The Diversity and Taxonomy of Thelephoraceae (Basidiomycota) with Descriptions of Four Species from Southwestern China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Herbarium Specimen Preparation
2.2. Morphology
2.3. DNA Extraction and PCR Sequencing
2.4. Phylogenetic Analyses
2.5. Pairwise Homoplasy Test
3. Results
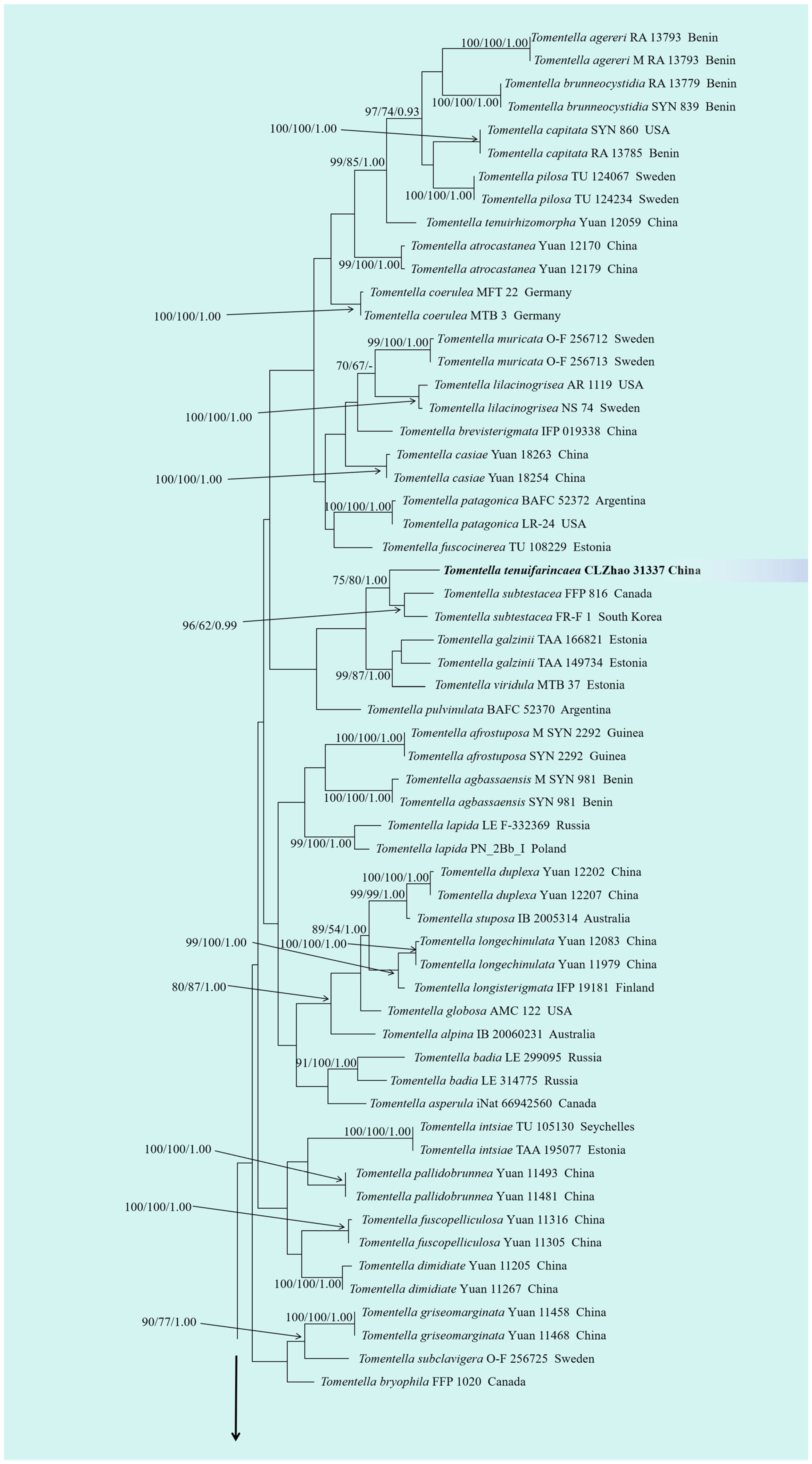
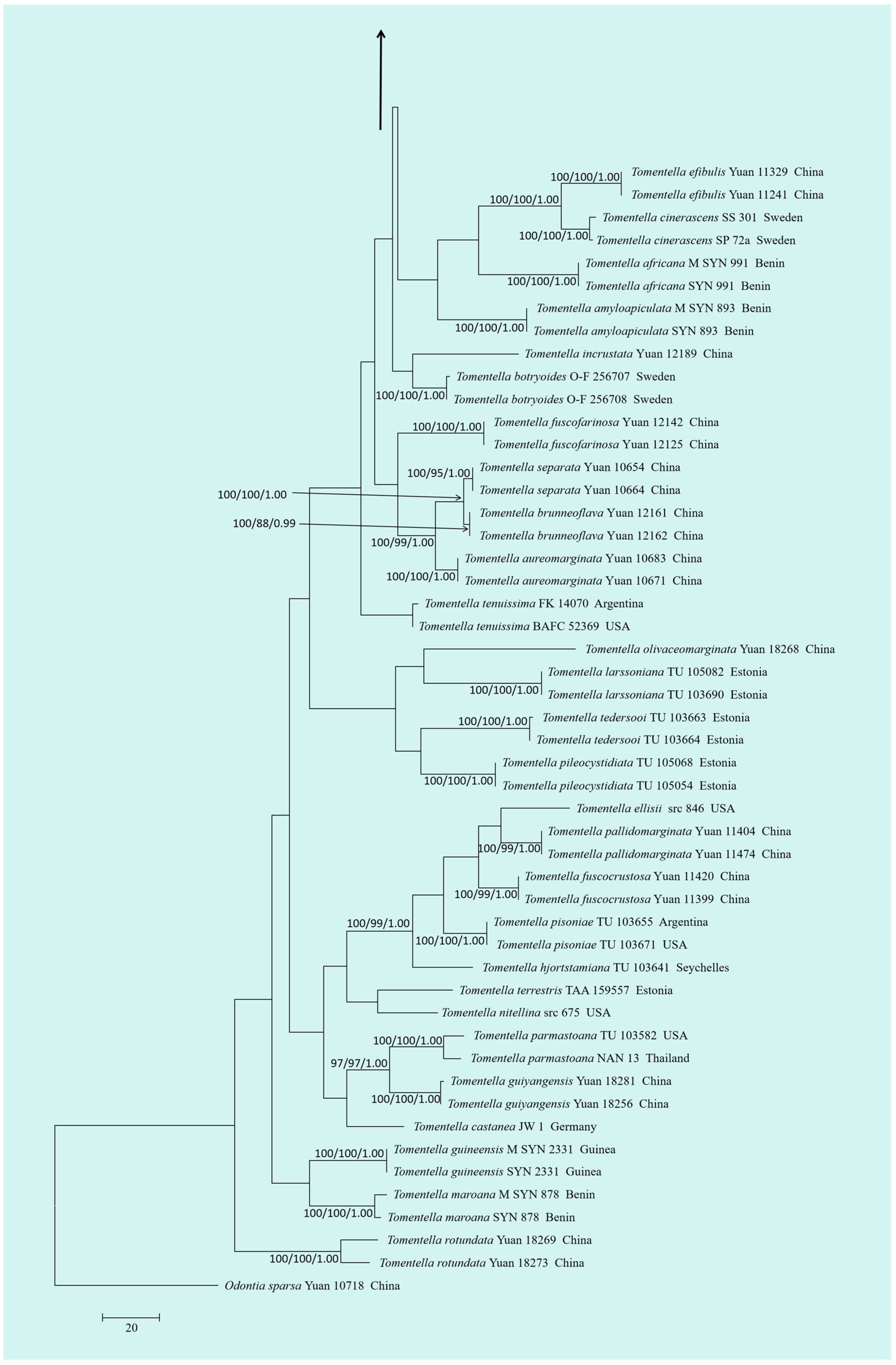
3.1. Molecular Phylogeny
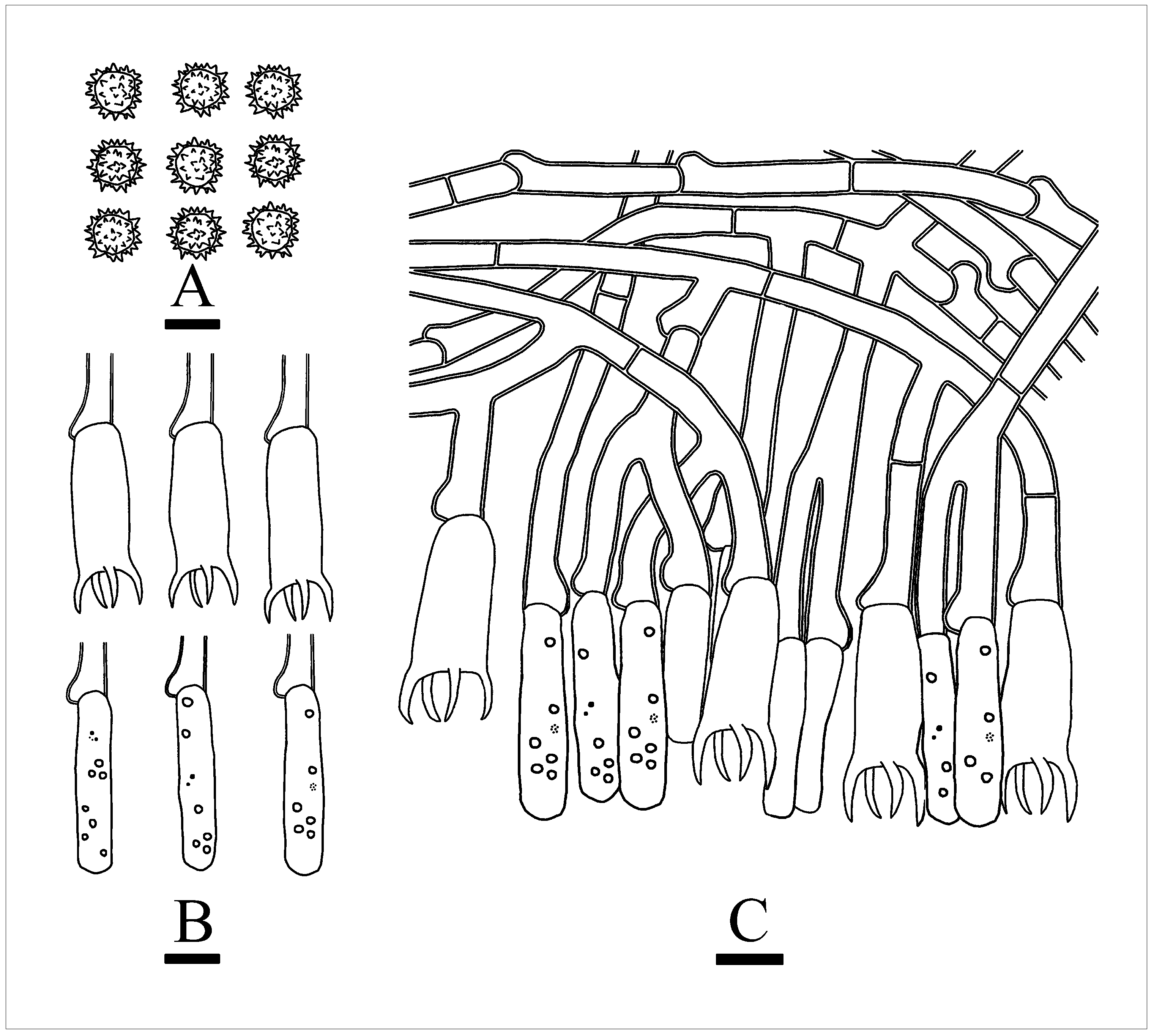
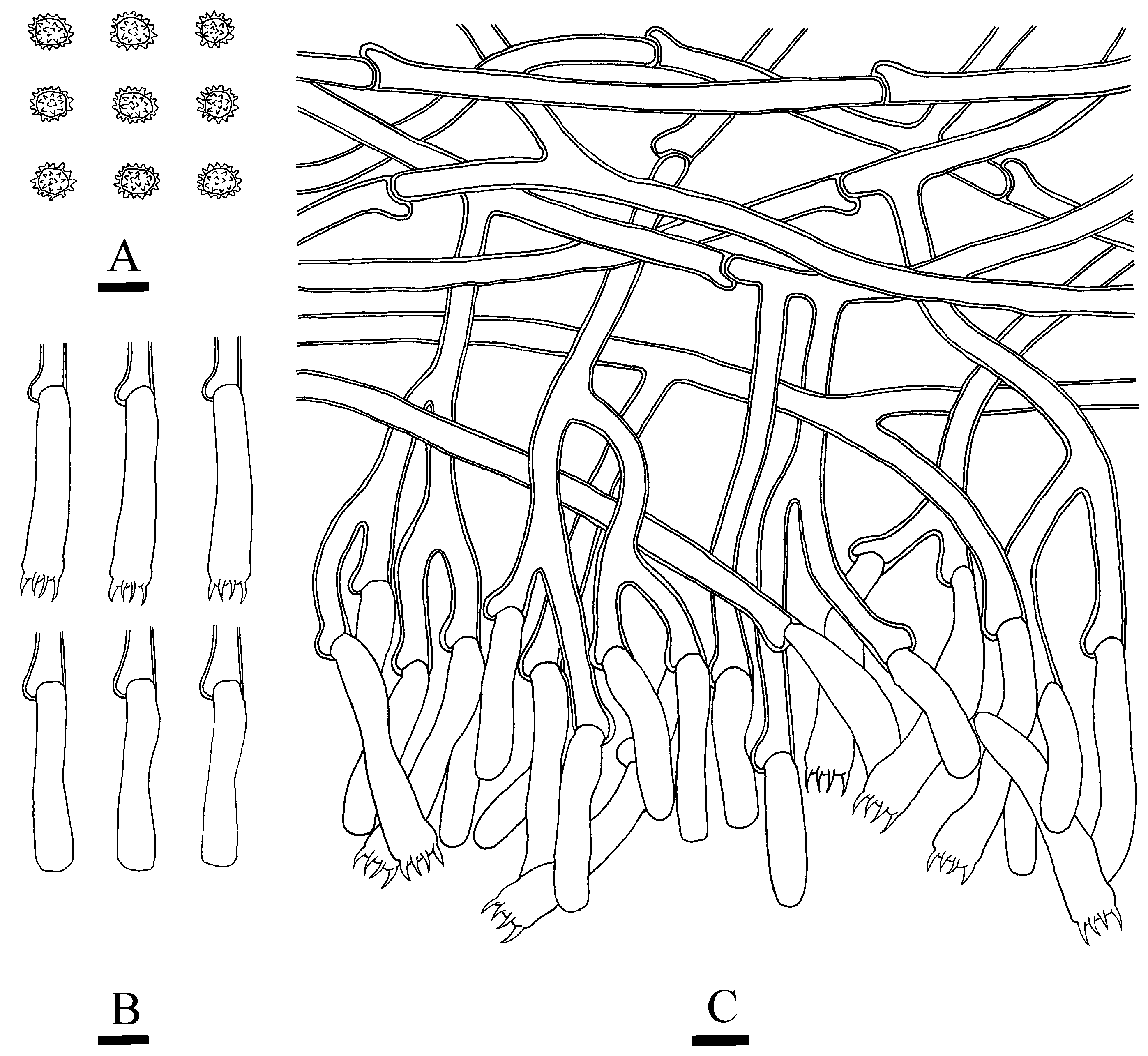
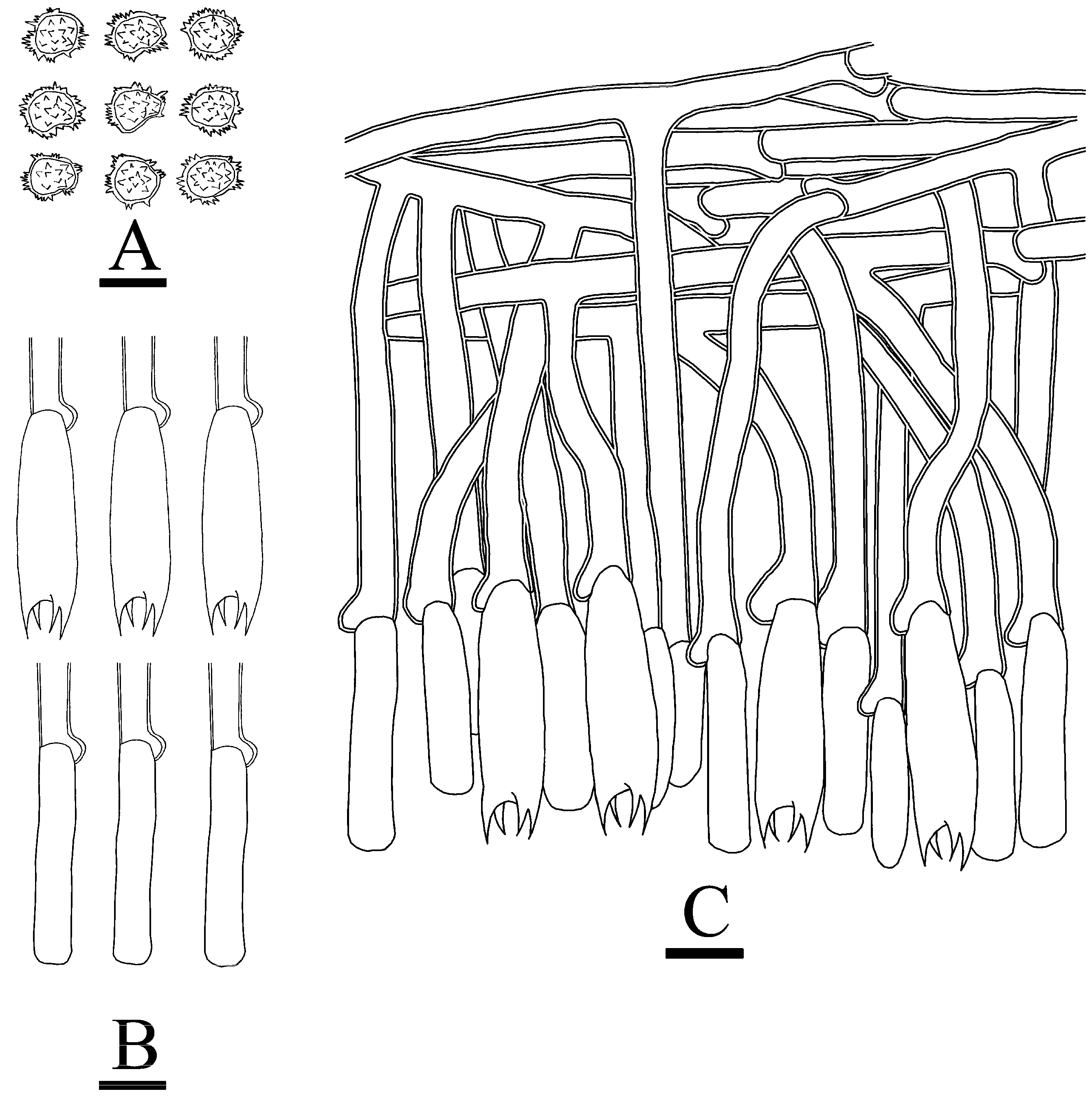
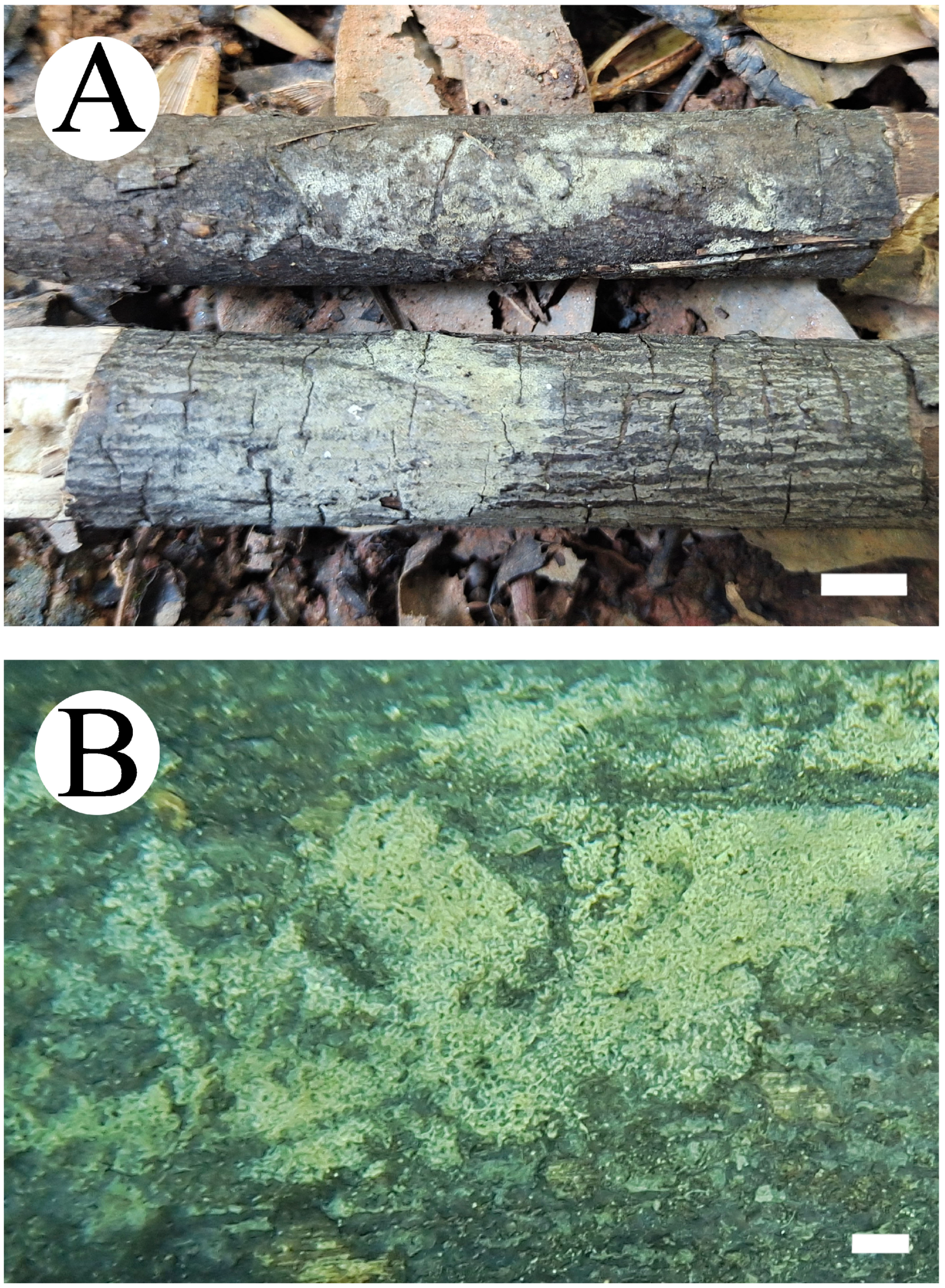
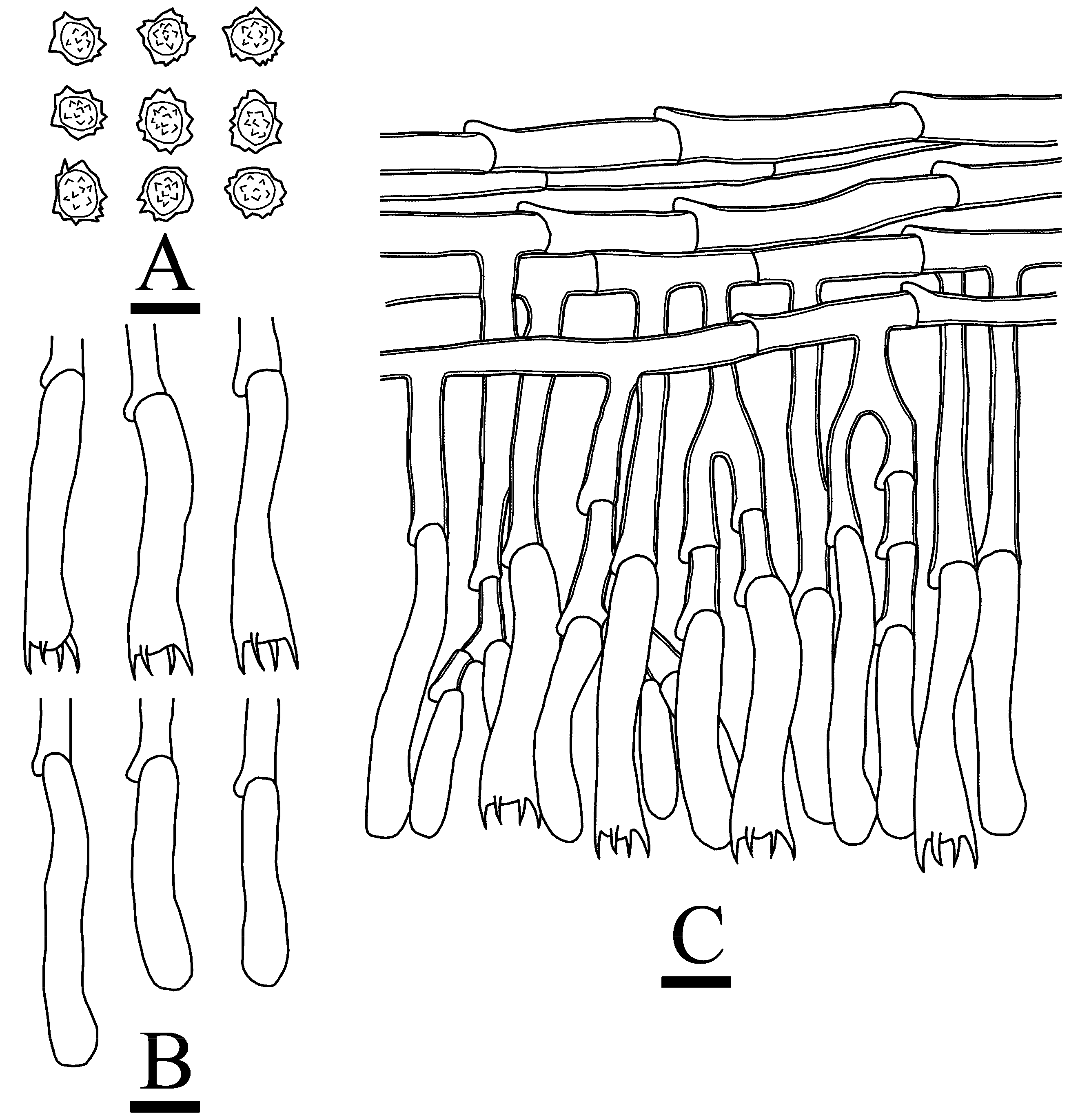
3.2. Taxonomy
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reid, D.A.; Larsen, M.J. A contribution to the taxonomy of the genus Tomentella. Kew Bull. 1976, 31, 195. [Google Scholar] [CrossRef]
- Kõljalg, U.; Dahlberg, A.; Taylor, A.F.S.; Larsson, E.; Hallenberg, N.; Stenlid, J.; Larsson, K.H.; Fransson, P.M.; Kårén, O.; Jonsson, L. Diversity and abundance of resupinate thelephoroid fungi as ectomycorrhizal symbionts in Swedish boreal forests. Mol. Ecol. 2000, 9, 1985–1996. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D. The numbers of fungi. Fungal Divers. 2022, 114, 1. [Google Scholar] [CrossRef]
- Coker, W.C. Notes on the Thelephoraceae of North Carolina. J. Elisha Mitchell Sci. Soc. 1921, 36, 146–196. [Google Scholar]
- Hilszczanska, D.; Sierota, Z. Persistence of ectomycorrhizas by Thelephora terrestris on outplanted Scots pine seedlings. Acta Mycol. 2006, 41, 313–318. [Google Scholar] [CrossRef]
- Ramírez-López, I.; Villegas-Ríos, M.; Salas-Lizana, R. Thelephora versatilis and Thelephora pseudoversatilis: Two new cryptic species with polymorphic basidiomes inhabiting tropical deciduous and sub-perennial forests of the Mexican Pacific coast. Mycologia 2015, 107, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Ehrhart, J.F. 1785–1795: Plantae Cryptogamae Linn., Quas in Locis Earum Natalibus Collegit et Exsiccavit Fridericus Ehrhart; Hannover: Lower Saxony, Germany, 2020; pp. 1–320. [Google Scholar]
- Li, T.; LI, T.H.; Song, B.; Hosen, M.I. Thelephora austrosinensis (Thelephoraceae), a new species close to T. ganbajun from southern China. Phytotaxa 2020, 471, 208–220. [Google Scholar] [CrossRef]
- Tian, M.Z.; Xia, H.B.; Gao, Z.L.; Zhao, C.Y.; Ma, D.; Yang, Z.L.; Li, Y.C. Four new species and one new record of Thelephora from China. J. Fungi 2024, 10, 300. [Google Scholar] [CrossRef]
- Bi, Z.S.; Zheng, G.Y.; Li, T.H. The Macrofungus Flora of China’s Guangdong Province; Chinese University Press: Hong Kong, China, 1993; p. 734. [Google Scholar]
- Iwanski, M.; Rudawski, M.; Leski, T. Mycorrhizal associations of nursery grown scots pine (Pinus sylvestris L.) seedlings in poland. Ann. For. Sci. 2006, 63, 715–723. [Google Scholar] [CrossRef]
- Flykt, E.; Timonen, S.; Pennanen, T. Variation of ectomycorrhizal colonisation in Norway spruce seedlings in Finnish forest nurseries. Silva Fenn. 2008, 42, 571–585. [Google Scholar] [CrossRef]
- Sha, T.; Xu, J.; Palanichamy, M.G.; Zhang, H.B.; Li, T.; Zhao, Z.W.; Zhang, Y.P. Genetic diversity of the endemic gourmet mushroom Thelephora ganbajun from south-western China. Microbiology 2008, 154, 3460–3468. [Google Scholar] [CrossRef]
- Yorou, N.S. Miscellaneous Contributions to the Anatomy and Molecular Phylogeny of Tropical African Resupinate Thelephorales. Ph.D. Thesis, University of Munich, Munich, Germany, 2008; p. 120. [Google Scholar]
- He, J.; Zhou, Z.; Yang, H.; Xu, J. Integrative management of commercialized wild mushroom: A case study of Thelephora ganbajun in Yunnan, southwest China. Environ. Manag. 2011, 48, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Yangdo, R.; Kumar, S.; Sharma, Y.P. Three hitherto unreported macrofungi from cold arid region of Ladakh Province, Jammu and Kashmir, India. Kavaka 2015, 44, 42–44. [Google Scholar]
- Ezhov, O.N.; Zmitrovich, I.V. Lignotrophic basidiomycetes from pioneering microsites in boreal forests of the White Sea Region. Byulleten Moskovskogo Obshchestva Ispytateley Prirody. Otd. Biol. 2017, 122, 44–50. [Google Scholar]
- Zmitrovich, I.V.; Shchepin, O.N.; Malysheva, V.F.; Kalinovskaya, N.I.; Volobuev, S.V.; Myasnikov, A.G.; Ezhov, O.N. Basidiome reduction in litter-inhabiting Thelephorales in boreal forest environments: Morphological and molecular evidence. Curr. Res. Environ. Appl. Mycol. 2018, 8, 360–371. [Google Scholar] [CrossRef]
- Danielson, R.M.; Visser, S.; Parkinson, D. Microbial activity and mycorrhizal potential of four overburden types used in the reclamation of extracted oil sands. Can. J. Soil. Sci. 1983, 63, 363–375. [Google Scholar] [CrossRef]
- Tedersoo, L.; Harend, H.; Buegger, F.; Pritsch, K.; Saar, I.; Kõljalg, U. Stable isotope analysis, field observations and synthesis experiments suggest that Odontia is a non-mycorrhizal sister genus of Tomentella and Thelephora. Fungal Ecol. 2014, 11, 80–90. [Google Scholar] [CrossRef]
- Jakucs, E.; Eros-Honti, Z.; Seress, D.; Kovács, G.M. Enhancing our understanding of anatomical diversity in Tomentella ectomycorrhizas: Characterization of six new morphotypes. Mycorrhiza 2015, 25, 419–429. [Google Scholar] [CrossRef]
- Nouhra, E.; Pastor, N.; Becerra, A.; Areitio, E.S.; Geml, J. Greenhouse seedlings of alnus showed low host intrageneric specificity and a strong preference for some Tomentella ectomycorrhizal associates. Microb. Ecol. 2015, 69, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Willdenow, C.L. Florae Berolinensis Prodromus: Secundum Systema Linneanum Ab Illustr. Viro Ac Eq. C. P. Thunbergio Emendatum Conscriptus (Classic Reprint); Impensis Wilhelmi Viewegii Berolini: Berlin, Germany, 1787; pp. 1–476. [Google Scholar]
- Cunningham, G.H. The Thelephoraceae of Australia and New Zealand. Mycologia 1963, 55, 824–825. [Google Scholar] [CrossRef]
- Corner, E.J.H. A monograph of Thelephora (Basidiomycetes). Nova Hedwig. 1968, 27, 1–110. [Google Scholar] [CrossRef]
- Corner, E.J.H. Futher notes on cantharelloid fungi and Thelephora. Nova Hedwig. 1976, 27, 325–342. [Google Scholar]
- Stalpers, J.A. The Aphyllophoraceous fungi I. Keys to the species of the Thelephorales. Stud. Mycol. 1993, 35, 1–168. [Google Scholar]
- Xu, D.P.; Zheng, J.; Zhou, Y.; Li, Y.; Li, S.; Li, H.B. Extraction of natural antioxidants from the Thelephora ganbajun mushroom by an ultrasound-assisted extraction technique and evaluation of antiproliferative activity of the extract against human cancer cells. Int. J. Mol. Sci. 2016, 17, 1664. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.F.; Sha, T.; Zhang, Y.R.; Cao, Y.; Mi, F.; Liu, C.L.; Yang, D.; Tang, X.Z.; He, X.X.; Dong, J.Y.; et al. Frequent heteroplasmy and recombination in the mitochondrial genomes of the basidiomycete mushroom Thelephora ganbajun. Sci. Rep. 2017, 7, 1626. [Google Scholar] [CrossRef]
- Zheng, L.; Ma, Y.H.; Zhang, Y.J.; Meng, Q.J.; Yang, H.J.; Gong, W.L.; Liu, Q.G.; Cai, L.; Hao, L.J.; Wang, B.L.; et al. Distribution of Zinc in mycelial cells and antioxidant and anti-inflammatory activities of mycelia Zinc polysaccharides from Thelephora ganbajun TG-01. Oxid. Med. Cell. Long. 2020, 2308017, 1–17. [Google Scholar] [CrossRef]
- Lu, X.; Cao, T.; Nguyên, T.T.T.; Yuan, H.S. Six new species of Tomentella (Thelephorales, Basidiomycota) from tropical pine forests in Central Vietnam. Front. Microbiol. 2022, 13, 864128. [Google Scholar] [CrossRef]
- Yorou, N.S.; Agerer, R. Tomentella furcata, a new species from Benin (West Africa) with basidia forming internal hyphae. Mycol. Prog. 2007, 6, 239–247. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Andre, A.; Janusz, B.; Nattawut, B.; Gladstone, A.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- James, T.Y.; Stajich, J.E.; Hittinger, C.T.; Rokas, A. Toward a fully resolved fungal tree of life. Annu. Rev. Microbiol. 2020, 74, 291–313. [Google Scholar] [CrossRef]
- Larsson, K.H.; Larsson, E.; Kljalg, U. High phylogenetic diversity among corticioid homobasidiomycetes. Mycol. Res. 2004, 108, 983–1002. [Google Scholar] [CrossRef]
- Kuhar, F.; Barroetaveña, C.; Rajchenberg, M. New species of Tomentella (Thelephorales) from the patagonian andes forests. Mycologia 2016, 108, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Vizzini, A.; Angelini, C.; Losi, C.; Ercole, E. Thelephora dominicana (Basidiomycota, Thelephorales), a new species from the dominican republic, and preliminary notes on thelephoroid genera. Phytotaxa 2016, 265, 27–38. [Google Scholar] [CrossRef]
- Patouillard, N. Les Hyménomycètes d’Europe; Paul Klincksieck: Paris, France, 1887; p. 166. [Google Scholar]
- Stalpers, J.A. The aphyllophoraceous fungi II. Keys to the species of the Hericiales. Stud. Mycol. 1996, 40, 1–183. [Google Scholar]
- Zecchin, G. Ⅱ genere Thelephora in Friuli-Ottavo contributo. Riv. Di Micol. 2008, 51, 117–125. [Google Scholar]
- Das, K.; Hembrom, M.E.; Ghosh, A.; Parihar, A.; Kuhar, F. Thelephora sikkimensis sp. nov. (Thelephoraceae) from the Eastern Himalayas (India). Nova Hedwig. 2018, 107, 337–347. [Google Scholar] [CrossRef]
- Yang, Y.; Li, R.; Jiang, Q.Q.; Zhou, H.M.; Muhammad, A.; Wang, H.J.; Zhao, C.L. Phylogenetic and Taxonomic Analyses Reveal Three New Wood-Inhabiting Fungi (Polyporales, Basidiomycota) in China. J. Fungi 2024, 10, 55. [Google Scholar] [CrossRef]
- Guan, Q.X.; Zhao, C.L. Two new corticioid species, Hyphoderma sinense and H. floccosum (Hyphodermataceae, Polyporales), from southern China. Mycosyserma 2021, 40, 447–461. [Google Scholar]
- Zhou, Q.; Jiang, Q.Q.; Yang, X.; Zhao, C.L.; Zhao, J. Phylogenetic and taxonomic analyses of five new wood-Inhabiting fungi of Botryobasidium, Coltricia and Coltriciella (Basidiomycota) from China. J. Fungi 2024, 10, 205. [Google Scholar] [CrossRef]
- Deng, Y.L.; Li, J.F.; Zhao, C.L.; Zhao, J. Four new fungal species in forest ecological system from southwestern China. J. Fungi 2024, 10, 194. [Google Scholar] [CrossRef]
- Yang, S.R.; Wei, Y.L.; Yuan, H.S. Molecular phylogeny and morphology reveal four new species of Thelephora (Thelephorales, Basidiomycota) from subtropical China, closely related to T. ganbajun. Front. Microbiol. 2023, 14, 1109924. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Karunarathna, S.C.; Li, H.; Galappaththi, M.C.; Zhao, C.L.; Kakumyan, P.; Mortimer, P.E. The impact of drying temperature on basidiospore size. Diversity 2022, 14, 239. [Google Scholar] [CrossRef]
- Petersen, J.H. The Danish Mycological Society’s Colour-Chart; Foreningen til Svampekundskabens Fremme: Greve, Germany, 1996. [Google Scholar]
- Zhao, C.L.; Qu, M.H.; Huang, R.X.; Karunarathna, S.C. Multi-gene phylogeny and taxonomy of the wood-rotting fungal genus Phlebia sensu lato (Polyporales, Basidiomycota). J. Fungi 2023, 9, 320. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.; Innis, M.A.; Gelfand, D.H.; Sninsky, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Rehner, S.A.; Samuels, G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 1994, 98, 625–634. [Google Scholar] [CrossRef]
- Miettinen, O.; Kõljalg, U. Amaurodon sumatranus (Thelephorales, Basidiomycota), a new species from Indo- nesia. Mycotaxon 2007, 100, 51–59. [Google Scholar]
- Zhou, L.W.; Kõljalg, U. A new species of Lenzitopsis (Thelephorales, Basidiomycota) and its phylogenetic placement. Mycoscience 2013, 54, 87–92. [Google Scholar] [CrossRef]
- Yuan, Y.; Wu, F.; Dai, Y.C.; Qin, W.M.; Yuan, H.S. Odontia aculeata and O. sparsa, two new species of tomentelloid fungi (Thelephorales, Basidiomycota) from the secondary forests of northeast China. Phytotaxa 2018, 372, 183–192. [Google Scholar] [CrossRef]
- Salvador-Montoya, C.A.; Elias, S.G.; Popoff, O.F.; Robledo, G.L.; Urcelay, C.; Martínez, A.G.S.; Drechsler-Santos, E.R. Neotropical Studies on Hymenochaetaceae: Unveiling the diversity and endemicity of Phellinotus. J. Fungi 2022, 8, 216. [Google Scholar] [CrossRef]
- Song, C.G.; Sun, Y.F.; Liu, S.; Chen, Y.Y.; Cui, B.K. Phylogenetic Analyses and Morphological Studies Reveal Four New Species of Phellodon (Bankeraceae, Thelephorales) from China. J. Fungi 2023, 9, 30. [Google Scholar] [CrossRef]
- Voitk, A.; Saar, I.; Trudell, S.; Spirin, V.; Beug, M.; Kõljalg, U. Polyozellus multiplex (Thelephorales) is a species complex containing four new species. Mycologia 2018, 109, 975–992. [Google Scholar] [CrossRef]
- Liu, X.F.; Tibpromma, S.; Xu, J.C.; Kumla, J.; Karunarathna, S.C.; Zhao, C.L. Taxonomy and phylogeny reveal two new potential edible ectomycorrhizal mushrooms of Thelephora from East Asia. Diversity 2021, 13, 646. [Google Scholar] [CrossRef]
- Borovička, J.; Braeuer, S.; Walenta, M.; Hršelová, H.; Leonhardt, T.; Sácký, J.; Kaňa, A.; Goessler, W. A new mushroom hyperaccumulator: Cadmium and arsenic in the ectomycorrhizal basidiomycete Thelephora penicillata. Sci. Total Environ. 2022, 826, 154227. [Google Scholar] [CrossRef] [PubMed]
- Yorou, N.S.; Gardt, S.; Guissou, M.L.; Diabaté, M.; Agerer, R. Three new Tomentella species from West Africa identified by anatomical and molecular data. Mycol. Prog. 2012, 11, 449–462. [Google Scholar] [CrossRef]
- Yorou, N.S.; Diabaté, M.; Agerer, R. Phylogenetic placement and anatomical characterisation of two new West African Tomentella (Basidiomycota, Fungi) species. Mycol.Prog. 2012, 11, 171–180. [Google Scholar] [CrossRef]
- Yorou, N.S.; Guelly, A.K.; Agerer, R. Anatomical and ITS rDNA-based phylogenetic identification of two new West African resupinate thelephoroid species. Mycoscience 2011, 52, 363–375. [Google Scholar] [CrossRef]
- Yuan, H.S.; Lu, X.; Dai, Y.C.; Hyde, K.D.; Kan, Y.H.; Kušan, I.; He, S.H.; Liu, N.G.; Sarma, V.V.; Zhao, C.L.; et al. Fungal diversity notes 1277–1386: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2020, 1, 1–260. [Google Scholar] [CrossRef]
- Svantesson, S.; Larsson, K.; Larsson, E. Pseudotomentella badjelanndana, Pseudotomentella sorjusensis and Tomentella viridibasidia—Three new corticioid Thelephorales species from the Scandes Mountains. Phytotaxa 2021, 2, 497. [Google Scholar] [CrossRef]
- Jones, M.D.; Phillips, L.A.; Treu, R.; Ward, V.; Berch, S.M. Functional responses of ectomycorrhizal fungal communities to long-term fertilization of lodgepole pine (Pinus contorta Dougl. ex Loud. var. Latifolia Engelm.) stands in central British Columbia. Appl. Soil Ecol. 2012, 60, 29–40. [Google Scholar] [CrossRef]
- Smith, M.E.; Douhan, G.W.; Rizzo, D.M. Ectomycorrhizal community structure in a xeric Quercus woodland based on rDNA sequence analysis of sporocarps and pooled roots. New Phytol. 2007, 174, 847–863. [Google Scholar] [CrossRef]
- Zhu, Y.Q.; Li, X.L.; Zhao, D.X.; Wei, Y.L.; Yuan, H.S. Four new species of Tomentella (Thelephorales, Basidiomycota) from subtropical forests in Southwestern China. J. Fungi 2024, 10, 440. [Google Scholar] [CrossRef] [PubMed]
- Suvi, T.; Tedersoo, L.; Abarenkov, K.; Beaver, K.; Gerlach, J.; Kõljalg, U. Mycorrhizal symbionts of Pisonia grandis and P. sechellarum in Seychelles: Identification of mycorrhizal fungi and description of new Tomentella species. Mycologia 2010, 102, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Suvi, T.; Beaver, K.; Kõljalg, U. Ectomycorrhizal fungi of the Seychelles: Diversity patterns and host shifts from the native Vateriopsis seychellarum (Dipterocarpaceae) and Intsia bijuga (Caesalpiniaceae) to the introduced Eucalyptus robusta (Myrtaceae), but not Pinus caribea (Pinaceae). New Phytol. 2007, 175, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Ivanushenko, Y.Y.; Volobuev, S. Species of Odontia and Tomentella (Thelephorales, Basidiomycota) new to Dagestan, Russia. S. Russ. Ecol. Dev. 2020, 15, 165–173. [Google Scholar] [CrossRef]
- Hrynkiewicz, K.; Toljander, Y.K.; Baum, C.; Fransson, P.; Taylor, A.F.S.; Weih, M. Correspondence of ectomycorrhizal diversity and colonisation of willows (Salix spp.) grown in short rotation coppice on arable sites and adjacent natural stands. Mycorrhiza 2012, 22, 603–613. [Google Scholar] [CrossRef]
- Menkis, A.; Vasiliauskas, R.; Taylor, A.F.S.; Stenlid, J.; Finlay, R. Fungal communities in mycorrhizal roots of conifer seedlings in forest nurseries under different cultivation systems, assessed by morphotyping, direct sequencing and mycelial isolation. Mycorrhiza 2005, 16, 33–41. [Google Scholar] [CrossRef]
- Timling, I.; Dahlberg, A.; Walker, D.; Gardes, M.; Charcosset, J.; Welker, J.; Taylor, D. Distribution and drivers of ectomycorrhizal fungal communities across the North American Arctic. Ecosphere 2012, 3, 1–25. [Google Scholar] [CrossRef]
- Lu, X.; Mu, Y.H.; Yuan, H.S. Two new species of Tomentella (Thelephorales, Basidiomycota) from Lesser Xingan Mts., northeastern China. Phytotaxa 2018, 369, 080–092. [Google Scholar] [CrossRef]
- Zhang, X.J.; Shi, F.L.; Yang, K.; Zhao, C.L. The diversity and taxonomy of Tomentella (Thelephoraceae, Thelephorales) with descriptions of four new species from Southwestern China. MycoKeys 2024, 109, 1–29. [Google Scholar] [CrossRef]
- Kuhar, F.; Nouhra, E.; Smith, M.E.; Caiafa, M.V.; Greslebin, A. Tomentellopsis rosannae sp. nov. (Basidiomycota, Thelephorales), first species in the genus described from the Southern Hemisphere. Fund. Miguel Lillo 2022, 59, 115–123. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence intervals on phylogenetics: An approach using bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffe, W.; Schwartz, T. The CIPRES science gateway. In Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the Extreme to the Campus and Beyond, Chicago, IL, USA, 16–19 July 2012; pp. 1–39. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModeltest V.2. Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Bret, L.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Bruen, T.C.; Philippe, H.; Bryant, D. A simple and robust statistical test for detecting the presence of recombination. Genetics 2006, 172, 2665–2681. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Quaedvlieg, W.; Binder, M.; Groenewald, J.Z.; Summerell, B.A.; Carnegie, A.J.; Burgess, T.I.; Crous, P.W. Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia 2014, 33, 1–40. [Google Scholar] [CrossRef]
- Luo, K.Y.; Zhao, C.L. A Molecular Systematics and Taxonomy Research on Trechispora (Hydnodontaceae, Trechisporales): Concentrating on Three New Trechispora Species from East Asia. J. Fungi 2022, 8, 1020. [Google Scholar] [CrossRef]
- Kõljalg, U. Tomentella (Basidiomycota) and related genera in temperate Eurasia. Synop. Fungorum 1996, 9, 1–213. [Google Scholar]
- Zang, M. Some new and noteworthy higher fungi from eastern Himalayas. Acta Bot. Yunnanica 1987, 9, 81–88. [Google Scholar]
- Cunningham, G.H. Thelephoraceae of New Zealand (parts XII and XIII). Part XII: The genera Thelephora and Tomentella. Trans. R. Soc. N. Z. 1957, 84, 479–496. [Google Scholar]
- Ramirez-Lópezl, I.; Villegas-Ríos, M.; Cano-Santana, Z. Phenotypic plasticity of the basidiomata of Thelephora sp. (Thelephoraceae) in tropical forest habitats. Rev. Biol. Trop. 2013, 61, 343–350. [Google Scholar] [CrossRef][Green Version]
- Tai, F.L. Sylloge Fungorum Sinicorum; Science Press, Academia Sinica: Beijing, China, 1979; pp. 740–741. [Google Scholar]
- Erland, S.; Taylor, A.F.S. Resupinate ectomycorrhizal fungal genera. In Ectomycorrhizal Fungi Key Genera in Profile; Springer: Berlin/Heidelberg, Germany, 1999; pp. 347–363. [Google Scholar] [CrossRef]
- Vidal, J.M.; Siquier, J.L.; Constantino, C. Alguns macromicets nous o interessants de l’illa de Mallorca (Balears). Rev. Catalana Micol. 1994, 16, 135–144. [Google Scholar]
- Mleczko, P. Mycorrhizal and saprobic macrofungi of two zinc wastes in southern poland. Acta Biol. Cracoviensia Ser. Bot. 2004, 46, 25–38. [Google Scholar] [CrossRef]
- Abrahão, M.C.; Gugliotta, A.D.M.; Bonon, V.L.R. Xylophilous Agaricomycetes (Basidiomycota) of the Brazilian Cerrado. Check List 2012, 8, 1102–1116. [Google Scholar] [CrossRef]
- Czernyadjeva, I.V.; Afonina, O.M.; Davydov, E.A.; Doroshina, G.Y.; Zyatnina, V. New cryptogamic records. 5. Nov. Sist. Nizshikh Rastenii 2020, 54, 261–286. [Google Scholar] [CrossRef]
- Lentz, P.L. The genus Thelephora in Iowa. Proc. Iowa Acad. Sci. 1942, 49, 175–184. [Google Scholar]
- Roberts, P.J.; Spooner, B.M. Cantharelloid, clavarioid and thelephoroid fungi from Brunei Darussalam. Kew Bull. 2000, 55, 843–851. [Google Scholar] [CrossRef]
- Budathoki, U.S.H.A. A new species of Periconiella from Kathmandu valley, Nepal. Mycol. Res. 2009, 47, 77–80. [Google Scholar]
- Dai, Y.C. A revised checklist of corticioid and hydnoid fungi in China for 2010. Mycoscience 2011, 52, 69–79. [Google Scholar] [CrossRef]
- Chacón, S.; Tapia, F.; Jarvio, D. Four interesting aphyllophoroid species in the tropical northern region of Veracruz, Mexico. Mycotaxon 2018, 133, 153–163. [Google Scholar] [CrossRef]
- Liu, S.; Shen, L.L.; Xu, T.M.; Song, C.G.; Gao, N.; Wu, D.M.; Sun, Y.F.; Cui, B.K. Global diversity, molecular phylogeny and divergence times of the brown-rot fungi within the Polyporales. Mycosphere 2023, 14, 1564–1664. [Google Scholar] [CrossRef]
- Cui, B.K.; Li, H.J.; Ji, X.; Zhou, J.L.; Song, J.; Si, J.; Yang, Z.L.; Dai, Y.C. Species diversity, taxonomy and phylogeny of Polyporaceae (Basidiomycota) in China. Fungal Divers. 2019, 97, 137–392. [Google Scholar] [CrossRef]
- Wu, F.; Man, X.W.; Tohtirjap, A.; Dai, Y.C. A comparison of polypore funga and species composition in forest ecosystems of China, North America, and Europe. For. Ecosyst. 2022, 9, 100051. [Google Scholar] [CrossRef]
- Dai, Y.C.; Yang, Z.L.; Cui, B.K.; Wu, G.; Yuan, H.S.; Zhou, L.W.; He, S.H.; Ge, Z.W.; Wu, F.; Wei, Y.L.; et al. Diversity and systematics of the important macrofungi in Chinese forests. Mycosystema 2021, 40, 770–805. [Google Scholar]
- Wu, F.; Zhou, L.W.; Yang, Z.L.; Bau, T.; Li, T.H.; Dai, Y.C. Resource diversity of Chinese macrofungi: Edible, medicinal and poisonous species. Fungal Divers. 2019, 98, 1–76. [Google Scholar] [CrossRef]
- He, M.Q.; Zhao, R.L.; Hyde, K.D.; Begerow, D.; Kemler, M.; Yurkov, A.M.; McKenzie, E.; Raspé, O.; Kakishima, M.; Sánchez-Ramírez, S.; et al. Notes, outline and divergence times of Basidiomycota. Fungal Divers. 2019, 99, 105–367. [Google Scholar] [CrossRef]
- Svantesson, S.; Kõljalg, U.; Wurzbacher, C.; Saar, I.; Larsson, K.H.; Larsson, E. Polyozellus vs. Pseudotomentella: Generic delimitation with a multi-gene dataset. Fungal Syst. Evol. 2021, 8, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Mikryukov, V.; Anslan, S.; Bahram, M.; Khalid, A.N.; Corrales, A.; Agan, A.; Vasco-Palacios, A.M.; Saitta, A.; Antonelli, A.; et al. The Global Soil Mycobiome consortium dataset for boosting fungal diversity research. Fungal Divers. 2021, 111, 573–588. [Google Scholar] [CrossRef]
- Mikryukov, V.; Dulya, O.; Zizka, A.; Bahram, M.; Hagh-Doust, N.; Anslan, S.; Prylutskyi, O.; Delgado-Baquerizo, M.; Maestre, F.T.; Nilsson, R.H.; et al. Connecting the multiple dimensions of global soil fungal diversity. Sci. Adv. 2023, 9, eadj8016. [Google Scholar] [CrossRef]
- Kõljalg, U.; Saar, I.; Svantesson, S. Merging the genus Tomentella with Thelephora (Fungi, Thelephorales). Folia Cryptog. Est. Fasc. 2024, 61, 67–86. [Google Scholar] [CrossRef]
- Matthew, L.; Smith, M.E. Lineages of ectomycorrhizal fungi revisited: Foraging strategies and novel lineages revealed by sequences from belowground. Fungal Biol. Rev. 2013, 87, 83–99. [Google Scholar] [CrossRef]








| Species Name | Sample No. | GenBank Accession No. | Country | References | ||
|---|---|---|---|---|---|---|
| ITS | nLSU | mtSSU | ||||
| Amaurodon aquicoeruleus | TU100989 | AM490944 | — | — | Australia | [53] |
| Amaurodon caeruleocaseus | PERTH:066707 | MT565478 | — | — | Australia | [53] |
| Amaurodon hydnoides | TU108407 | AM490941 | — | — | Venezuela | [53] |
| Lenzitopsis daii | Yuan2952 | JN169798 | — | — | China | [54] |
| Lenzitopsis oxycedri | KHLarsson15304 | MK602774 | — | — | Sweden | [54] |
| Odontia fibrosa | LE F-332368 | MT981502 | MT981502 | — | Russia | [20] |
| Odontia sparsa | Yuan 10718 | MG719980 | — | — | China | [55] |
| Phellinotus neoaridus | URM 83203 | MZ92858 | MZ964977 | — | Brazil | [56] |
| Phellodon atroardesiacus | Cui 18449 | MZ221189 | MZ225598 | MZ225636 | China | [57] |
| Phellodon atroardesiacus | Cui 18457 | MZ225577 | MZ225599 | — | China | [57] |
| Phellodon cinereofuscus | Cui 16962 | MZ225583 | MZ225605 | MZ225643 | China | [57] |
| Phellodon cinereofuscus | Cui 16963 | MZ225584 | MZ225606 | MZ225644 | China | [57] |
| Phellodon melaleucus | Cui 18614 | OL449262 | OL439032 | OL439022 | China | [57] |
| Phellodon melaleucus | Cui 18620 | OL449263 | OL439033 | OL439023 | China | [57] |
| Phellodon yunnanensis | Cui 17129 | MZ225594 | MZ225614 | MZ225652 | China | [57] |
| Phellodon yunnanensis | Cui 17131 | MZ225595 | MZ225615 | MZ225653 | China | [57] |
| Polyozellus atrolazulinus | TU117477 | MF100839 | — | — | Canada | [58] |
| Polyozellus atrolazulinus | TU117559 | MG214657 | — | — | Canada | [58] |
| Polyozellus mariae | TU117235 | MF100826 | — | — | Canada | [58] |
| Polyozellus purpureoniger | TU103000 | MF100821 | — | — | USA | [58] |
| Thelephora americana | BMJ01 | MT196971 | — | — | USA | Unpublished |
| Thelephora anthocephala | NSK101420 | MT773612 | — | — | Russia | Unpublished |
| Thelephora anthocephala | src614 | DQ974771 | — | — | USA | Unpublished |
| Thelephora aquila | Wei 8833 | OP793744 | — | — | China | [46] |
| Thelephora aquila | Wei 8831 | OP793743 | — | — | China | [46] |
| Thelephora aurantiotincta | 520625MF420 | MZ057686 | — | — | China | Unpublished |
| Thelephora austrosinensis | GDGM 48867 | MF593265 | — | — | China | [8] |
| Thelephora caryophyllea | BSI 13/103 | KR606030 | — | — | Switzerland | Unpublished |
| Thelephora caryophyllea | GO-2010-163 | KC152242 | — | — | Mexico | Unpublished |
| Thelephora dactyliophora | KUN-HKAS131941 | OR940521 | — | — | China | [9] |
| Thelephora dactyliophora | KUN-HKAS131943 | OR940523 | — | — | China | [9] |
| Thelephora dominicana | JBSD126510 | KX216400 | — | — | Italy | [37] |
| Thelephora ganbajun | Yuan 16756 | OP793761 | OP793790 | OP793718 | China | [46] |
| Thelephora ganbajun | Yuan16817 | OP793762 | OP793687 | OP793721 | China | [46] |
| Thelephora glaucoflora | Dai 13627A | OP793752 | — | — | China | [46] |
| Thelephora glaucoflora | Dai 13623A | OP793751 | — | — | China | [46] |
| Thelephora grandinioides | CLZhao 3406 | MZ400677 | MZ400671 | — | China | [59] |
| Thelephora grandinioides | CLZhao 3408 | MZ400674 | MZ400672 | — | China | [59] |
| Thelephora iqbalii | MH810 | JX241471 | — | — | Pakistan | [60] |
| Thelephora lacunosa | KUN-HKAS128966 | OR512336 | — | — | China | [9] |
| Thelephora lacunosa | KUN-HKAS128967 | OR512337 | — | — | China | [9] |
| Thelephora nebula | Yuan 11516 | OP793746 | — | — | China | [46] |
| Thelephora nebula | Yuan 11515 | OP793745 | — | — | China | [46] |
| Thelephora palmata | TU115271 | MH310778 | — | — | Sweden | Unpublished |
| Thelephora penicillata | X619 | OL469899 | — | — | Czechia | [60] |
| Thelephora penicillata | X618 | OL469898 | — | — | Czechia | [60] |
| Thelephora petaloides | KUN-HKAS128969 | OR512332 | — | — | China | [11] |
| Thelephora petaloides | KUN-HKAS128970 | OR512333 | — | — | China | [9] |
| Thelephora pinnatiflda | KUN-HKAS131946 | OR940524 | — | — | China | [9] |
| Thelephora pinnatiflda | KUN-HKAS131947 | OR940525 | — | — | China | [9] |
| Thelephora pseudoganbajun | Yuan 16780 | OP793766 | — | — | China | [46] |
| Thelephora pseudoganbajun | Yuan 16771 | OP793765 | — | — | China | [46] |
| Thelephora pseudoversatilis | FCME 26152 | KJ462486 | — | — | Mexico | Unpublished |
| Thelephora pseudoversatilis | FCME 26232 | JX075890 | — | — | Mexico | Unpublished |
| Thelephora regularis | JMT17371 | U83485 | — | — | USA | Unpublished |
| Thelephora regularis | UBC F33227 | MG953966 | — | — | Canada | Unpublished |
| Thelephora resupinata | CLZhao 34548 | PP810222 | — | PQ060160 | China | Present study |
| Thelephora resupinata | CLZhao 34538 | PP810221 | PP809695 | PQ060159 | China | Present study |
| Thelephora scissilis | MUOB:324045 | OK376730 | — | — | USA | Unpublished |
| Thelephora sikkimensis | KUN-HKAS128972 | OR512330 | — | — | China | [9] |
| Thelephora sikkimensis | KUN-HKAS128965 | OR512331 | — | — | China | [9] |
| Thelephora sublilacina | UP161 | EF493288 | — | — | Sweden | Unpublished |
| Thelephora subtropica | CLZhao 30591 | PP810227 | — | PQ060160 | China | Present study |
| Thelephora subtropica | CLZhao 30590 | PP810226 | PP809695 | PQ060159 | China | Present study |
| Thelephora terrestris | Hilszczanska D. 1-IBL | FJ532478 | — | — | Poland | Unpublished |
| Thelephora terrestris | KGP22 | DQ822828 | — | — | USA | Unpublished |
| Thelephora versatilis | FCME26141 | KJ462504 | — | — | Mexico | Unpublished |
| Thelephora versatilis | FCME26146 | NR12492 | — | — | Mexico | Unpublished |
| Thelephora vialis | TENN-F- 072281H2 | MN121029 | — | — | USA | Unpublished |
| Thelephora vialis | TENN-F-072094 | MN121022 | — | — | USA | Unpublished |
| Thelephora wuliangshanensis | CLZhao 21020 | MZ400672 | — | — | China | [59] |
| Thelephora wuliangshanensis | CLZhao 4107 | MZ400671 | — | — | China | [59] |
| Thelephora yunnanensis | CLZhao 20929 | PP810224 | PP809696 | PQ060161 | China | Present study |
| Thelephora yunnanensis | CLZhao 20935 | PP810225 | PP809697 | PQ060162 | China | Present study |
| Thelephora yunnanensis | CLZhao 20926 | PP810223 | — | — | China | Present study |
| Tomentella africana | SYN 991 | EF50722 | — | — | Benin | [14] |
| Tomentella africana | M SYN 991 | NR_119637 | — | — | Benin | [14] |
| Tomentella afrostuposa | SYN 2292 | JF520431 | — | — | Guinea | [61] |
| Tomentella afrostuposa | M SYN 2292 | NR_11992 | — | — | Guinea | Unpublished |
| Tomentella agbassaensis | M SYN 981 | NR_119638 | — | — | Benin | Unpublished |
| Tomentella agbassaensis | SYN 981 | EF507257 | — | — | Benin | [62] |
| Tomentella agereri | RA 13793 | EF538424 | — | — | Benin | [62] |
| Tomentella agereri | M RA 13793 | NR_119641 | — | — | Benin | Unpublished |
| Tomentella alpina | IB 20060231 | NR_121330 | — | — | Australia | Unpublished |
| Tomentella amyloapiculata | SYN 893 | EF507263 | — | — | Benin | [63] |
| Tomentella amyloapiculata | M SYN 893 | NR_119639 | — | — | Benin | Unpublished |
| Tomentella asperula | iNat66942560 | ON943290 | — | — | Canada | Unpublished |
| Tomentella atrocastanea | Yuan 12170 | MK211742 | MK446337 | — | China | [64] |
| Tomentella atrocastanea | Yuan 12179 | MK211743 | MK446338 | — | China | [64] |
| Tomentella aureomarginata | Yuan 10683 | MK211745 | MK878395 | — | China | [64] |
| Tomentella badia | LE 299095 | MT981507 | — | — | Russia | Unpublished |
| Tomentella badia | LE 314775 | MT981499 | — | — | Russia | Unpublished |
| Tomentella botryoides | O-F256708 | MT146455 | — | — | Sweden | [65] |
| Tomentella botryoides | O-F256707 | MT14642 | — | — | Sweden | [65] |
| Tomentella brevisterigmata | IFP 019338 | NR_185567 | — | — | China | Unpublished |
| Tomentella brunneocystidia | SYN 839 | DQ848613 | — | — | Benin | [32] |
| Tomentella brunneocystidia | RA 13779 | DQ848610 | — | — | Benin | [32] |
| Tomentella brunneoflava | Yuan 12162 | MK211749 | MK850194 | — | China | [64] |
| Tomentella brunneoflava | Yuan 12161 | MK211748 | — | — | China | [64] |
| Tomentella bryophila | FFP1020 | JQ711917 | — | — | Canada | [65] |
| Tomentella capitata | SYN 860 | DQ848612 | — | — | USA | [32] |
| Tomentella capitata | RA 13785 | DQ848611 | — | — | Benin | [32] |
| Tomentella casiae | Yuan 18263 | PP479638 | PP486302 | — | China | [62] |
| Tomentella casiae | Yuan 18254 | PP479637 | PP486299 | — | China | [62] |
| Tomentella castanea | JW1 | KC952674 | — | — | Germany | Unpublished |
| Tomentella cinerascens | SS301 | MT146467 | — | — | Sweden | [66] |
| Tomentella cinerascens | SP72a | OQ418570 | — | — | Sweden | [66] |
| Tomentella coerulea | MFT22 | MK431005 | — | — | Germany | Unpublished |
| Tomentella coerulea | MTB3 | MN947340 | — | — | Germany | Unpublished |
| Tomentella dimidiata | Yuan 11205 | MK211704 | MK446355 | — | China | [64] |
| Tomentella dimidiata | Yuan 11267 | MK211705 | MK446356 | — | China | [64] |
| Tomentella duplexa | Yuan 12202 | MK211706 | MK446357 | — | China | [64] |
| Tomentella duplexa | Yuan 12207 | MK211707 | MK446358 | — | China | [64] |
| Tomentella efibulis | Yuan 11241 | MK211708 | MK446361 | — | China | [64] |
| Tomentella efibulis | Yuan 11329 | MK211709 | MK446362 | — | China | [64] |
| Tomentella ellisii | src846 | DQ974775 | — | — | USA | [67] |
| Tomentella fuscocinerea | TU108229 | GU214810 | — | — | Estonia | Unpublished |
| Tomentella fuscocrustosa | Yuan 11420 | MK211713 | MK446367 | — | China | [64] |
| Tomentella fuscocrustosa | Yuan 11399 | MK211712 | MK446366 | — | China | [64] |
| Tomentella fuscofarinosa | Yuan 12142 | MK211715 | MK446369 | — | China | [64] |
| Tomentella fuscofarinosa | Yuan 12125 | MK211714 | MK446368 | — | China | [64] |
| Tomentella fuscopelliculosa | Yuan 11316 | MK211717 | — | — | China | [64] |
| Tomentella fuscopelliculosa | Yuan 11305 | MK211716 | MK446372 | — | China | [64] |
| Tomentella galzinii | TAA166821 | AF272932 | — | — | Estonia | [67] |
| Tomentella galzinii | TAA149734 | AF272928 | — | — | Estonia | [67] |
| Tomentella globosa | AMC122 | OP413006 | — | — | USA | Unpublished |
| Tomentella globospora | Yuan 10748 | MK446375 | — | — | China | [64] |
| Tomentella globospora | Yuan 10668 | MK446374 | — | — | China | [64] |
| Tomentella griseomarginata | Yuan 11458 | MK211720 | MK446382 | — | China | [64] |
| Tomentella griseomarginata | Yuan 11468 | MK211721 | MK446383 | — | China | [64] |
| Tomentella guineensis | M SYN 2331 | NR_119955 | — | — | Guinea | [63] |
| Tomentella guineensis | SYN 2331 | JF520432 | — | — | Guinea | [63] |
| Tomentella guiyangensis | Yuan 18281 | PP479645 | PP486306 | — | China | [68] |
| Tomentella guiyangensis | Yuan 18256 | PP479643 | PP486300 | — | China | [68] |
| Tomentella hjortstamiana | TU103641 | NR_121290 | — | — | Seychelles | [69] |
| Tomentella incrustata | Yuan 12189 | MK211723 | MK446387 | — | China | [64] |
| Tomentella intsiae | TAA195077 | AM412296 | — | — | Estonia | [70] |
| Tomentella intsiae | TU105130 | NR_121286 | — | — | Seychelles | [69] |
| Tomentella lapida | LE F-332369 | MT981496 | — | — | Russia | [71] |
| Tomentella lapida | PN_2Bb_I | JQ724049 | — | — | Poland | [72] |
| Tomentella larssoniana | TU103690 | AM412294 | — | — | Estonia | [70] |
| Tomentella larssoniana | TU105082 | NR_119738 | — | — | Estonia | [71] |
| Tomentella lilacinogrisea | NS74 | DQ068972 | — | — | Sweden | [73] |
| Tomentella lilacinogrisea | AR1119 | JX630832 | — | — | USA | [74] |
| Tomentella longechinulata | Yuan 12083 | MK211727 | MK446394 | — | China | [64] |
| Tomentella longiaculeifera | Yuan 10744 | MK446391 | — | China | [64] | |
| Tomentella longisterigmata | IFP 19181 | NR_161037 | — | — | Finland | [75] |
| Tomentella maroana | M SYN 878 | NR_119636 | — | — | Benin | [40] |
| Tomentella maroana | SYN 878 | EF507250 | — | — | Benin | [40] |
| Tomentella muricata | O-F256712 | MT146462 | — | — | Sweden | [68] |
| Tomentella muricata | O-F256713 | MT146461 | — | — | Sweden | [68] |
| Tomentella nitellina | src675 | DQ974778 | — | — | USA | [66] |
| Tomentella olivaceobasidiosa | CLZhao 14051 | PP810228 | — | — | China | [76] |
| Tomentella olivaceobasidiosa | CLZhao 14056 | PP810229 | PP809698 | — | China | [76] |
| Tomentella olivaceomarginata | Yuan 18268 | PP479639 | PP486303 | — | China | [68] |
| Tomentella pallidobrunnea | Yuan 11493 | MK211731 | MK446402 | — | China | [64] |
| Tomentella pallidobrunnea | Yuan 11481 | MK211730 | MK446401 | — | China | [64] |
| Tomentella pallidomarginata | Yuan 11474 | MK211733 | MK446404 | — | China | [64] |
| Tomentella pallidomarginata | Yuan 11404 | MK211732 | MK446403 | — | China | [64] |
| Tomentella parmastoana | NAN13 | MN075506 | — | — | Thailand | Unpublished |
| Tomentella parmastoana | TU103582 | NR_121289 | — | — | USA | [70] |
| Tomentella patagonica | BAFC52372 | NR_159018 | — | — | Argentina | [36] |
| Tomentella patagonica | LR-24 | MT366710 | — | — | USA | Unpublished |
| Tomentella pileocystidiata | TU105068 | NR_119739 | — | — | Estonia | [69] |
| Tomentella pileocystidiata | TU10502 | FM955845 | — | — | Estonia | [69] |
| Tomentella pilosa | TU124067 | MT146459 | MT52521 | — | Sweden | [65] |
| Tomentella pilosa | TU124234 | MT146458 | — | — | Sweden | [65] |
| Tomentella pisoniae | TU103671 | NR_121358 | — | — | USA | [70] |
| Tomentella pisoniae | TU103655 | FN185986 | — | — | Argentina | [36] |
| Tomentella pulvinulata | BAFC52370 | NR_159017 | — | — | Argentina | [36] |
| Tomentella rotundata | Yuan 18269 | PP479641 | PP486304 | — | China | [68] |
| Tomentella rotundata | Yuan 18273 | PP479642 | PP486305 | — | China | [68] |
| Tomentella separata | Yuan 10664 | MK211737 | MK850196 | — | China | [64] |
| Tomentella separata | Yuan 1062 | MK211736 | MK850197 | — | China | [64] |
| Tomentella stuposa | IB2005314 | EF644117 | — | — | Australia | [77] |
| Tomentella subclavigera | O-F256725 | MT146460 | — | — | Sweden | [65] |
| Tomentella subtestacea | FR-F10 | MW546915 | — | — | South Korea | Unpublished |
| Tomentella subtestacea | FFP816 | JQ711878 | — | — | Canada | [66] |
| Tomentella tedersooi | TU103663 | NR_121359 | — | — | Estonia | [70] |
| Tomentella tedersooi | TU103664 | FN185989 | — | — | Estonia | [70] |
| Tomentella tenuifarinacea | CLZhao 31337 | PQ276703 | PQ276704 | — | China | Present study |
| Tomentella tenuirhizomorpha | Yuan 12059 | MG799185 | MN684327 | — | China | [75] |
| Tomentella tenuissima | FK14070 | KT032087 | — | — | Argentina | [36] |
| Tomentella tenuissima | BAFC52369 | NR_159016 | — | — | USA | [69] |
| Tomentella terrestris | TAA159557 | AF272911 | — | — | Estonia | [69] |
| Tomentella velutina | CLZhao 25474 | PP645440 | PP809700 | — | China | [76] |
| Tomentella viridula | MTB37 | MN947374 | — | — | Estonia | [69] |
| Tomentella wumenshanensis | CLZhao 33775 | PP810230 | PP809699 | — | China | [76] |
| Tomentella yunnanensis | CLZhao 32532 | PP810231 | — | — | China | [76] |
| Tomentellopsis rosannae | MES-3338 | MT366690 | — | — | Chile | [77] |
| Tomentellopsis submollis | RS-22498 | AJ410774 | — | — | Finland | [77] |
| Tomentellopsis submollis | P24-F | AM086447 | — | — | Norway | [77] |
| Tomentellopsis zygodesmoides | JS-27216 | AJ410759 | — | — | Norway | [77] |
| Tomentellopsis zygodesmoides | KHL-8653 | AJ410761 | — | — | Norway | [77] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Shi, F.; Zhang, S.; Hosen, M.I.; Zhao, C. The Diversity and Taxonomy of Thelephoraceae (Basidiomycota) with Descriptions of Four Species from Southwestern China. J. Fungi 2024, 10, 775. https://doi.org/10.3390/jof10110775
Zhang X, Shi F, Zhang S, Hosen MI, Zhao C. The Diversity and Taxonomy of Thelephoraceae (Basidiomycota) with Descriptions of Four Species from Southwestern China. Journal of Fungi. 2024; 10(11):775. https://doi.org/10.3390/jof10110775
Chicago/Turabian StyleZhang, Xiaojie, Fulei Shi, Sicheng Zhang, Md. Iqbal Hosen, and Changlin Zhao. 2024. "The Diversity and Taxonomy of Thelephoraceae (Basidiomycota) with Descriptions of Four Species from Southwestern China" Journal of Fungi 10, no. 11: 775. https://doi.org/10.3390/jof10110775
APA StyleZhang, X., Shi, F., Zhang, S., Hosen, M. I., & Zhao, C. (2024). The Diversity and Taxonomy of Thelephoraceae (Basidiomycota) with Descriptions of Four Species from Southwestern China. Journal of Fungi, 10(11), 775. https://doi.org/10.3390/jof10110775






