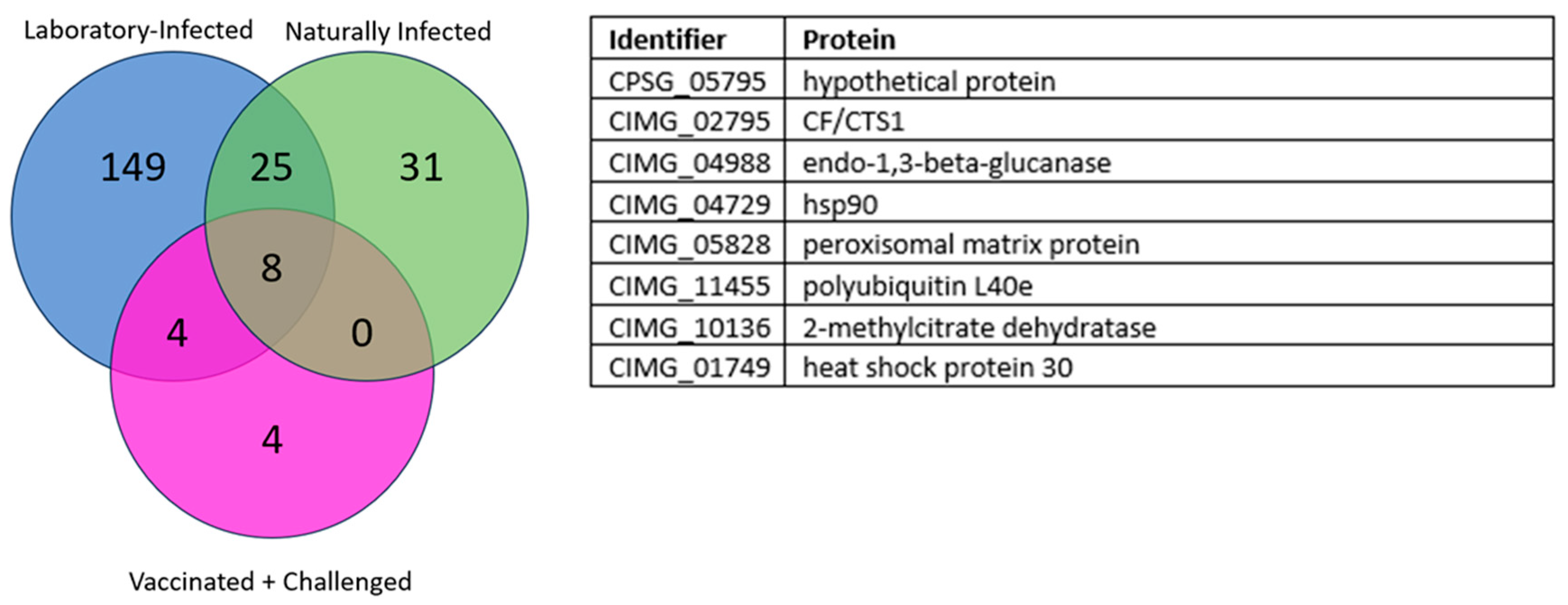
3. Results
3.1. Overlapping Proteins between the Different Groups
3.1.1. Laboratory and Naturally Infected Dogs
3.1.2. Laboratory-Infected, Naturally Infected, and Vaccinated and Challenged Dogs
3.2. Longitudinal Analysis
3.2.1. Laboratory-Infected Dogs
3.2.2. Vaccinated and Challenged Dogs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galgiani, J.N.; Kauffman, C.A. Coccidioidomycosis and Histoplasmosis in Immunocompetent Persons. N. Engl. J. Med. 2024, 390, 536–547. [Google Scholar] [CrossRef]
- Laniado-Laborin, R. Expanding Understanding of Epidemiology of Coccidioidomycosis in the Western Hemisphere. In Annals of the New York Academy of Sciences; Blackwell Publishing Inc.: Hoboken, NJ, USA, 2007; Volume 1111, pp. 19–34. [Google Scholar]
- Gorris, M.E.; Treseder, K.K.; Zender, C.S.; Randerson, J.T. Expansion of Coccidioidomycosis Endemic Regions in the United States in Response to Climate Change. Geohealth 2019, 3, 308–327. [Google Scholar] [CrossRef]
- Butkiewicz, C.D.; Shubitz, L.F.; Dial, S.M. Risk Factors Associated with Coccidioides Infection in Dogs. J. Am. Vet. Med. Assoc. 2005, 226, 1851–1854. [Google Scholar] [CrossRef]
- Pappagianis, D.; Zimmer, B.L. Serology of Coccidioidomycosis. Clin. Microbiol. Rev. 1990, 3, 247–268. [Google Scholar] [CrossRef]
- Cox, R.A.; Britt, L.A. Isolation of a Coccidioidin Component That Reacts with Immunoglobulin M Precipitin Antibody. Infect. Immun. 1986, 53, 449–453. [Google Scholar] [CrossRef]
- Dolan, M.J.; Cox, R.A.; Williams, V.; Woolley, S. Development and Characterization of a Monoclonal Antibody against the Tube Precipitin Antigen of Coccidioides Immitis. Infect. Immun. 1989, 57, 1035–1039. [Google Scholar] [CrossRef]
- Hung, C.Y.; Yu, J.J.; Lehmann, P.F.; Cole, G.T. Cloning and Expression of the Gene Which Encodes a Tube Precipitin Antigen and Wall-Associated Beta-Glucosidase of Coccidioides Immitis. Infect. Immun. 2001, 69, 2211–2222. [Google Scholar] [CrossRef]
- Johnson, S.M.; Zimmermann, C.R.; Pappagianis, D. Amino-Terminal Sequence Analysis of the Coccidioides Immitis Chitinase/Immunodiffusion-Complement Fixation Protein. Infect. Immun. 1993, 61, 3090–3092. [Google Scholar] [CrossRef]
- Yang, C.; Zhu, Y.; Magee, D.M.; Cox, R.A. Molecular Cloning and Characterization of the Coccidioides Immitis Complement Fixation/Chitinase Antigen. Infect. Immun. 1996, 64, 1992–1997. [Google Scholar] [CrossRef]
- Yang, M.C.; Magee, D.M.; Cox, R.A. Mapping of a Coccidioides Immitis-Specific Epitope That Reacts with Complement-Fixing Antibody. Infect. Immun. 1997, 65, 4068–4074. [Google Scholar] [CrossRef]
- Johnson, S.M.; Pappagianis, D. The Coccidioidal Complement Fixation and Immunodiffusion-Complement Fixation Antigen Is a Chitinase. Infect. Immun. 1992, 60, 2588–2592. [Google Scholar] [CrossRef]
- Mitchell, N.M.; Sherrard, A.L.; Dasari, S.; Magee, D.M.; Grys, T.E.; Lake, D.F. Proteogenomic Re-Annotation of Coccidioides Posadasii Strain Silveira. Proteomics 2018, 18, 1700173. [Google Scholar] [CrossRef]
- Song, L.; Wallstrom, G.; Yu, X.; Hopper, M.; Van Duine, J.; Steel, J.; Park, J.; Wiktor, P.; Kahn, P.; Brunner, A.; et al. Identification of Antibody Targets for Tuberculosis Serology Using High-Density Nucleic Acid Programmable Protein Arrays. Mol. Cell. Proteom. 2017, 16, S277–S289. [Google Scholar] [CrossRef]
- Shubitz, L.F.; Robb, E.J.; Powell, D.A.; Bowen, R.A.; Bosco-Lauth, A.; Hartwig, A.; Porter, S.M.; Trinh, H.; Moale, H.; Bielefeldt-Ohmann, H.; et al. Δcps1 Vaccine Protects Dogs against Experimentally Induced Coccidioidomycosis. Vaccine 2021, 39, 6894–6901. [Google Scholar] [CrossRef]
- Lal, D.; Song, L.; Brar, T.; Cope, E.K.; Keim, P.; Williams, S.; Chung, Y.; Murugan, V.; LaBaer, J.; Magee, D.M. Antibody Responses to the Host Microbiome in Chronic Rhinosinusitis. Int. Forum Allergy Rhinol. 2023, 13, 1503–1510. [Google Scholar] [CrossRef]
- Mitchell, N.M.; Dasari, S.; Grys, T.E.; Lake, D.F. Laser Capture Microdissection-Assisted Protein Biomarker Discovery from Coccidioides-Infected Lung Tissue. J. Fungi 2020, 6, 365. [Google Scholar] [CrossRef]
- Kruse, D.; Cole, G.T. A Seroreactive 120-Kilodalton Beta-1,3-Glucanase of Coccidioides Immitis Which May Participate in Spherule Morphogenesis. Infect. Immun. 1992, 60, 4350–4363. [Google Scholar] [CrossRef]
- Kellner, E.M.; Orsborn, K.I.; Siegel, E.M.; Mandel, M.A.; Orbach, M.J.; Galgiani, J.N. Coccidioides Posadasii Contains a Single 1,3-Beta-Glucan Synthase Gene That Appears to Be Essential for Growth. Eukaryot. Cell 2005, 4, 111–120. [Google Scholar] [CrossRef]
- Campuzano, A.; Zhang, H.; Ostroff, G.R.; Dos Santos Dias, L.; Wuthrich, M.; Klein, B.S.; Yu, J.J.; Lara, H.H.; Lopez-Ribot, J.L.; Hung, C.Y. CARD9-Associated Dectin-1 and Dectin-2 Are Required for Protective Immunity of a Multivalent Vaccine against Coccidioides Posadasii Infection. J. Immunol. 2020, 204, 3296–3306. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lee, T.-J.; Fites, S.J.; Merkhofer, R.; Zarnowski, R.; Brandhorst, T.; Galles, K.; Klein, B.; Wüthrich, M. Ligation of Dectin-2 with a Novel Microbial Ligand Promotes Adjuvant Activity for Vaccination. PLoS Pathog. 2017, 13, e1006568. [Google Scholar] [CrossRef] [PubMed]
- Delgado, N.; Xue, J.; Yu, J.J.; Hung, C.Y.; Cole, G.T. A Recombinant Beta-1,3-Glucanosyltransferase Homolog of Coccidioides Posadasii Protects Mice against Coccidioidomycosis. Infect. Immun. 2003, 71, 3010–3019. [Google Scholar] [CrossRef]
- Orsborn, K.I.; Shubitz, L.F.; Peng, T.; Kellner, E.M.; Orbach, M.J.; Haynes, P.A.; Galgiani, J.N. Protein Expression Profiling of Coccidioides Posadasii by Two-Dimensional Differential in-Gel Electrophoresis and Evaluation of a Newly Recognized Peroxisomal Matrix Protein as a Recombinant Vaccine Candidate. Infect. Immun. 2006, 74, 1865–1872. [Google Scholar] [CrossRef]
- Zhu, Y.; Yang, C.; Magee, D.M.; Cox, R.A. Molecular Cloning and Characterization of Coccidioides Immitis Antigen 2 CDNA. Infect. Immun. 1996, 64, 2695–2699. [Google Scholar] [CrossRef]
- Jiang, C.; Magee, D.M.; Quitugua, T.N.; Cox, R.A. Genetic Vaccination against Coccidioides Immitis: Comparison of Vaccine Efficacy of Recombinant Antigen 2 and Antigen 2 CDNA. Infect. Immun. 1999, 67, 630–635. [Google Scholar] [CrossRef]
- Peng, T.; Shubitz, L.; Simons, J.; Perrill, R.; Orsborn, K.I.; Galgiani, J.N. Localization within a Proline-Rich Antigen (Ag2/PRA) of Protective Antigenicity against Infection with Coccidioides Immitis in Mice. Infect. Immun. 2002, 70, 3330–3335. [Google Scholar] [CrossRef]
- Blair, J.E.; Currier, J.T. Significance of Isolated Positive IgM Serologic Results by Enzyme Immunoassay for Coccidioidomycosis. Mycopathologia 2008, 166, 77–82. [Google Scholar] [CrossRef]

| Weeks Post Infection | |||||
|---|---|---|---|---|---|
| Identifier: | Protein: | 2 | 4 | 6 | 8 |
| CIMG_06562 | nucleoside diphosphate kinase | 5 | 2 | 2 | 6 |
| CIMG_01267 | 26S proteosome regulatory subunit N7 | 3 | 2 | 1 | 2 |
| CPSG_05795 | Hypothetical | 9 | 30 | 37 | 18 |
| CIMG_04729 | Hsp90 | 1 | 4 | 4 | 4 |
| CIMG_02795 | CF/CTS1 | 1 | 41 | 96 | 96 |
| CIMG_04988 | Endo-1,3-Beta-Glucanase | 1 | 10 | 24 | 44 |
| CIMG_05828 | Peroxisomal Matrix Protein | 1 | 2 | 7 | 16 |
| Weeks Post Vaccine | Weeks Post Infection | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifier: | Protein: | 2 | 4 | 6 | 10 | 2 | 4 | 6 | 8 |
| CIMG_02795 | CF/CTS1 | 1 | 10 | 20 | 5 | 5 | 10 | 10 | 20 |
| CIMG_04988 | Endo-1,3-Beta-Glucanase | 2 | 10 | 20 | 20 | 5 | 10 | 20 | 10 |
| CPSG_05795 | Hypothetical | 10 | 5 | 50 | 20 | 50 | 20 | 50 | 20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koehler, M.A.; Song, L.; Grill, F.J.; Shubitz, L.F.; Powell, D.A.; Galgiani, J.N.; Orbach, M.J.; Robb, E.J.; Chung, Y.; Williams, S.A.; et al. Discovery of a Unique Set of Dog-Seroreactive Coccidioides Proteins Using Nucleic Acid Programmable Protein Array. J. Fungi 2024, 10, 307. https://doi.org/10.3390/jof10050307
Koehler MA, Song L, Grill FJ, Shubitz LF, Powell DA, Galgiani JN, Orbach MJ, Robb EJ, Chung Y, Williams SA, et al. Discovery of a Unique Set of Dog-Seroreactive Coccidioides Proteins Using Nucleic Acid Programmable Protein Array. Journal of Fungi. 2024; 10(5):307. https://doi.org/10.3390/jof10050307
Chicago/Turabian StyleKoehler, Megan A., Lusheng Song, Francisca J. Grill, Lisa F. Shubitz, Daniel A. Powell, John N. Galgiani, Marc J. Orbach, Edward J. Robb, Yunro Chung, Stacy A. Williams, and et al. 2024. "Discovery of a Unique Set of Dog-Seroreactive Coccidioides Proteins Using Nucleic Acid Programmable Protein Array" Journal of Fungi 10, no. 5: 307. https://doi.org/10.3390/jof10050307
APA StyleKoehler, M. A., Song, L., Grill, F. J., Shubitz, L. F., Powell, D. A., Galgiani, J. N., Orbach, M. J., Robb, E. J., Chung, Y., Williams, S. A., Murugan, V., Park, J.-g., LaBaer, J., Lake, D. F., & Magee, D. M. (2024). Discovery of a Unique Set of Dog-Seroreactive Coccidioides Proteins Using Nucleic Acid Programmable Protein Array. Journal of Fungi, 10(5), 307. https://doi.org/10.3390/jof10050307






