Environmental Factors Drive the Biogeographic Pattern of Hippophae rhamnoides Root Endophytic Fungal Diversity in the Arid Regions of Northwest China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Soil Physicochemical Analyses
2.3. DNA Extraction, PCR Amplification, and High-Throughput Sequencing
2.4. Statistical Analysis
3. Results
3.1. Analysis of Soil Physicochemical Properties
3.2. Quality Analysis of Fungal ITS Sequencing Results
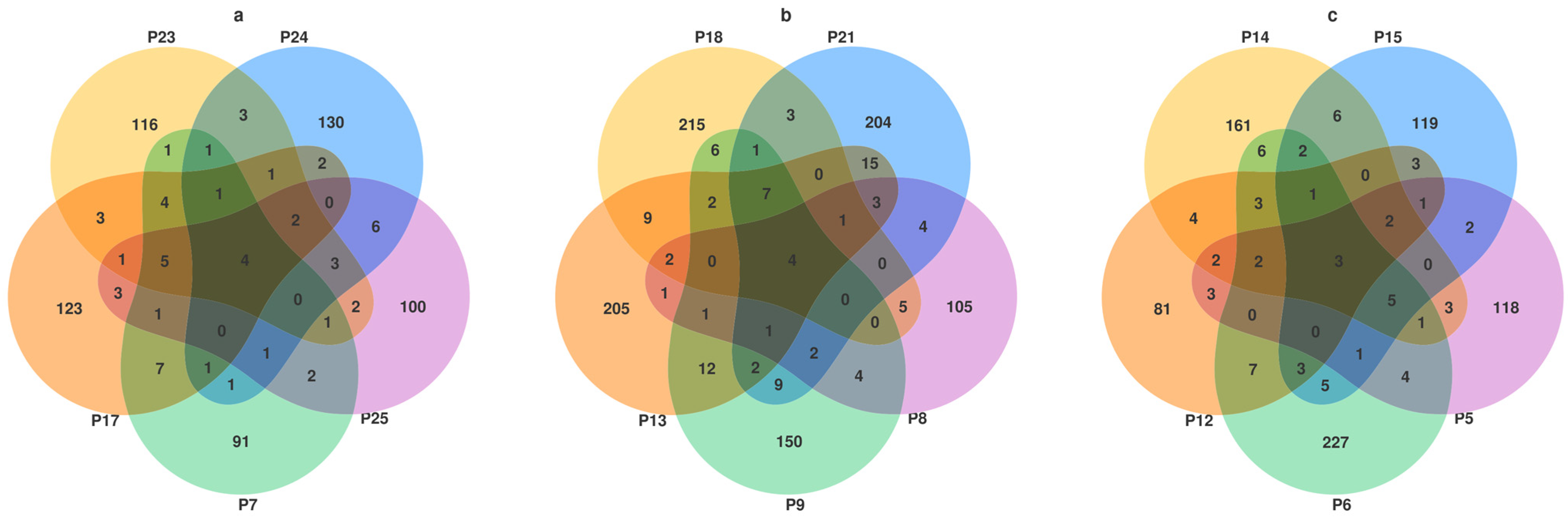
3.3. Venn Diagram Analysis
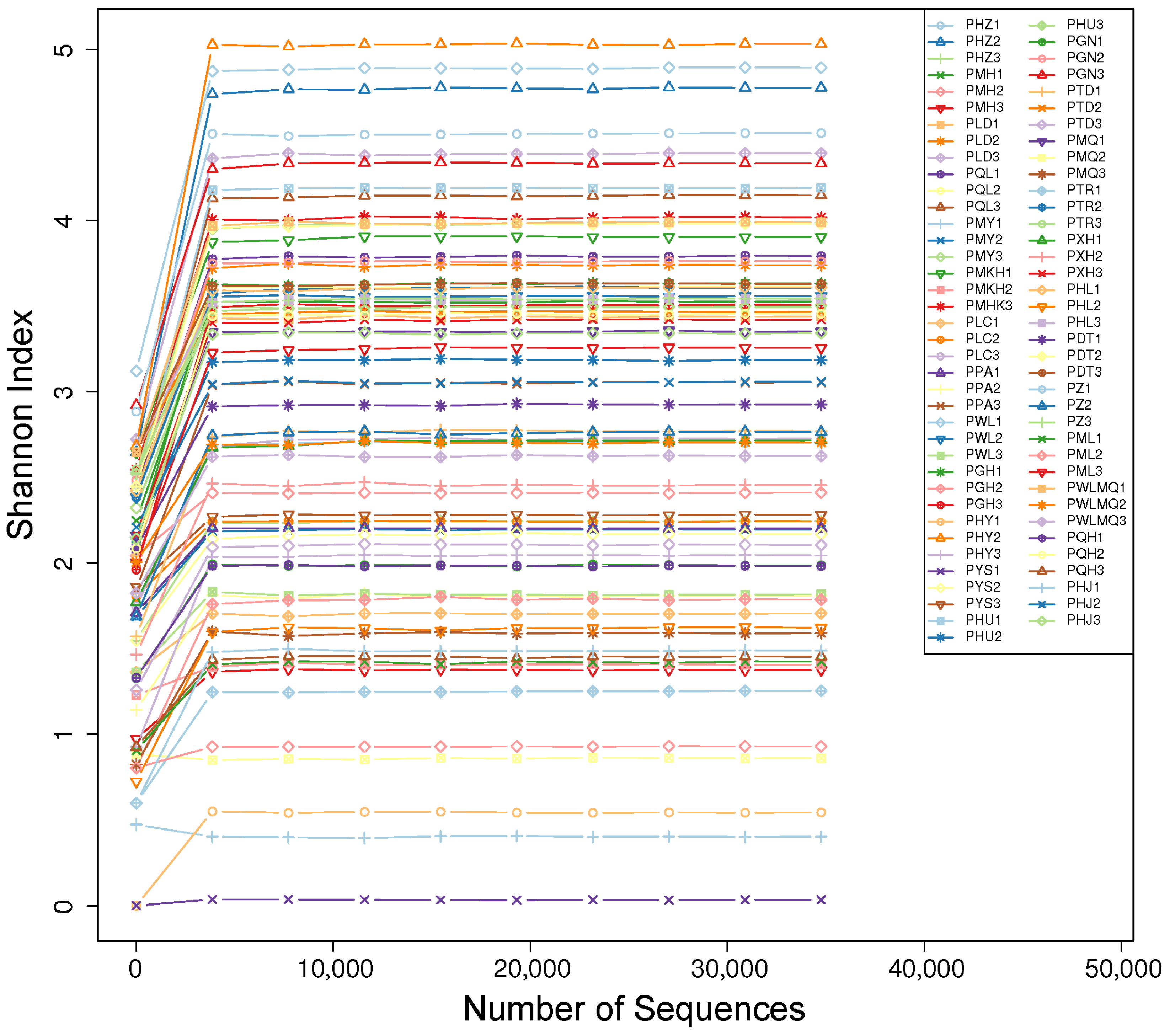
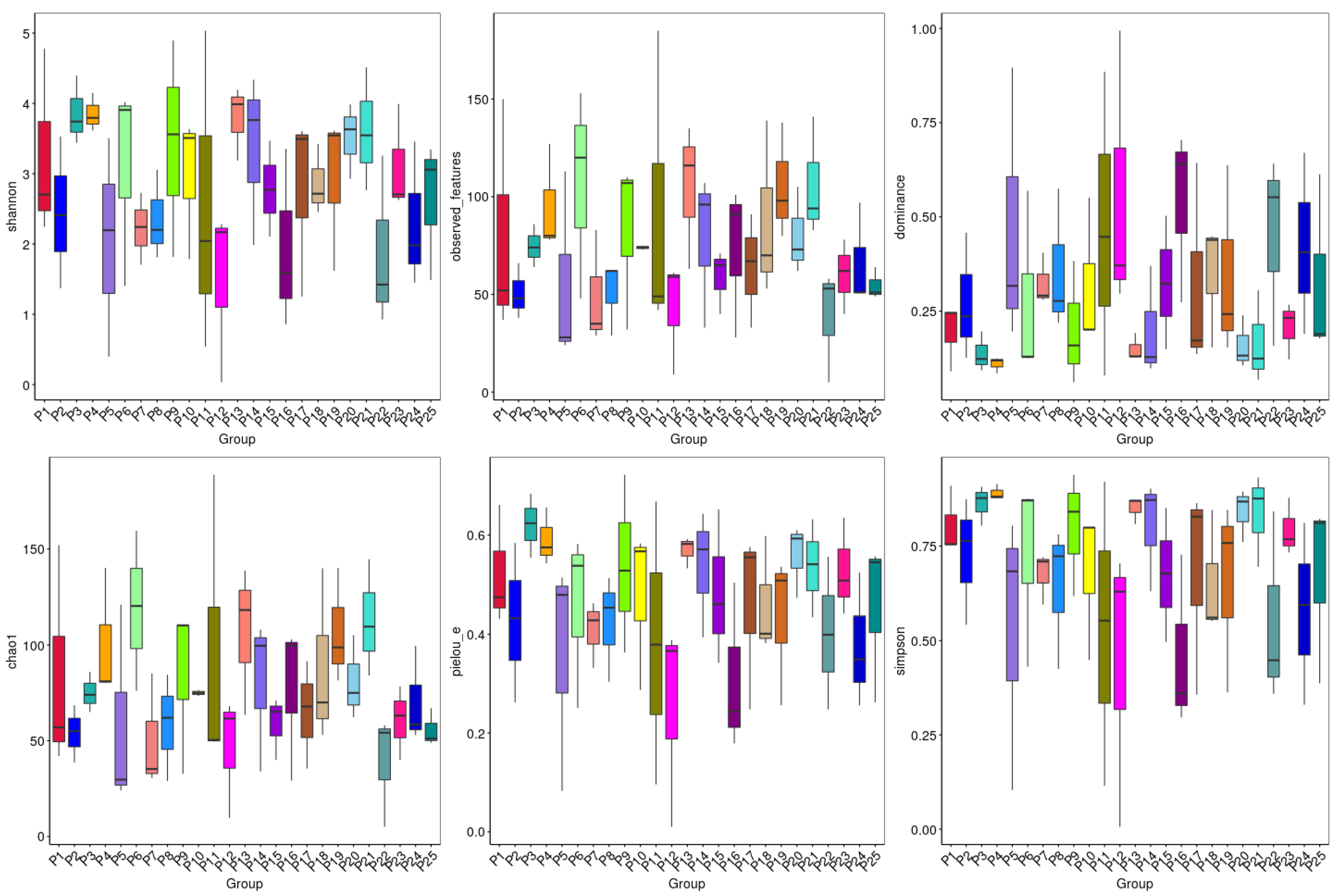
3.4. Alpha Diversity Analysis
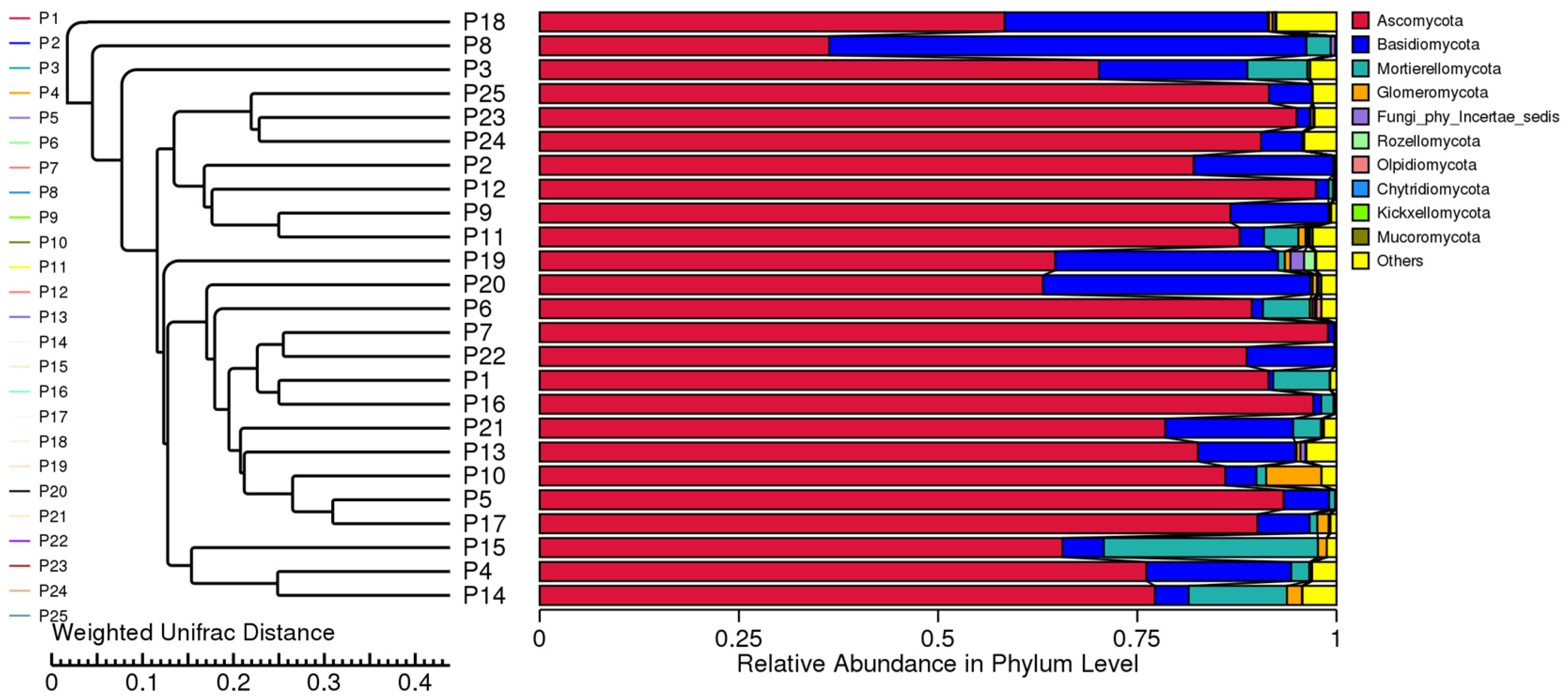
3.5. Beta Diversity Analysis
3.6. Species Composition Analysis
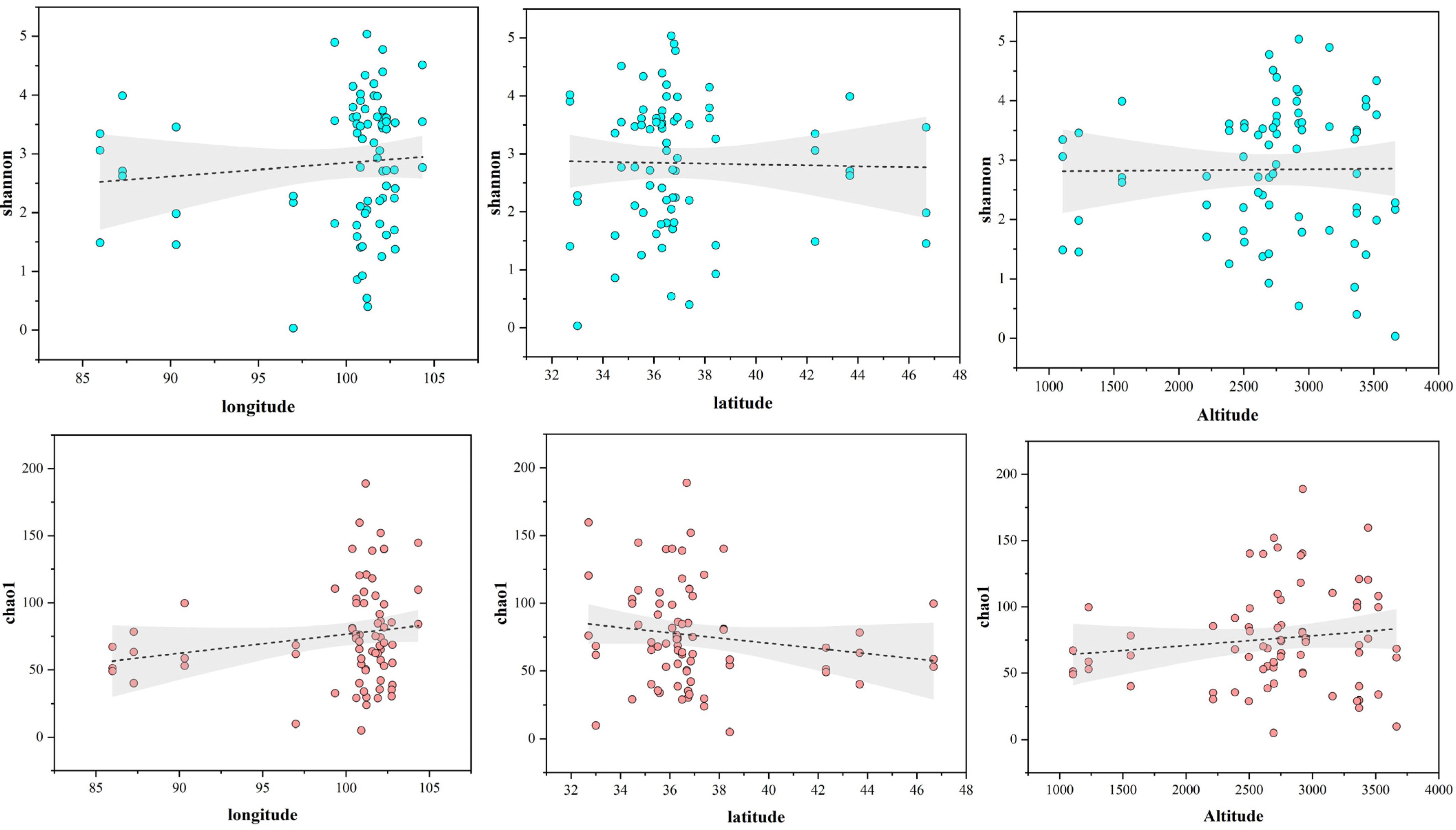
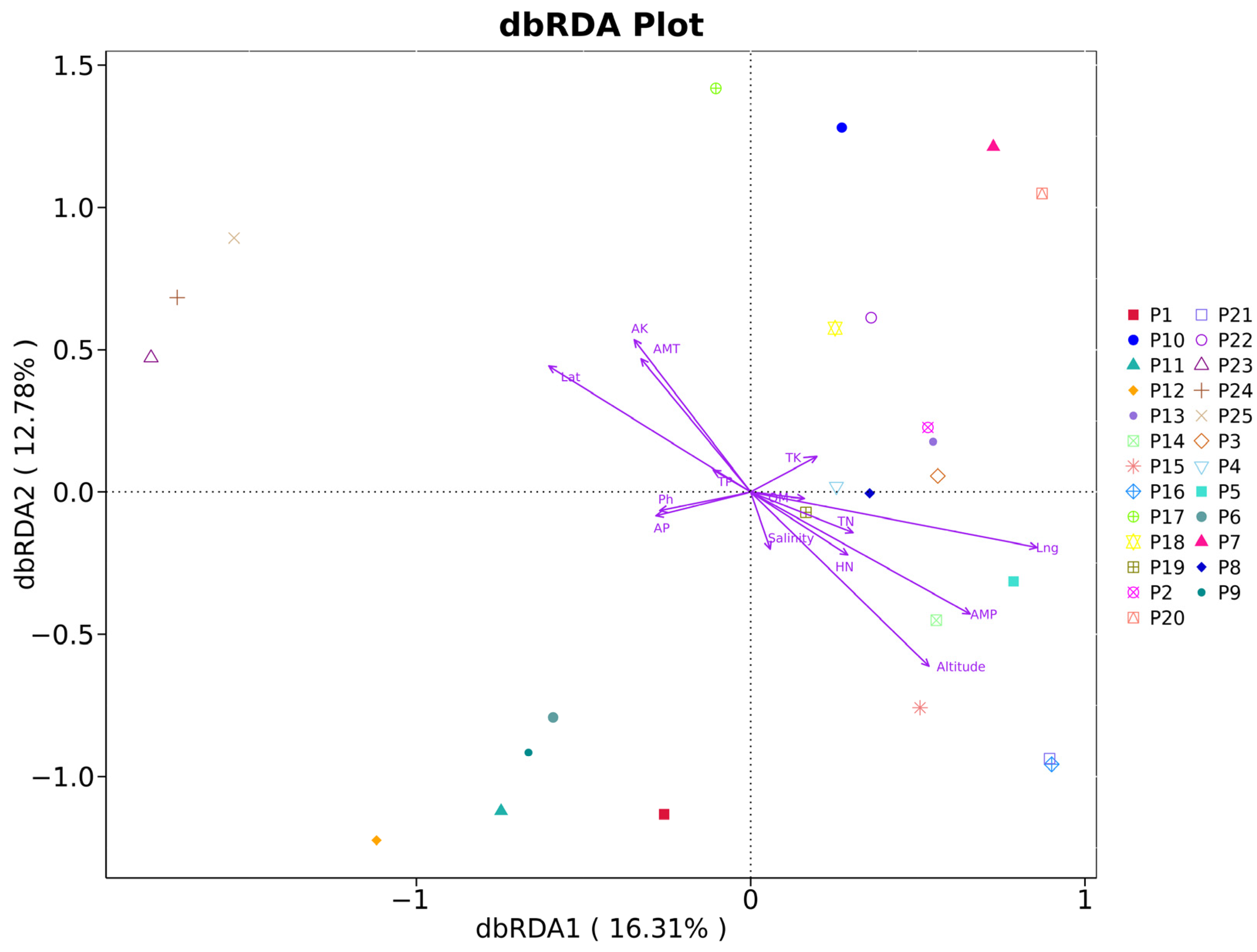
3.7. Origin Difference Analysis
3.8. Fungal Community Function Prediction
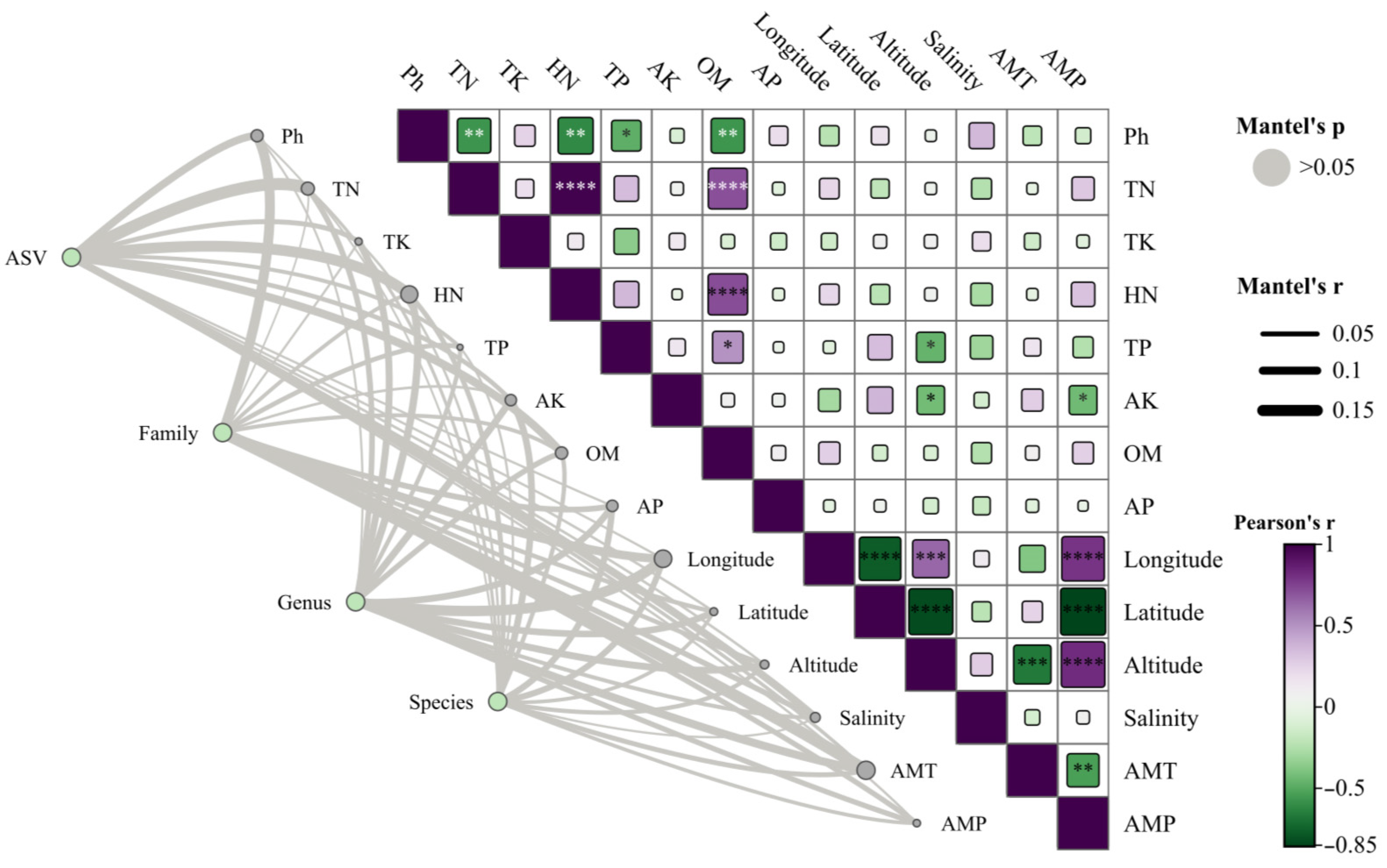
3.9. Fungal Molecular Network
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Sample | Sample Name | AddPess | Soil Type | Sample No. | Latitude (N°) | Longitude (E°) | Altitude /m | AMT | AMP |
|---|---|---|---|---|---|---|---|---|---|
| HZ | P1 | Huzhu County, Haidong City | Sand | PHZ1, PHZ2, PHZ3 | 36.977 | 102.077 | 2696.3 | 1.715 | 126.085 |
| MH | P2 | Minhe County, Haidong City | Loam | PMH1, PMH2, PMH3 | 36.376 | 102.782 | 2644.8 | 5.456 | 101.830 |
| LD | P3 | Ledu District, Haidong City | Loam | PLD1, PLD2, PLD3 | 36.978 | 102.077 | 2753.5 | 1.715 | 126.085 |
| QL | P4 | Qilian County, Haibei Tibetan Autonomous Prefecture | Sand | PQL1, PQL2, PQL3 | 38.076 | 100.378 | 2920.8 | −0.019 | 124.205 |
| MY | P5 | Menyuan County, Haibei Tibetan Autonomous Prefecture | Clay | PMY1, PMY2, PMY3 | 37.755 | 101.220 | 3369.0 | −1.769 | 134.440 |
| MKH | P6 | Make River, Luoguoluo Tibetan Autonomous Prefecture | Clay | PMKH1, PMKH2, PMKH3 | 32.874 | 100.820 | 3441.0 | 1.925 | 158.825 |
| LC | P7 | Liancheng, Lanzhou City | Loam | PLC1, PLC2, PLC3 | 36.661 | 102.736 | 2214.6 | 4.394 | 101.465 |
| PA | P8 | Ping’an District, Haidong City | Loam | PPA1, PPA2, PPA3 | 36.338 | 101.914 | 2497.8 | 3.779 | 114.900 |
| WL | P9 | Wulan County, Haixi Mongolian and Tibetan Autonomous Prefecture | Sand | PWL1, PWL2, PWL3 | 36.660 | 99.341 | 3159.0 | 1.356 | 76.410 |
| GH | P10 | Gonghe County, Hainan Tibetan Autonomous Prefecture | Clay | PGH1, PGH2, PGH3 | 36.311 | 100.595 | 2947.6 | 3.467 | 102.030 |
| HY | P11 | Huangyuan County, Xining City | Loam | PHY1, PHY2, PHY3 | 36.545 | 101.184 | 2923.1 | 1.756 | 122.610 |
| YS | P12 | Yushu Tibetan Autonomous Prefecture | Loam | PYS1, PYS2, PYS3 | 33.005 | 96.992 | 3665.0 | 1.927 | 132.345 |
| HU | P13 | Huangzhong District, Xining City | Clay | PHU1, PHU2, PHU3 | 36.400 | 101.575 | 2906.8 | 1.863 | 124.360 |
| GN | P14 | Guinan County, Hainan Tibetan Autonomous Prefecture | Loam | PGN1, PGN2, PGN3 | 35.707 | 101.081 | 3522.1 | 0.252 | 120.735 |
| TD | P15 | Tongde County, Hainan Tibetan Autonomous Prefecture | Loam | PTD1, PTD2, PTD3 | 34.736 | 100.806 | 3369.0 | 1.473 | 124.870 |
| MQ | P16 | Maqin County, Guoluo Tibetan Autonomous Prefecture | Loam | PMQ1, PMQ2, PMQ3 | 34.660 | 100.622 | 3353.5 | 0.615 | 125.880 |
| TR | P17 | Tongren City, Huangnan Tibetan Autonomous Prefecture | Clay | PTR1, PTR2, PTR3 | 35.518 | 102.019 | 2387.9 | 5.525 | 114.660 |
| XH | P18 | Xunhua Salar Autonomous County, Haidong City | Clay | PXH1, PXH2, PXH3 | 35.716 | 102.287 | 2610.6 | 2.383 | 121.885 |
| HL | P19 | Haidong Hualong Hui Autonomous County | Clay | PHL1, PHL2, PHL3 | 36.073 | 102.280 | 2503.9 | 3.667 | 114.750 |
| DT | P20 | Haidong Hualong Hui Autonomous County | Clay | PDT1, PDT2, PDT3 | 37.002 | 101.771 | 2748.7 | 2.596 | 125.640 |
| Z | P21 | Dingxizhang County, Gansu Province | Clay | PZ1, PZ2, PZ3 | 34.823 | 104.330 | 2725.2 | 4.510 | 124.825 |
| ML | P22 | Minle County, Gansu Province | Clay | PML1, PML2, PML3 | 38.265 | 100.915 | 2692.5 | 0.881 | 116.380 |
| WLMQ | P23 | urumqi county of xinjiang | Clay | PWLMQ1, PWLMQ2, PWLMQ3 | 43.451 | 87.267 | 1562.5 | 3.717 | 63.720 |
| QH | P24 | Qinghe County, Altay Prefecture, Xinjiang Uygur Autonomous Region | Sand | PQH1, PQH2, PQH3 | 46.532 | 90.319 | 1230.0 | 2.206 | 29.035 |
| HJ | P25 | Hejing County, Bayinguoleng Mongolian Autonomous Prefecture, Xinjiang | Sand | PHJ1, PHJ2, PHJ3 | 42.282 | 85.992 | 1106.3 | 10.763 | 31.560 |
References
- Bardgett, R.D.; Mommer, L.; De Vries, F.T. Going underground: Root traits as drivers of ecosystem processes. Trends Ecol. Evol. 2014, 29, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Davy, A.J. 12 establishment and manipulation of plant populations and communities in terrestrial systems. In Handbook of Ecological Restoration; Cambridge University: Cambridge, UK, 2008; p. 223. [Google Scholar] [CrossRef]
- Liang, S.; Yingning ZO, U.; Bo SH, U.; Qiangsheng, W.U. Arbuscular mycorrhizal fungi and endophytic fungi differentially modulate polyamines or proline of peach in response to soil flooding. Pedosphere 2024, 34, 460–472. [Google Scholar] [CrossRef]
- Sieber, T.N. Endophytic fungi in forest trees: Are they mutualists? Fungal Biol. Rev. 2007, 21, 75–89. [Google Scholar] [CrossRef]
- Mamangkey, J.; Mendes, L.W.; Harahap, A.; Briggs, D.; Kayacilar, C. Endophytic bacteria and fungi from indonesian medicinal plants with antibacterial, pathogenic antifungal and extracellular enzymes activities: A review. Int. J. Sci. Technol. Manag. 2022, 3, 245–255. [Google Scholar] [CrossRef]
- Yan, L.; Zhu, J.; Zhao, X.; Shi, J.; Jiang, C.; Shao, D. Beneficial effects of endophytic fungi colonization on plants. Appl. Microbiol. Biotechnol. 2019, 103, 3327–3340. [Google Scholar] [CrossRef] [PubMed]
- Bandara, W.M.M.S.; Seneviratne, G.; Kulasooriya, S.A. Interactions among endophytic bacteria and fungi: Effects and potentials. J. Biosci. 2006, 31, 645–650. [Google Scholar] [CrossRef]
- Rodriguez, R.J.; White, J.F., Jr.; Arnold, A.E.; Redman, R.S. Fungal endophytes: Diversity and functional roles. New Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef]
- Kariman, K.; Barker, S.J.; Tibbett, M. Structural plasticity in root-fungal symbioses: Diverse interactions lead to improved plant fitness. PeerJ 2018, 6, e6030. [Google Scholar] [CrossRef]
- Duan, X.; Xu, F.; Qin, D.; Gao, T.; Shen, W.; Zuo, S.; Yu, B.; Xu, J.; Peng, Y.; Dong, J. Diversity and bioactivities of fungal endophytes from Distylium chinense, a rare waterlogging tolerant plant endemic to the Three Gorges Reservoir. BMC Microbiol. 2019, 19, 278. [Google Scholar] [CrossRef]
- Caracciolo, A.B.; Terenzi, V. Rhizosphere microbial communities and heavy metals. Microorganisms 2021, 9, 1462. [Google Scholar] [CrossRef]
- Pang, Z.; Chen, J.; Wang, T.; Gao, C.; Li, Z.; Guo, L.; Xu, J.; Cheng, Y. Linking plant secondary metabolites and plant microbiomes: A review. Front. Plant Sci. 2021, 12, 621276. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pan, J.; Zhang, R.; Wang, J.; Tian, D.; Niu, S. Environmental factors, bacterial interactions and plant traits jointly regulate epiphytic bacterial community composition of two alpine grassland species. Sci. Total. Environ. 2022, 836, 155665. [Google Scholar] [CrossRef] [PubMed]
- Perotti, E.B.; Pidello, A. Plant-soil-microorganism interactions on nitrogen cycle: Azospirillum inoculation. In Advances in Selected Plant Physiology Aspects; IntechOpen: London, UK, 2012; pp. 189–208. [Google Scholar] [CrossRef][Green Version]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef]
- Classen, A.T.; Sundqvist, M.K.; Henning, J.A.; Newman, G.S.; Moore, J.A.M.; Cregger, M.A.; Moorhead, L.C.; Patterson, C.M. Direct and indirect effects of climate change on soil microbial and soil microbial-plant interactions: What lies ahead? Ecosphere 2015, 6, 1–21. [Google Scholar] [CrossRef]
- Bahram, M.; Hildebrand, F.; Forslund, S.K.; Anderson, J.L.; Soudzilovskaia, N.A.; Bodegom, P.M.; Bengtsson-Palme, J.; Anslan, S.; Coelho, L.P.; Harend, H.; et al. Structure and function of the global topsoil microbiome. Nature 2018, 560, 233–237. [Google Scholar] [CrossRef]
- Glynou, K.; Ali, T.; Buch, A.; Kia, S.H.; Ploch, S.; Xia, X.; Çelik, A.; Thines, M.; Maciá-Vicente, J.G. The local environment determines the assembly of root endophytic fungi at a continental scale. Environ. Microbiol. 2016, 18, 2418–2434. [Google Scholar] [CrossRef]
- Johnson, D.; Martin, F.; Cairney, J.W.G.; Anderson, I.C. The importance of individuals: Intraspecific diversity of mycorrhizal plants and fungi in ecosystems. New Phytol. 2012, 194, 614–628. [Google Scholar] [CrossRef]
- Bahram, M.; Põlme, S.; Kõljalg, U.; Zarre, S.; Tedersoo, L. Regional and local patterns of ectomycorrhizal fungal diversity and community structure along an altitudinal gradient in the Hyrcanian forests of northern Iran. New Phytol. 2012, 193, 465–473. [Google Scholar] [CrossRef]
- Li, X.; Gai, J.; Cai, X.; Christie, P.; Zhang, F.; Zhang, J. Molecular diversity of arbuscular mycorrhizal fungi associated with two co-occurring perennial plant species on a Tibetan altitudinal gradient. Mycorrhiza 2014, 24, 95–107. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef]
- Guo, X.; Gong, J. Differential effects of abiotic factors and host plant traits on diversity and community composition of root-colonizing arbuscular mycorrhizal fungi in a salt-stressed ecosystem. Mycorrhiza 2014, 24, 79–94. [Google Scholar] [CrossRef] [PubMed]
- He, C.Y.; Zhang, G.Y.; Zhang, J.G.; Duan, A.G.; Luo, H.M. Physiological, biochemical, and proteome profiling reveals key pathways underlying the drought stress responses of Hippophae rhamnoides. Proteomics 2016, 16, 2688–2697. [Google Scholar] [CrossRef] [PubMed]
- Dagar, J.C.; Tewari, V.P. Evolution of agroforestry as a modern science. In Agroforestry: Anecdotal to Modern Science; Springer: Berlin/Heidelberg, Germany, 2017; pp. 13–90. [Google Scholar] [CrossRef]
- Padulosi, S.; Thompson, J.; Rudebjer, P.G. Fighting Poverty, Hunger and Malnutrition with Neglected and Underuti-Lized Species: Needs, Challenges and the Way Forward; Bioversity International: Rome, Italy, 2013. [Google Scholar]
- Yin, R.; Zhao, M. Ecological restoration programs and payments for ecosystem services as integrated biophysical and socioeconomic processes—China’s experience as an example. Ecol. Econ. 2012, 73, 56–65. [Google Scholar] [CrossRef]
- Zhou, X.; Tian, L.; Zhang, J.; Ma, L.; Li, X.; Tian, C. Rhizospheric fungi and their link with the nitrogen-fixing Frankia harbored in host plant Hippophae rhamnoides L. J. Basic Microbiol. 2017, 57, 1055–1064. [Google Scholar] [CrossRef]
- Zhang, J.; Nasir, F.; Tian, L.; Bahadur, A.; Batool, A.; Ma, L.; Zhou, X.; Zhao, S.; Tian, C. Impact of ecological factors on the diversity and community assemblage of the bacteria harbored in the rhizosphere of Hippophae rhamnoides. Int. J. Agric. Biol. 2018, 20, 1632–1640. [Google Scholar]
- Luo, Y.J.; Sun, H.M.; He, N.; Yuan, L.J.; Xie, Y.Y. Isolation and antibacterial activity of actinomycetes from the nodules and rhizosphere soil of Hippophae rhamnoides in tibet. Biotechnol. Bull. 2021, 37, 225. [Google Scholar] [CrossRef]
- Wang, Z.; Zou, J.; Shi, Y.; Zhang, X.; Zhai, B.; Guo, D.; Sun, J.; Luan, F. Extraction techniques, structural features and biological functions of Hippophae rhamnoides polysaccharides: A review. Int. J. Biol. Macromol. 2024, 263, 130206. [Google Scholar] [CrossRef]
- Ma, Q.-G.; He, N.-X.; Huang, H.-L.; Fu, X.-M.; Zhang, Z.-L.; Shu, J.-C.; Wang, Q.-Y.; Chen, J.; Wu, G.; Zhu, M.-N.; et al. Hippophae rhamnoides L.: A comprehensive review on the botany, traditional uses, phytonutrients, health benefits, quality markers, and applications. J. Agric. Food Chem. 2023, 71, 4769–4788. [Google Scholar] [CrossRef]
- Krejcarová, J.; Straková, E.; Suchý, P.; Herzig, I.; Karásková, K. Sea buckthorn (Hippophae rhamnoides L.) as a potential source of nutraceutics and its therapeutic possibilities—A review. Acta Veter. Brno 2015, 84, 257–268. [Google Scholar] [CrossRef]
- Zapala, M.A.; Schork, N.J. Multivariate regression analysis of distance matrices for testing associations between gene expression patterns and related variables. Proc. Natl. Acad. Sci. USA 2006, 103, 19430–19435. [Google Scholar] [CrossRef] [PubMed]
- Algina, J.; Keselman, H.J. Comparing squared multiple correlation coefficients: Examination of a confidence interval and a test significance. Psychol. Methods 1999, 4, 76–83. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Bever, J.D.; Platt, T.G.; Morton, E.R. Microbial population and community dynamics on plant roots and their feedbacks on plant communities. Annu. Rev. Microbiol. 2012, 66, 265–283. [Google Scholar] [CrossRef] [PubMed]
- Hartman, K.; Tringe, S.G. Interactions between plants and soil shaping the root microbiome under abiotic stress. Biochem. J. 2019, 476, 2705–2724. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Batista, B.D.; Bazany, K.E.; Singh, B.K. Plant–microbiome interactions under a changing world: Responses, consequences and perspectives. New Phytol. 2022, 234, 1951–1959. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.; Law, A.D.; Sahib, M.R.; Pervaiz, Z.H.; Zhang, Q. Impact of root system architecture on rhizosphere and root microbiome. Rhizosphere 2018, 6, 47–51. [Google Scholar] [CrossRef]
- Martin, F.M.; Perotto, S.; Bonfante, P. Mycorrhizal fungi: A fungal community at the interface between soil and roots. In The Rhizosphere; CRC Press: Boca Raton, FL, USA, 2000; pp. 279–312. [Google Scholar]
- Maestre, F.T.; Benito, B.M.; Berdugo, M.; Concostrina-Zubiri, L.; Delgado-Baquerizo, M.; Eldridge, D.J.; Guirado, E.; Gross, N.; Kéfi, S.; Le Bagousse-Pinguet, Y.; et al. Biogeography of global drylands. New Phytol. 2021, 231, 540–558. [Google Scholar] [CrossRef]
- Hernández-Cáceres, D.; Stokes, A.; Angeles-Alvarez, G.; Abadie, J.; Anthelme, F.; Bounous, M.; Freschet, G.T.; Roumet, C.; Weemstra, M.; Merino-Martín, L.; et al. Vegetation creates microenvironments that influence soil microbial activity and functional diversity along an elevation gradient. Soil Biol. Biochem. 2022, 165, 108485. [Google Scholar] [CrossRef]
- Gray, S.B.; Brady, S.M. Plant developmental responses to climate change. Dev. Biol. 2016, 419, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P.; St. Clair, S.B. Mineral stress: The missing link in understanding how global climate change will affect plants in real world soils. Field Crop. Res. 2004, 90, 101–115. [Google Scholar] [CrossRef]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Liu, B.; Hu, Y.; Wang, Y.; Xue, H.; Li, Z.; Li, M. Effects of saline-alkali stress on bacterial and fungal community diversity in Leymus chinensis rhizosphere soil. Environ. Sci. Pollut. Res. 2022, 29, 70000–70013. [Google Scholar] [CrossRef]
- Xu, X.; Chen, C.; Zhang, Z.; Sun, Z.; Chen, Y.; Jiang, J.; Shen, Z. The influence of environmental factors on communities of arbuscular mycorrhizal fungi associated with Chenopodium ambrosioides revealed by MiSeq sequencing investigation. Sci. Rep. 2017, 7, srep45134. [Google Scholar] [CrossRef]
- Bai, X.; Zhang, E.; Wu, J.; Ma, D.; Zhang, C.; Zhang, B.; Liu, Y.; Zhang, Z.; Tian, F.; Zhao, H.; et al. Soil fungal community is more sensitive than bacterial community to modified materials application in saline–alkali land of Hetao Plain. Front. Microbiol. 2024, 15, 1255536. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wei, Z.; Yang, T.; Zhao, X.; Dang, Q.; Chen, X.; Wu, J.; Zhao, Y. Core microorganisms promote the transformation of DOM fractions with different molecular weights to improve the stability during composting. Bioresour. Technol. 2020, 299, 122575. [Google Scholar] [CrossRef] [PubMed]

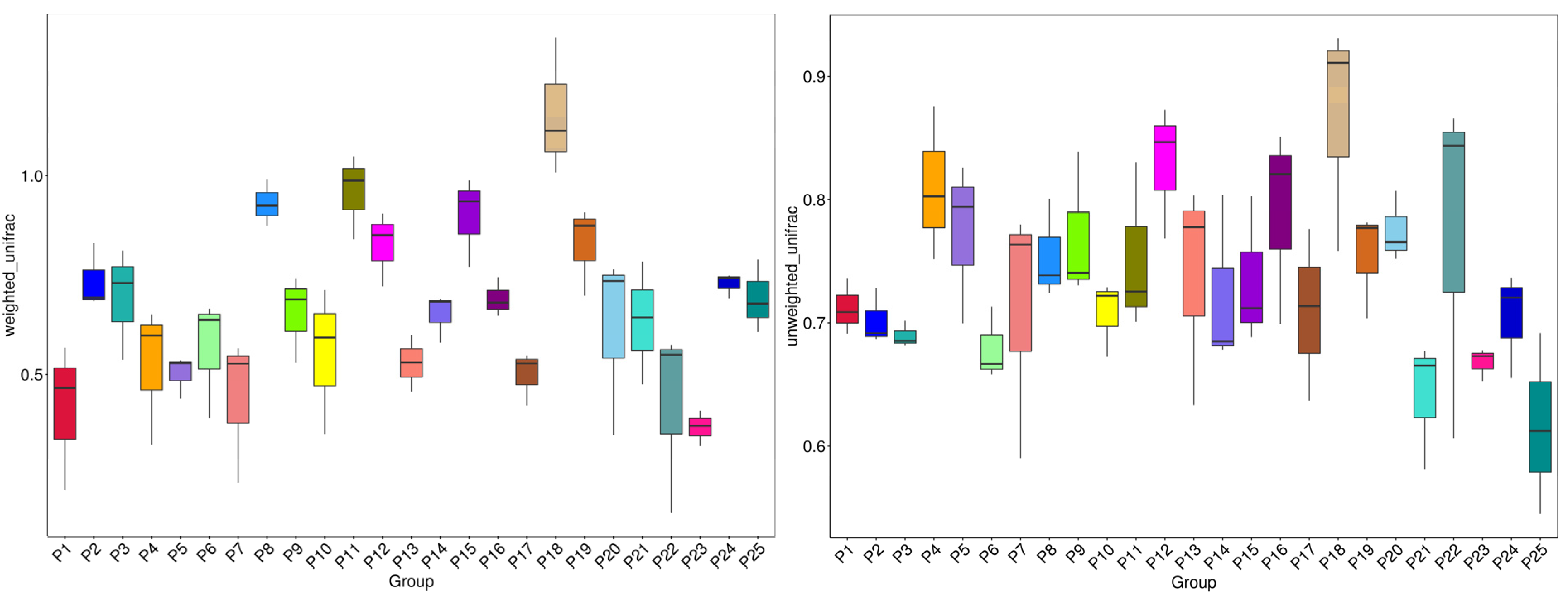
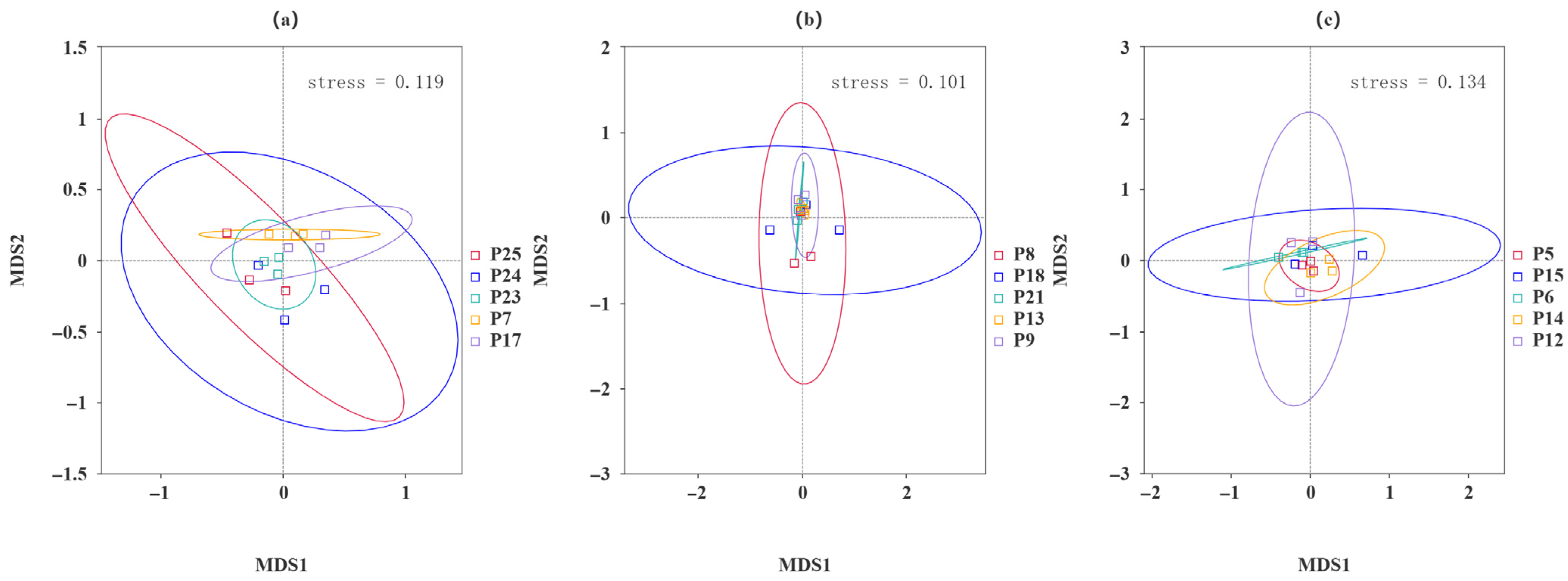
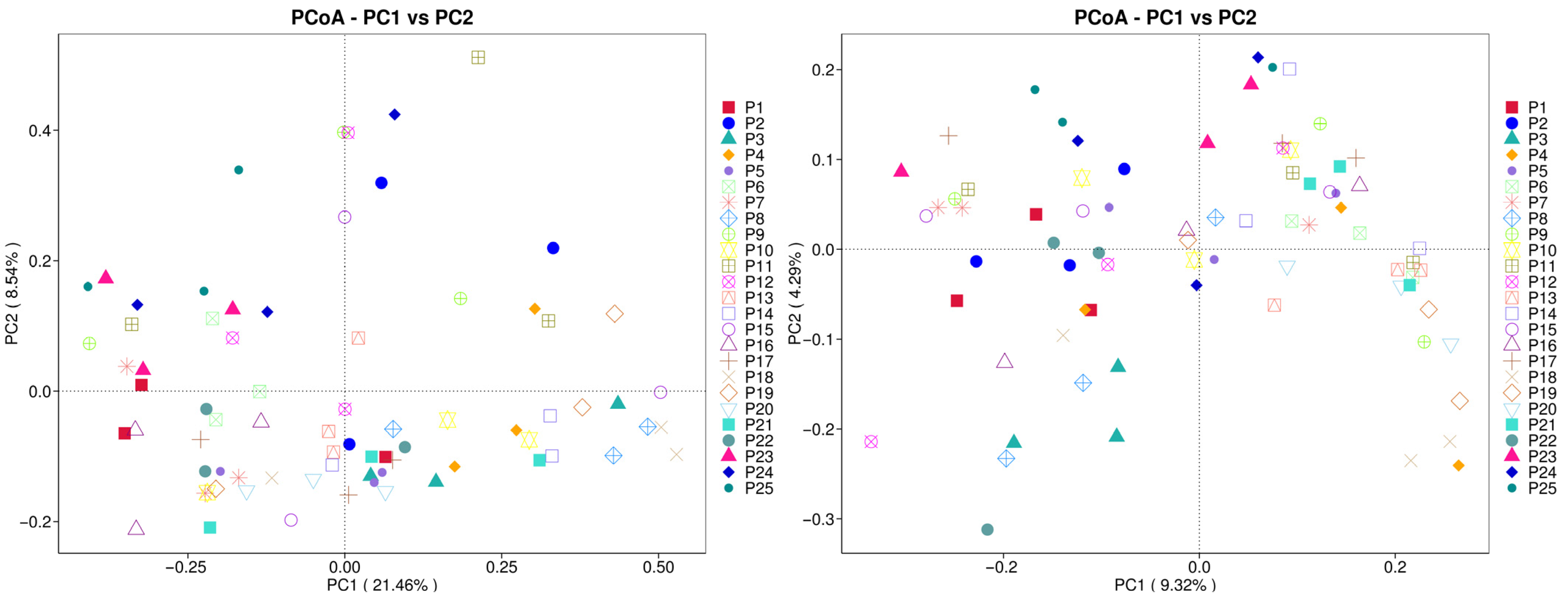
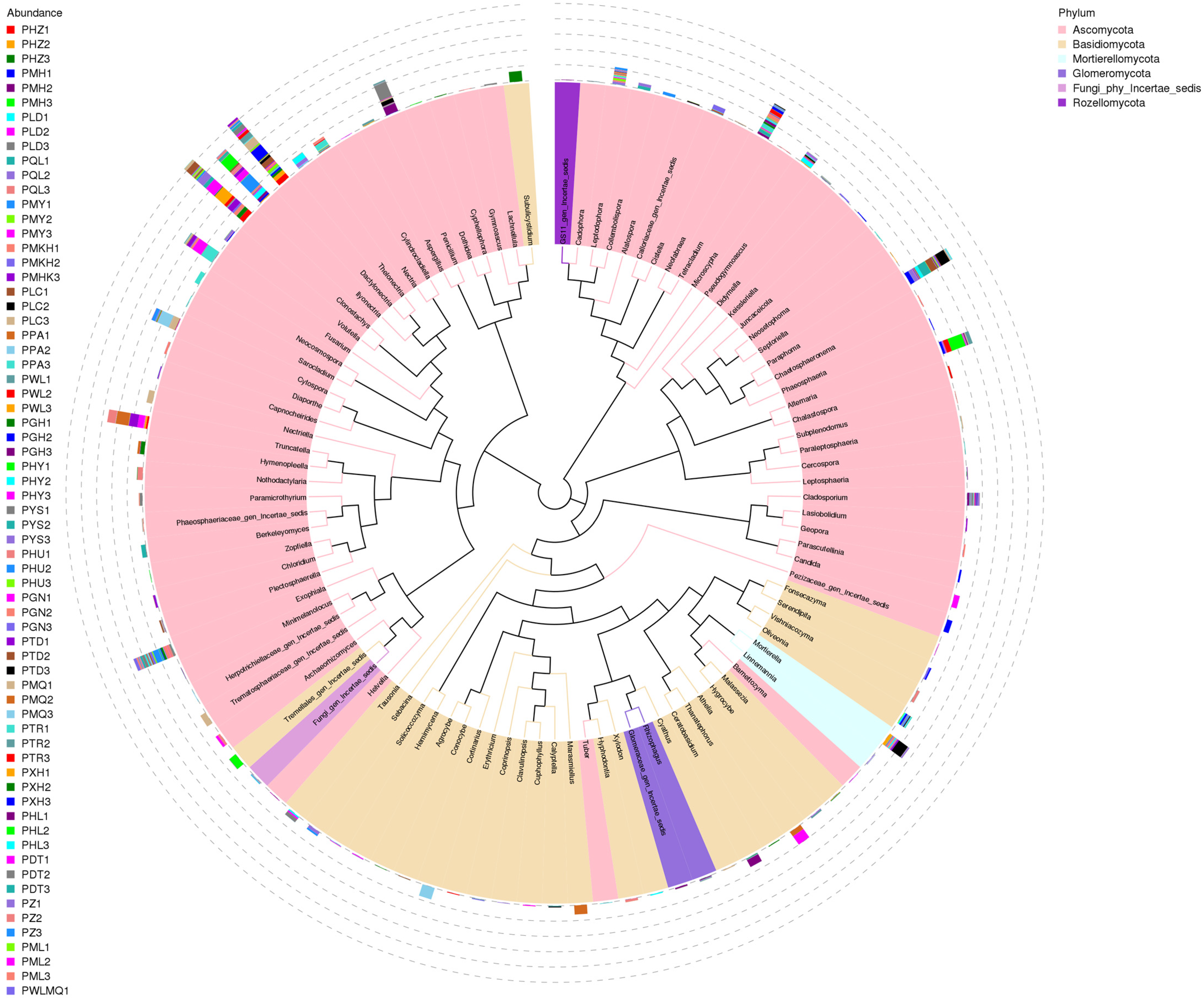




| Sample | pH | Moisture | Salinity | TNg/kg | TKg/kg | HNmg/kg | TPg/kg | AKmg/kg | OMg/kg | APmg/kg |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 7.600 | 14.519 | 3.270 | 3.050 | 15.489 | 225.554 | 0.753 | 39.065 | 56.772 | 6.735 |
| P2 | 8.070 | 11.093 | 4.327 | 1.787 | 19.045 | 106.049 | 0.636 | 67.980 | 26.704 | 2.097 |
| P3 | 7.530 | 32.745 | 1.613 | 5.972 | 20.600 | 413.271 | 0.991 | 157.386 | 95.352 | 2.847 |
| P4 | 7.990 | 7.209 | 2.433 | 2.550 | 18.337 | 181.034 | 0.635 | 47.380 | 41.769 | 1.204 |
| P5 | 8.080 | 18.195 | 4.180 | 1.321 | 19.339 | 89.088 | 0.516 | 21.388 | 22.782 | 0.929 |
| P6 | 7.650 | 23.879 | 1.067 | 2.612 | 17.951 | 198.994 | 0.586 | 21.164 | 42.655 | 3.069 |
| P7 | 7.920 | 10.702 | 4.637 | 2.634 | 19.223 | 158.822 | 0.721 | 89.629 | 44.497 | 2.832 |
| P8 | 6.850 | 17.726 | 1.457 | 2.645 | 13.982 | 199.854 | 1.464 | 19.008 | 44.269 | 0.795 |
| P9 | 7.920 | 4.222 | 5.257 | 0.903 | 18.325 | 46.434 | 0.560 | 87.477 | 9.041 | 3.696 |
| P10 | 6.850 | 17.632 | 0.843 | 1.649 | 17.965 | 98.343 | 0.591 | 215.698 | 22.729 | 1.254 |
| P11 | 8.390 | 16.960 | 0.910 | 2.140 | 16.081 | 157.388 | 0.747 | 85.091 | 38.031 | 7.981 |
| P12 | 8.080 | 3.721 | 5.050 | 1.832 | 18.272 | 127.150 | 0.532 | 90.198 | 27.458 | 6.109 |
| P13 | 8.010 | 19.759 | 0.967 | 1.913 | 18.256 | 124.927 | 0.615 | 28.365 | 31.179 | 2.090 |
| P14 | 8.360 | 7.813 | 0.883 | 0.476 | 18.374 | 21.179 | 0.366 | 18.443 | 3.652 | 0.511 |
| P15 | 8.720 | 7.616 | 5.940 | 0.613 | 20.679 | 18.010 | 0.551 | 36.399 | 3.604 | 0.636 |
| P16 | 7.750 | 22.009 | 1.943 | 5.187 | 20.630 | 373.019 | 0.571 | 33.415 | 3.863 | 3.306 |
| P17 | 8.050 | 7.439 | 3.607 | 2.222 | 17.206 | 124.072 | 0.676 | 139.983 | 40.595 | 2.699 |
| P18 | 8.280 | 11.779 | 0.797 | 0.853 | 18.201 | 44.853 | 0.613 | 54.288 | 32.450 | 17.454 |
| P19 | 7.970 | 12.272 | 1.780 | 1.981 | 17.660 | 136.579 | 0.679 | 152.801 | 9.588 | 3.972 |
| P20 | 7.480 | 12.288 | 1.750 | 3.092 | 19.510 | 213.332 | 0.514 | 66.059 | 30.930 | 3.160 |
| P21 | 6.460 | 28.396 | 0.697 | 5.003 | 18.552 | 397.373 | 1.167 | 42.630 | 98.584 | 3.389 |
| P22 | 7.840 | 12.830 | 1.570 | 2.536 | 19.443 | 158.571 | 0.923 | 135.010 | 43.617 | 1.182 |
| P23 | 7.890 | 7.220 | 1.480 | 3.299 | 19.760 | 232.105 | 0.793 | 153.395 | 49.156 | 7.157 |
| P24 | 8.310 | 8.249 | 1.213 | 0.770 | 18.392 | 29.043 | 1.252 | 144.198 | 9.455 | 5.728 |
| P25 | 7.950 | 6.948 | 1.247 | 1.124 | 18.582 | 70.907 | 0.608 | 84.829 | 16.131 | 0.365 |
| Sample | Phylum | Class | Order | Family | Genus | Species | ASV |
|---|---|---|---|---|---|---|---|
| P1 | 6 | 13 | 25 | 39 | 52 | 53 | 216 |
| P2 | 5 | 13 | 29 | 51 | 65 | 65 | 126 |
| P3 | 6 | 18 | 32 | 58 | 73 | 78 | 197 |
| P4 | 6 | 21 | 41 | 73 | 91 | 84 | 250 |
| P5 | 5 | 17 | 31 | 48 | 57 | 58 | 146 |
| P6 | 8 | 18 | 40 | 62 | 84 | 91 | 271 |
| P7 | 6 | 13 | 28 | 45 | 53 | 54 | 120 |
| P8 | 6 | 17 | 30 | 37 | 54 | 51 | 132 |
| P9 | 6 | 15 | 33 | 57 | 82 | 83 | 200 |
| P10 | 5 | 11 | 24 | 37 | 57 | 62 | 196 |
| P11 | 8 | 17 | 36 | 61 | 86 | 91 | 236 |
| P12 | 7 | 13 | 27 | 37 | 46 | 51 | 116 |
| P13 | 7 | 19 | 39 | 62 | 100 | 101 | 266 |
| P14 | 5 | 15 | 34 | 51 | 67 | 70 | 202 |
| P15 | 4 | 12 | 26 | 44 | 58 | 55 | 154 |
| P16 | 6 | 17 | 37 | 64 | 96 | 88 | 190 |
| P17 | 5 | 12 | 28 | 54 | 68 | 63 | 159 |
| P18 | 7 | 17 | 36 | 56 | 70 | 68 | 256 |
| P19 | 8 | 21 | 45 | 66 | 88 | 93 | 281 |
| P20 | 8 | 19 | 39 | 57 | 70 | 72 | 210 |
| P21 | 8 | 19 | 43 | 74 | 104 | 99 | 257 |
| P22 | 3 | 12 | 26 | 35 | 47 | 47 | 95 |
| P23 | 7 | 13 | 30 | 49 | 53 | 54 | 149 |
| P24 | 6 | 15 | 29 | 46 | 59 | 61 | 157 |
| P25 | 5 | 10 | 26 | 33 | 44 | 47 | 132 |
| Sample | Chao1 | Dominance | Goods_Coverage | Observed_Features | Pielou_e | Shannon | Simpson |
|---|---|---|---|---|---|---|---|
| P1 | 232.167 | 0.1 | 1 | 217 | 0.561 | 4.357 | 0.9 |
| P2 | 134.5 | 0.096 | 1 | 127 | 0.551 | 3.85 | 0.904 |
| P3 | 199.2 | 0.07 | 1 | 198 | 0.638 | 4.868 | 0.93 |
| P4 | 266.545 | 0.051 | 1 | 251 | 0.63 | 5.021 | 0.949 |
| P5 | 156.545 | 0.253 | 1 | 147 | 0.416 | 2.997 | 0.747 |
| P6 | 280.462 | 0.111 | 1 | 270 | 0.528 | 4.262 | 0.889 |
| P7 | 125.5 | 0.191 | 1 | 121 | 0.438 | 3.031 | 0.809 |
| P8 | 149.5 | 0.121 | 1 | 133 | 0.548 | 3.866 | 0.879 |
| P9 | 206.077 | 0.093 | 1 | 201 | 0.58 | 4.438 | 0.907 |
| P10 | 199.1 | 0.158 | 1 | 197 | 0.519 | 3.953 | 0.842 |
| P11 | 242.111 | 0.16 | 1 | 236 | 0.503 | 3.968 | 0.84 |
| P12 | 123.273 | 0.187 | 1 | 115 | 0.441 | 3.018 | 0.813 |
| P13 | 272.091 | 0.065 | 1 | 265 | 0.619 | 4.984 | 0.935 |
| P14 | 204.6 | 0.077 | 1 | 201 | 0.614 | 4.696 | 0.923 |
| P15 | 153.083 | 0.11 | 1 | 153 | 0.585 | 4.247 | 0.89 |
| P16 | 200.4 | 0.197 | 1 | 189 | 0.431 | 3.257 | 0.803 |
| P17 | 161 | 0.134 | 1 | 158 | 0.534 | 3.901 | 0.866 |
| P18 | 256.5 | 0.117 | 1 | 255 | 0.548 | 4.379 | 0.883 |
| P19 | 285.833 | 0.118 | 1 | 280 | 0.536 | 4.354 | 0.882 |
| P20 | 212.273 | 0.077 | 1 | 209 | 0.592 | 4.567 | 0.923 |
| P21 | 274.4 | 0.062 | 1 | 256 | 0.614 | 4.914 | 0.938 |
| P22 | 95.667 | 0.238 | 1 | 94 | 0.432 | 2.831 | 0.762 |
| P23 | 150.545 | 0.147 | 1 | 148 | 0.522 | 3.763 | 0.853 |
| P24 | 165.067 | 0.186 | 1 | 156 | 0.473 | 3.444 | 0.814 |
| P25 | 132.5 | 0.135 | 1 | 131 | 0.544 | 3.824 | 0.865 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Ye, G.; Liu, W.; Liu, R.; Liu, Z.; Ma, Y. Environmental Factors Drive the Biogeographic Pattern of Hippophae rhamnoides Root Endophytic Fungal Diversity in the Arid Regions of Northwest China. J. Fungi 2024, 10, 679. https://doi.org/10.3390/jof10100679
Guo S, Ye G, Liu W, Liu R, Liu Z, Ma Y. Environmental Factors Drive the Biogeographic Pattern of Hippophae rhamnoides Root Endophytic Fungal Diversity in the Arid Regions of Northwest China. Journal of Fungi. 2024; 10(10):679. https://doi.org/10.3390/jof10100679
Chicago/Turabian StyleGuo, Siyu, Guisheng Ye, Wenjie Liu, Ruoqi Liu, Zhehao Liu, and Yuhua Ma. 2024. "Environmental Factors Drive the Biogeographic Pattern of Hippophae rhamnoides Root Endophytic Fungal Diversity in the Arid Regions of Northwest China" Journal of Fungi 10, no. 10: 679. https://doi.org/10.3390/jof10100679
APA StyleGuo, S., Ye, G., Liu, W., Liu, R., Liu, Z., & Ma, Y. (2024). Environmental Factors Drive the Biogeographic Pattern of Hippophae rhamnoides Root Endophytic Fungal Diversity in the Arid Regions of Northwest China. Journal of Fungi, 10(10), 679. https://doi.org/10.3390/jof10100679






