Whole-Genome Sequence Analysis of Flammulina filiformis and Functional Validation of Gad, a Key Gene for γ-Aminobutyric Acid Synthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Strains and Plasmids
2.2. Culture Medium
2.3. F. filiformis Fruiting Body Cultivation
2.4. Determination of GABA Content in the Fruiting Body of F. filiformis
2.5. F. filiformis Monokaryon Genome Sequencing and Analysis
2.5.1. Clamp Connection Observation Under Fluorescence Microscope
2.5.2. Mycelium Collection
2.5.3. Genome Sequencing and Assembly
2.5.4. Gene Prediction and Annotation
2.5.5. Gene Family and Species Tree Construction
2.5.6. Identification of CAZymes
2.5.7. Cytochrome P450 Gene Family Identification and Phylogenetic Tree Construction for F. filiformis
2.6. Mining and Bioinformatics Analysis of Key GABA Synthesis Gene Gad
2.7. Construction of Heterologous Ff-gad2 Expression Plasmid in F. filiformis
2.8. Transformation and Validation of Ff-gad2 in H. marmoreus
2.9. Determination of T-DNA Integration Sites in H. marmoreus
2.10. Real-Time Fluorescence Quantitative PCR of Exogenous Ff-gad2 in H. marmoreus
2.11. Determination of GABA Content of Transformations
2.12. Mycelial Growth Rate and Aerial Mycelial Fresh Weight Determination in Transformations
2.13. Data Analysis
3. Results and Analysis
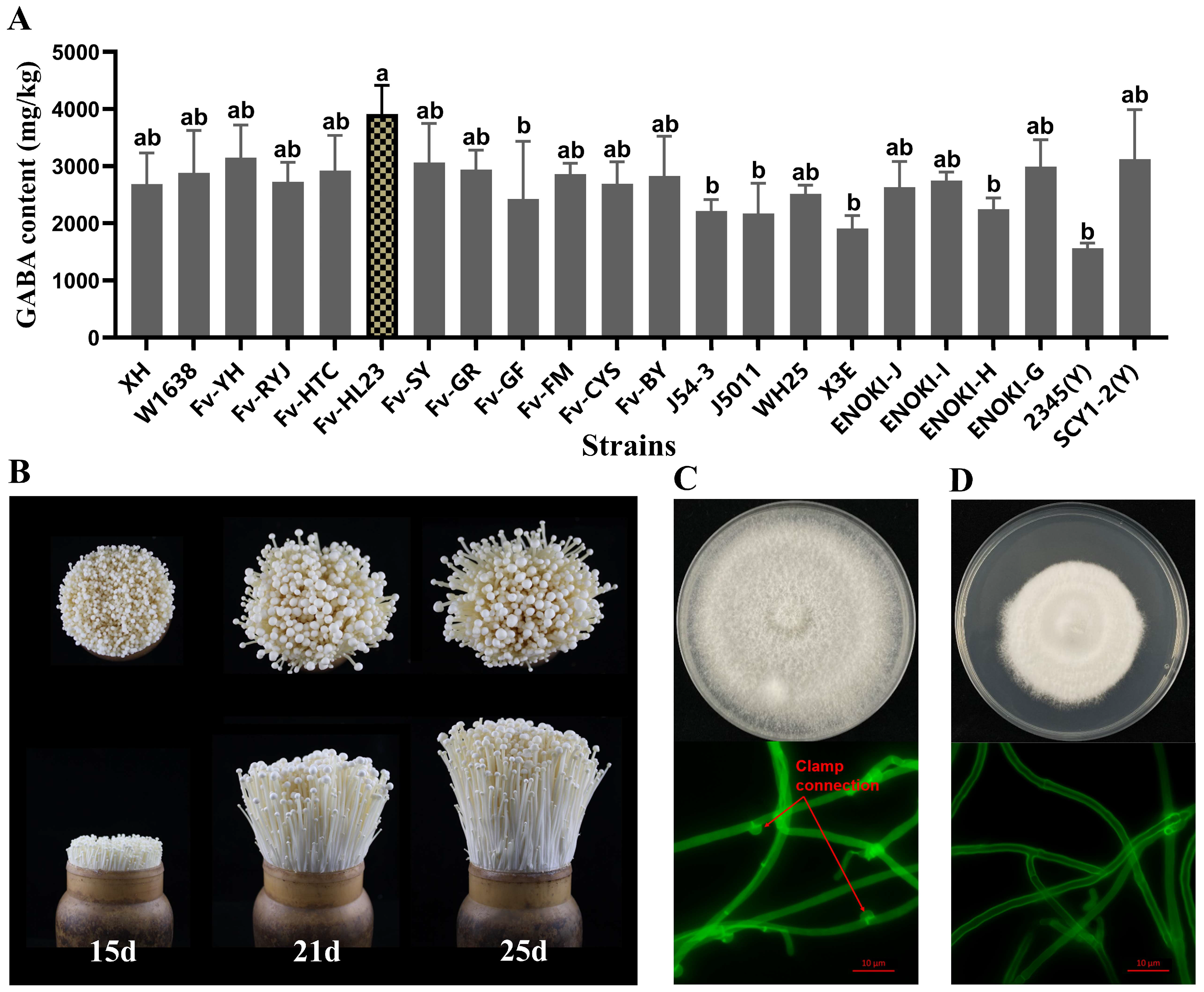
3.1. Screening of F. filiformis Strains with High GABA Content and Isolation of Monocytic Strains
3.2. Gene Assembly and Analysis of Monosomal Strain Fv-HL23-1
3.3. Phylogenetic Analysis
3.4. CAZyme Analysis
3.5. Analysis of Cytochrome P450 in F. filiformis
3.6. Bioinformatics Analysis of Gad in F. filiformis and H. marmoreus
3.7. Functional Validation of GABA Synthesis Gene Ff-gad2 in F. filiformis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Royse, D.J.; Baars, J.; Tan, Q. Current Overview of Mushroom Production in the World: Technology and Applications; Wiley: Hoboken, NJ, USA, 2017; pp. 5–13. [Google Scholar]
- Association, C.E.F. Analysis of the results of the 2022 national statistical survey on edible fungi. Edible Fungi China 2024, 43, 118–126. [Google Scholar]
- Ma, S.; Zhang, H.C.; Xu, J.X. Characterization, Antioxidant and Anti-Inflammation Capacities of Fermented Flammulina velutipes Polyphenols. Molecules 2021, 26, 6205. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xu, D.Z.; Hu, Q.H.; Ma, N.; Pei, F.; Su, A.X.; Ma, G.X. Immune regulatory functions of biologically active proteins from edible fungi. Front. Immunol. 2022, 13, 1034545. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Zhang, H. Advances in the extraction, purification, structural-property relationships and bioactive molecular mechanism of Flammulina velutipes polysaccharides: A review. Int. J. Biol. Macromol. 2021, 167, 528–538. [Google Scholar] [CrossRef]
- Hao, Y.T.; Wang, X.D.; Yuan, S.J.; Wang, Y.Y.; Liao, X.S.; Zhong, M.L.; He, Q.N.; Shen, H.B.; Liao, W.Z.; Shen, J. Flammulina velutipes polysaccharide improves C57BL/6 mice gut health through regulation of intestine microbial metabolic activity. Int. J. Biol. Macromol. 2021, 167, 1308–1318. [Google Scholar] [CrossRef]
- Li, W.Y.; Zou, G.; Bao, D.P.; Wu, Y.Y. Current Advances in the Functional Genes of Edible and Medicinal Fungi: Research Techniques, Functional Analysis, and Prospects. J. Fungi 2024, 10, 311. [Google Scholar] [CrossRef]
- Cao, L.P.; Zhang, Q.; Miao, R.Y.; Lin, J.B.; Feng, R.C.; Ni, Y.Q.; Li, W.S.; Yang, D.L.; Zhao, X. Application of omics technology in the research on edible fungi. Curr. Res. Food Sci. 2023, 6, 100430. [Google Scholar] [CrossRef]
- Park, Y.J.; Baek, J.H.; Lee, S.; Kim, C.; Rhee, H.; Kim, H.; Seo, J.S.; Park, H.R.; Yoon, D.E.; Nam, J.Y.; et al. Whole genome and global gene expression analyses of the model mushroom Flammulina velutipes reveal a high capacity for lignocellulose degradation. PLoS ONE 2014, 9, e93560. [Google Scholar] [CrossRef]
- Fu, Y.; Tan, H.; Wang, B.; Peng, W.H.; Sun, Q.; Yu, Y. Integrated multi-omic analyses on yellow Flammulina filiformis cultivar reveal postharvest oxidative damage responses. Postharvest Biol. Technol. 2023, 195, 112111. [Google Scholar] [CrossRef]
- Chen, J.; Li, J.M.; Tang, Y.J.; Ma, K.; Li, B.; Zeng, X.; Liu, X.B.; Li, Y.; Yang, Z.L.; Xu, W.N.; et al. Genome-wide analysis and prediction of genes involved in the biosynthesis of polysaccharides and bioactive secondary metabolites in high-temperature-tolerant wild Flammulina filiformis. BMC Genom. 2020, 21, 719. [Google Scholar] [CrossRef]
- Xu, B.; Long, Y.; Feng, X.Y.; Zhu, X.J.; Sai, N.; Chirkova, L.; Betts, A.; Herrmann, J.; Edwards, E.J.; Okamoto, M.; et al. GABA signalling modulates stomatal opening to enhance plant water use efficiency and drought resilience. Nat. Commun. 2021, 12, 1952. [Google Scholar] [CrossRef] [PubMed]
- Koh, W.; Kwak, H.; Cheong, E.; Lee, C.J. GABA tone regulation and its cognitive functions in the brain. Nat. Rev. Neurosci. 2023, 24, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.L.; Chen, H.Y.; Bao, D.P.; Tan, Y.S.; Zhong, Y.J.; Zhang, X.; Wu, Y.Y. Recent advances of γ-aminobutyric acid: Physiological and immunity function, enrichment, and metabolic pathway. Front. Nutr. 2022, 9, 1076223. [Google Scholar]
- Mozzarelli, A.; Bettati, S. Exploring the pyridoxal 5′-phosphate-dependent enzymes. Chem. Rec. 2006, 6, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Takayama, M.; Koike, S.; Kusano, M.; Matsukura, C.; Saito, K.; Ariizumi, T.; Ezura, H. Tomato Glutamate Decarboxylase Genes SlGAD2 and SlGAD3 Play Key Roles in Regulating γ-Aminobutyric Acid Levels in Tomato (Solanum lycopersicum). Plant Cell Physiol. 2015, 56, 1533–1545. [Google Scholar] [CrossRef]
- Turano, F.J.; Fang, T.K. Characterization of Two Glutamate Decarboxylase cDNA Clones from Arabidopsis. Plant Physiol. 1998, 117, 1411–1421. [Google Scholar] [CrossRef]
- Wen, Q.; Zhao, H.Y.; Shao, Y.H.; Hu, Y.R.; Qi, Y.C.; Wang, F.Q.; Shen, J.W. Content determination and factors influencing production of γ-aminobutyric acid content in fruiting bodies of main edible fungi in China. Mycosystema 2023, 42, 231–243. [Google Scholar]
- Lin, S.Y.; Chen, Y.K.; Yu, H.T.; Barseghyan, G.S.; Asatiani, M.D.; Wasser, S.P.; Mau, J.L. Comparative study of contents of several bioactive components in fruiting bodies and mycelia of culinary-medicinal mushrooms. Int. J. Med. Mushrooms 2013, 15, 315–323. [Google Scholar] [CrossRef]
- Chen, S.Y.; Ho, K.J.; Hsieh, Y.J.; Wang, L.T.; Mau, J.L. Contents of lovastatin, γ-aminobutyric acid and ergothioneine in mushroom fruiting bodies and mycelia. LWT-Food Sci. Technol. 2012, 47, 274–278. [Google Scholar] [CrossRef]
- Cohen, N.; Cohen, J.; Asatiani, M.D.; Varshney, V.K.; Yu, H.T.; Yang, Y.C.; Li, Y.H.; Mau, J.L.; Wasser, S.P. Chemical composition and nutritional and medicinal value of fruit bodies and submerged cultured mycelia of culinary-medicinal higher Basidiomycetes mushrooms. Int. J. Med. Mushrooms 2014, 16, 273–291. [Google Scholar] [CrossRef]
- Zhao, J.; Chang, S.T. Monokaryotization by protoplasting heterothallic species of edible mushrooms. World J. Microbiol. Biotechnol. 1993, 9, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Morgenstern, B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005, 33, W465–W467. [Google Scholar] [CrossRef]
- Majoros, W.H.; Pertea, M.; Salzberg, S.L. TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics 2004, 20, 2878–2879. [Google Scholar] [CrossRef]
- Ter-Hovhannisyan, V.; Lomsadze, A.; Chernoff, Y.O.; Borodovsky, M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008, 18, 1979–1990. [Google Scholar] [CrossRef]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef]
- Flynn, J.M.; Hubley, R.; Goubert, C.; Rosen, J.; Clark, A.G.; Feschotte, C.; Smit, A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 2020, 117, 9451–9457. [Google Scholar] [CrossRef]
- Tempel, S. Using and understanding RepeatMasker. Methods Mol. Biol. 2012, 859, 29–51. [Google Scholar]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Attwood, T.K.; Babbitt, P.C.; Bateman, A.; Bork, P.; Bridge, A.J.; Chang, H.Y.; Dosztányi, Z.; El-Gebali, S.; Fraser, M.; et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017, 45, D190–D199. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Shang, J.J.; Li, Y.; Yang, R.H.; Wang, Y.; Mao, W.J.; Tang, L.H.; Wu, Y.Y.; Nakazawa, T.; Honda, Y.; Li, Y.; et al. Efficient transformation of Pleurotus eryngii with a safe selective marker mutated from the Pesdi1 gene. J. Microbiol. Methods 2018, 152, 7–9. [Google Scholar] [CrossRef]
- Bao, D.P.; Huang, Z.H.; Li, Y.; Zhou, C.L.; Li, Y.; Wan, J.N.; Tang, L.H.; Mao, W.J.; Wang, Y.; Gong, M.; et al. Agrobacterium-mediated transformation of arthroconidia obtained from the edible mushroom Hypsizygus marmoreus. J. Microbiol. Methods 2020, 171, 105878. [Google Scholar] [CrossRef]
- Du, S.S.; Bao, D.P.; Ma, L.J.; Li, X.L.; Shang, J.J. Expression analyses of exogenous genes controlled by different promoters in Hypsizygus marmoreus. Mycosystema 2021, 40, 2065–2073. [Google Scholar]
- Qi, Y.C.; Zhang, R.X.; Zhang, M.X.; Wen, Q.; Shen, J.W. Effects of exogenous ascorbic acid on the mycelia growth and primordia formation of Pleurotus ostreatus. J. Basic Microbiol. 2021, 61, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Chen, H.Y.; Zhou, C.L.; Gong, M.; Li, Y.; Shao, Y.R.; Wu, Y.Y.; Bao, D.P. Exogenous γ-Aminobutyric Acid (GABA) Enhanced Response to Abiotic Stress in Hypsizygus marmoreus by Improving Mycelial Growth and Antioxidant Capacity. Metabolites 2024, 14, 94. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.Y.; Lu, X.; Lin, Q.; Wu, X.P. Study on the differences of monokaryotic mycelium and dikaryotic mycelium of Ganoderma lucidum. Sci. Technol. Food Ind. 2016, 37, 211–215. [Google Scholar]
- Hou, D.; Zhou, C.L.; Li, Y.; Yang, R.H.; Bao, D.P. The gene expression profiles of promoting mycelial growth of monokaryotic strains of Lentinula edodes cultured on PDA with additional sawdust. Mycosystema 2023, 42, 507–519. [Google Scholar]
- Petersen, R.H.; Hughes, K.W. Species and Speciation in Mushrooms: Development of a species concept poses difficulties. BioScience 1999, 49, 440–452. [Google Scholar] [CrossRef]
- Dymond, J.; Boeke, J. The Saccharomyces cerevisiae SCRaMbLE system and genome minimization. Bioeng. Bugs 2012, 3, 168–171. [Google Scholar] [CrossRef]
- Li, W.C.; Huang, C.H.; Chen, C.L.; Chuang, Y.C.; Tung, S.Y.; Wang, T.F. Trichoderma reesei complete genome sequence, repeat-induced point mutation, and partitioning of CAZyme gene clusters. Biotechnol. Biofuels 2017, 10, 170. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, Y.J.; Xiao, G.H.; Zheng, P.; Xia, Y.L.; Zhang, X.Y.; St Leger, R.J.; Liu, X.Z.; Wang, C.S. Genome survey uncovers the secrets of sex and lifestyle in caterpillar fungus. Chin. Sci. Bull. 2013, 58, 2846–2854. [Google Scholar] [CrossRef]
- Xia, E.H.; Yang, D.R.; Jiang, J.J.; Zhang, Q.J.; Liu, Y.; Liu, Y.L.; Zhang, Y.; Zhang, H.B.; Shi, C.; Tong, Y.; et al. The caterpillar fungus, Ophiocordyceps sinensis, genome provides insights into highland adaptation of fungal pathogenicity. Sci. Rep. 2017, 7, 1806. [Google Scholar] [CrossRef]
- Murat, C.; Payen, T.; Noel, B.; Kuo, A.; Morin, E.; Chen, J.; Kohler, A.; Krizsán, K.; Balestrini, R.; Da Silva, C.; et al. Pezizomycetes genomes reveal the molecular basis of ectomycorrhizal truffle lifestyle. Nat. Ecol. Evol. 2018, 2, 1956–1965. [Google Scholar] [CrossRef]
- Kellis, M.; Patterson, N.; Endrizzi, M.; Birren, B.; Lander, E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 2003, 423, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.L.; Yang, W.J.; Qiu, T.M.; Gao, X.; Zhang, H.Y.; Zhang, S.L.; Cui, H.; Guo, L.Z.; Yu, H.L.; Yu, H. Complete genome sequences and comparative secretomic analysis for the industrially cultivated edible mushroom Lyophyllum decastes reveals insights on evolution and lignocellulose degradation potential. Front. Microbiol. 2023, 14, 1137162. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.L.; Zhang, M.Y.; Sun, Y.T.; Li, Q.Z.; Liu, J.Y.; Song, C.Y.; Shang, X.D.; Tan, Q.T.; Zhang, L.J.; Yu, H. Whole-genome sequence of a high-temperature edible mushroom Pleurotus giganteus (zhudugu). Front. Microbiol. 2022, 13, 941889. [Google Scholar] [CrossRef]
- Wei, J.; Cheng, M.; Zhu, J.F.; Zhang, Y.L.; Cui, K.; Wang, X.J.; Qi, J.Z. Comparative Genomic Analysis and Metabolic Potential Profiling of a Novel Culinary-Medicinal Mushroom, Hericium rajendrae (Basidiomycota). J. Fungi 2023, 9, 1018. [Google Scholar] [CrossRef]
- Zhao, C.H.; Feng, X.L.; Wang, Z.X.; Qi, J.Z. The First Whole Genome Sequencing of Agaricus bitorquis and Its Metabolite Profiling. J. Fungi 2023, 9, 485. [Google Scholar] [CrossRef]
- Gavande, P.V.; Goyal, A.; Fontes, C.M.G.A. Chapter 1—Carbohydrates and Carbohydrate-Active enZymes (CAZyme): An overview. In Glycoside Hydrolases; Goyal, A., Sharma, K., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 1–23. [Google Scholar]
- Geng, Y.Y.; Zhang, S.X.; Yang, N.X.; Qin, L.K. Whole-Genome Sequencing and Comparative Genomics Analysis of the Wild Edible Mushroom (Gomphus purpuraceus) Provide Insights into Its Potential Food Application and Artificial Domestication. Genes 2022, 13, 1628. [Google Scholar] [CrossRef]
- Chen, B.Z.; Gui, F.; Xie, B.G.; Deng, Y.; Sun, X.Y.; Lin, M.Y.; Tao, Y.X.; Li, S.J. Composition and expression of genes encoding carbohydrate-active enzymes in the straw-degrading mushroom Volvariella volvacea. PLoS ONE 2013, 8, e58780. [Google Scholar]
- Chettri, D.; Verma, A.K. Biological significance of carbohydrate active enzymes and searching their inhibitors for therapeutic applications. Carbohydr. Res. 2023, 529, 108853. [Google Scholar] [CrossRef]
- Zhao, Z.T.; Liu, H.Q.; Wang, C.F.; Xu, J.R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genom. 2013, 14, 274. [Google Scholar] [CrossRef]
- Alshareef, S.A. Metabolic analysis of the CAZy class glycosyltransferases in rhizospheric soil fungiome of the plant species Moringa oleifera. Saudi J. Biol. Sci. 2024, 31, 103956. [Google Scholar] [CrossRef] [PubMed]
- Lv, G.B.; Liu, X.P.; Zeng, B.; He, B. Genome-Wide Analysis of the Cytochromes P450 Gene Family in Cordyceps militaris. J. Phys. Conf. Ser. 2020, 1549, 032069. [Google Scholar] [CrossRef]
- Yang, H.C.; Yu, F.; Qian, Z.H.; Huang, T.W.; Peng, T.; Hu, Z. Cytochrome P450 for environmental remediation: Catalytic mechanism, engineering strategies and future prospects. World J. Microbiol. Biotechnol. 2023, 40, 33. [Google Scholar] [CrossRef] [PubMed]
- Črešnar, B.; Petrič, Š. Cytochrome P450 enzymes in the fungal kingdom. Biochim. Biophys. Acta 2011, 1814, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Xu, J.; Liu, C.; Zhu, Y.J.; Nelson, D.R.; Zhou, S.G.; Li, C.F.; Wang, L.Z.; Guo, X.; Sun, Y.Z.; et al. Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat. Commun. 2012, 3, 913. [Google Scholar] [CrossRef]
- Liu, X.N.; Zhu, X.X.; Wang, H.; Liu, T.; Cheng, J.; Jiang, H.F. Discovery and modification of cytochrome P450 for plant natural products biosynthesis. Synth. Syst. Biotechnol. 2020, 5, 187–199. [Google Scholar] [CrossRef]
- Li, H.X.; Li, W.M.; Liu, X.H.; Cao, Y.S. gadA gene locus in Lactobacillus brevis NCL912 and its expression during fed-batch fermentation. FEMS Microbiol. Lett. 2013, 349, 108–116. [Google Scholar] [CrossRef][Green Version]
- Hsin, K.Y.; Ghosh, S.; Kitano, H. Combining Machine Learning Systems and Multiple Docking Simulation Packages to Improve Docking Prediction Reliability for Network Pharmacology. PLoS ONE 2014, 8, e83922. [Google Scholar] [CrossRef]
- Wang, P.M.; Liu, X.B.; Dai, Y.C.; Horak, E.; Steffen, K.; Yang, Z.L. Phylogeny and species delimitation of Flammulina: Taxonomic status of winter mushroom in East Asia and a new European species identified using an integrated approach. Mycol. Prog. 2018, 17, 1013–1030. [Google Scholar] [CrossRef]
- Liu, F.; Wang, S.H.; Jia, D.H.; Tan, H.; Wang, B.; Zhao, R.L. Development of Multiple Nucleotide Polymorphism Molecular Markers for Enoki Mushroom (Flammulina filiformis) Cultivars Identification. J. Fungi 2023, 9, 330. [Google Scholar] [CrossRef]
- Dai, Y.C.; Yang, Z.L. Notes on the nomenclature of five important edible fungi in China. Mycosystema 2018, 37, 1572–1577. [Google Scholar]
- Dai, Y.C.; Yang, Z.L.; Cui, B.K.; Wu, G.; Yuan, H.S.; Zhou, L.W.; He, S.H.; GE, Z.W.; Wu, F.; Wei, Y.L.; et al. Diversity and systematics of the important macrofungi in Chinese forests. Mycosystema 2021, 40, 770–805. [Google Scholar]
- Yang, C.S.; Li, W.C.; Li, C.; Zhou, Z.H.; Xiao, Y.L.; Yan, X. Metabolism of ganoderic acids by a Ganoderma lucidum cytochrome P450 and the 3-keto sterol reductase ERG27 from yeast. Phytochemistry 2018, 155, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.F.; Xiao, H.; Zhong, J.J. Biosynthesis of a ganoderic acid in Saccharomyces cerevisiae by expressing a cytochrome P450 gene from Ganoderma lucidum. Biotechnol. Bioeng. 2018, 115, 1842–1854. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Jiang, C.J.; Wang, Q.; Fang, Y.B.; Wang, J.; Wang, M.; Xiao, H. Biosynthesis of mushroom-derived type II ganoderic acids by engineered yeast. Nat. Commun. 2022, 13, 7740. [Google Scholar] [CrossRef]
- Zhang, J.X.; Li, L.P.; Lv, Q.Z.; Yan, L.; Wang, Y.; Jiang, Y.Y. The Fungal CYP51s: Their Functions, Structures, Related Drug Resistance, and Inhibitors. Front. Microbiol. 2019, 10, 691. [Google Scholar] [CrossRef]
- Han, H.Y.; Zhang, P.; Xie, Z.K.; Qi, J.Z.; Wang, P.C.; Li, C.; Xue, Z.Y.; Wu, R.B.; Liu, C.W. Functional Characterization of Sesquiterpene Synthases and P450 Enzymes in Flammulina velutipes for Biosynthesis of Spiro [4.5] Decane Terpene. J. Agric. Food Chem. 2024, 72, 9227–9235. [Google Scholar] [CrossRef]
- Huang, J.; Mei, L.H.; Sheng, Q.; Yao, S.J.; Lin, D.Q. Purification and Characterization of Glutamate Decarboxylase of Lactobacillus brevis CGMCC 1306 Isolated from Fresh Milk. Chin. J. Chem. Eng. 2007, 15, 157–161. [Google Scholar] [CrossRef]
- Mehta, P.K.; Christen, P. The molecular evolution of pyridoxal-5′-phosphate-dependent enzymes. Adv. Enzymol. Relat. Areas Mol. Biol. 2000, 74, 129–184. [Google Scholar]
- Lan, S.X.; Zhai, T.K.; Zhang, X.Y.; Xu, L.Z.; Gao, J.; Lai, C.W.; Chen, Y.; Lai, Z.X.; Lin, Y.L. Genome-wide identification and expression analysis of the GAD family reveal their involved in embryogenesis and hormones responses in Dimocarpus longan Lour. Gene 2024, 927, 148698. [Google Scholar] [CrossRef]
- Akter, N.; Kulsum, U.; Moniruzzaman, M.; Yasuda, N.; Akama, K. Truncation of the calmodulin binding domain in rice glutamate decarboxylase 4 (OsGAD4) leads to accumulation of γ-aminobutyric acid and confers abiotic stress tolerance in rice seedlings. Mol. Breed. 2024, 44, 21. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.T.; Ji, J.; Chen, W.; Yue, J.Y.; Du, C.J.; Sun, J.C.; Chen, L.Z.; Jiang, Z.P.; Shi, S.Q. GABA negatively regulates adventitious root development in poplar. J. Exp. Bot. 2020, 71, 1459–1474. [Google Scholar] [CrossRef] [PubMed]
- Shaul, O. How introns enhance gene expression. Int. J. Biochem. Cell Biol. 2017, 91, 145–155. [Google Scholar] [CrossRef] [PubMed]







| Strain Name | Origin | Cap/Stipe Colour | Source Information |
|---|---|---|---|
| XH, W1638, Fv-YH, Fv-RYJ, Fv-HTC, Fv-HL23, Fv-SY, Fv-GR, Fv-GF, Fv-FM, Fv-CYS, Fv-BY | Shanghai, China | Snow white/Snow white | Industrial cultivation |
| J54-3, J5011, WH25 | Shanghai, China | Snow white/Snow white | SAAS-self-selection breeding |
| X3E | Shanghai, China | Snow white/Snow white | Enterprise-self-selection breeding |
| ENOKI-J, ENOKI-I, ENOKI-H, ENOKI-G | Malaysia | Snow white/Snow white | Industrial cultivation |
| 2345(Y) | Shanghai, China | Light yellow/Snow white | Conventional cultivation |
| SCY1-2(Y) | Shanghai, China | Solid yellow/Solid yellow | Industrial cultivation |
| Primers | Sequences (5′-3′) |
|---|---|
| Ff-gad2-gDNA-F | CTTCTGACTGACTTGAGGTAAATAGGTTAACATGCTCTCCAAGGTGACGAC |
| Ff-gad2-gDNA-R1 | TCTAGACTACTTGTCATCGTCGTCCTTGTAATCACACGGCTTCGCATATGT |
| Ff-gad2-gDNA-R2 | CTTCACTTCAAGTGCACACAACATATTCTAGACTACTTGTCATCGTCGTC |
| Ff-gad2-cDNA-F | CTTCTGACTGACTTGAGGT |
| Ff-gad2-cDNA-R | CTTCACTTCAAGTGCACACAA |
| Pesdi1-F | CTCATCTGGAAGGTGGCAGG |
| Pesdi1-R | TGGAGCGACGAGGATACAAC |
| q-Ff-gad2-F | TTTGAGCTGCACTACCTGGG |
| q-Ff-gad2-R | GTTCAAAGCGATTTGCCGGT |
| q-ACT1-F | CCGAGCGGAAGTACTCTGTG |
| q-ACT1-R | ATGCTATCTTGCCTCCAGCC |
| Assembly Statistics | Scaffolds |
|---|---|
| Genome size (Mb) | 40.96 |
| Total sequence number | 140 |
| Total sequenced length (bp) | 35,904,424 |
| Maximum sequence length (bp) | 2,467,745 |
| Minimum sequence length (bp) | 5510 |
| GC (%) | 49.62 |
| N20 length (bp) | 1,658,848 |
| N50 length (bp) | 917,125 |
| N90 length (bp) | 141,141 |
| Total gene length (bp) | 23,522,018 |
| Gene Percentage of Genome (%) | 65.51 |
| Total number of genes | 14,256 |
| Average gene length (bp) | 1649.9 |
| Total exon length (bp) | 19,949,251 |
| Exon percentage of genome (%) | 55.56 |
| Average Exon Length (bp) | 250.7 |
| Total coding sequence (CDS) length (bp) | 19,949,251 |
| Average CDS length (bp) | 1399.3 |
| CDS percentage of genome (%) | 55.56 |
| Average intron length (bp) | 54.7 |
| Sequencing platform | PacBio CLR, Illumina |
| Repeat Elements | Copies (Numbers) | Repeat Size (bp) | Percentage of the Assembled Genome |
|---|---|---|---|
| LTR elements | 1164 | 375,774 | 1.05% |
| LINEs | 276 | 28,892 | 0.08% |
| SINEs | 6 | 316 | 0.00% |
| DNA transposons | 563 | 49,884 | 0.14% |
| Simple repeats | 58 | 6577 | 0.02% |
| Low complexity | 4 | 467 | 0.00% |
| Satellites | 38 | 4626 | 0.01% |
| Unclassified | 104 | 22,885 | 0.06% |
| Total | 2213 | 487,753 | 1.36% |
| Name | Subfamily | Name | Subfamily |
|---|---|---|---|
| scaffold29.t59 | CYP1A1 | scaffold18.t145 | CYP4F2_3 |
| scaffold8.t249 | CYP1A2 | scaffold12.t307 | CYP4F22 |
| scaffold2.t316 | CYP1B1 | scaffold29.t79 | CYP4Z |
| scaffold3.t391 | CYP1B1 | scaffold35.t90 | CYP6 |
| scaffold17.t69 | CYP2AA_D | scaffold5.t196 | CYP6 |
| scaffold23.t115 | CYP2D | scaffold1.t1046 | CYP7A1 |
| scaffold16.t165 | CYP2E1 | scaffold25.t36 | CYP8B1 |
| scaffold42.t66 | CYP2E1 | scaffold11.t117 | CYP12 |
| scaffold72.t19 | CYP2E1 | scaffold3.t216 | CYP12 |
| scaffold18.t148 | CYP2J | scaffold25.t16 | CYP17A |
| scaffold104.t2 | CYP2R1 | scaffold25.t74 | CYP17A |
| scaffold30.t26 | CYP2R1 | scaffold3.t405 | CYP17A |
| scaffold33.t10 | CYP2R1 | scaffold47.t5 | CYP21A |
| scaffold11.t370 | CYP2U1 | scaffold2.t564 | CYP27A1 |
| scaffold6.t140 | CYP2U1 | scaffold29.t22 | CYP28 |
| scaffold13.t35 | CYP3A | scaffold24.t126 | CYP46A1 |
| scaffold3.t743 | CYP3A | scaffold7.t254 | CYP49A |
| scaffold52.t36 | CYP3A | scaffold3.t565 | CYP51 |
| scaffold29.t51 | CYP3A | scaffold4.t127 | CYP61A |
| scaffold24.t85 | CYP3A4 | scaffold1.t136 | CYP67 |
| scaffold24.t122 | CYP4 | scaffold20.t23 | CYP78A |
| scaffold1.t848 | CYP4A | scaffold9.t338 | CYP81F |
| scaffold29.t81 | CYP4B1 | scaffold1.t915 | CYP82G1 |
| scaffold45.t48 | CYP4B1 | scaffold37.t21 | CYP86A4S |
| scaffold10.t426 | CYP4F | scaffold8.t190 | CYP94C1 |
| scaffold20.t35 | CYP4F | scaffold2.t703 | CYP98A9 |
| scaffold20.t51 | CYP4F | scaffold31.t101 | CYP98A9 |
| scaffold20.t52 | CYP4F | scaffold12.t157 | CYP313 |
| scaffold3.t722 | CYP4F | scaffold51.t26 | CYP708A2 |
| scaffold34.t18 | CYP4F | scaffold41.t68 | CYP735A |
| scaffold38.t90 | CYP4F | scaffold18.t186 | CYPD |
| scaffold52.t22 | CYP4F | scaffold6.t375 | CYPH |
| Name | Number of Amino Acids | Formula | Molecular Weight (kDa) | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| Ff-GAD1 | 554 | C2786H4310N750O812S18 | 61.89 | 6.32 | 37.17 | 88.07 | −0.238 | mitochondrion |
| Ff-GAD2 | 536 | C2687H4173N725O782S14 | 59.60 | 6.27 | 36.60 | 91.23 | −0.216 | mitochondrion |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Shang, J.; Bao, D.; Wan, J.; Zhou, C.; Feng, Z.; Li, H.; Shao, Y.; Wu, Y. Whole-Genome Sequence Analysis of Flammulina filiformis and Functional Validation of Gad, a Key Gene for γ-Aminobutyric Acid Synthesis. J. Fungi 2024, 10, 862. https://doi.org/10.3390/jof10120862
Li W, Shang J, Bao D, Wan J, Zhou C, Feng Z, Li H, Shao Y, Wu Y. Whole-Genome Sequence Analysis of Flammulina filiformis and Functional Validation of Gad, a Key Gene for γ-Aminobutyric Acid Synthesis. Journal of Fungi. 2024; 10(12):862. https://doi.org/10.3390/jof10120862
Chicago/Turabian StyleLi, Wenyun, Junjun Shang, Dapeng Bao, Jianing Wan, Chenli Zhou, Zhan Feng, Hewen Li, Youran Shao, and Yingying Wu. 2024. "Whole-Genome Sequence Analysis of Flammulina filiformis and Functional Validation of Gad, a Key Gene for γ-Aminobutyric Acid Synthesis" Journal of Fungi 10, no. 12: 862. https://doi.org/10.3390/jof10120862
APA StyleLi, W., Shang, J., Bao, D., Wan, J., Zhou, C., Feng, Z., Li, H., Shao, Y., & Wu, Y. (2024). Whole-Genome Sequence Analysis of Flammulina filiformis and Functional Validation of Gad, a Key Gene for γ-Aminobutyric Acid Synthesis. Journal of Fungi, 10(12), 862. https://doi.org/10.3390/jof10120862






