Genetic and Pathogenic Variability among Isolates of Sporisorium reilianum Causing Sorghum Head Smut
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Esele, J.P. Sorghum and pearl millet diseases in the horn of Africa. In Sorghum and Millets Diseases; Leslie, J.F., Ed.; Iowa State Press, A Blackwell Publishing Company: Ames, IA, USA, 2002; pp. 383–387. ISBN 978-0-8138-0389-0. [Google Scholar]
- Marley, P.S.; Diourté, M.; Neya, A.; Nutsugah, S.K.; Sérémé, P.; Katilé, S.; Hess, D.E.; Mbaye, D.F.; Ngoko, Z. Sorghum and pearl millet diseases in West and Central Africa. In Sorghum and Millets Diseases; Leslie, J.F., Ed.; Iowa State Press, A Blackwell Publishing Company: Ames, IA, USA, 2002; pp. 419–425. ISBN 978-0-8138-0389-0. [Google Scholar]
- Ngugi, H.K.; King, S.B.; Abayo, G.O.; Reddy, Y.V.R. Prevalence, incidence, and severity of sorghum diseases in Western Kenya. Plant Dis. 2002, 86, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Prom, L.K.; Perumal, R.; Erattaimuthu, S.R.; Erpelding, J.E.; Montes, N.; Odvody, G.N.; Greenwald, C.; Jin, Z.; Frederiksen, R.; Magill, C.W. Virulence and molecular genotyping studies of Sporisorium reilianum isolates in sorghum. Plant Dis. 2011, 95, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ping, J.; Du, Z.; Cheng, Q.; Huang, Y. Identification of a new race of Sporisorium reilianum and characterization of the reaction of sorghum lines to four races of the head smut pathogen. J. Phytopathol. 2011, 159, 342–346. [Google Scholar] [CrossRef]
- Naidoo, G.; Torres-Montalvo, H. Genetic variability among and within host specialized isolates of Sporisorium reilianum. In Sorghum and Millets Diseases; Leslie, J.F., Ed.; Iowa State Press, A Blackwell Publishing Company: Ames, IA, USA, 2002; pp. 221–225. ISBN 978-0-8138-0389-0. [Google Scholar]
- Poloni, A.; Schirawski, J. Host Specificity in Sporisorium reilianum is determined by distinct mechanisms in Maize and Sorghum. Mol. Plant Pathol. 2016, 17, 741–754. [Google Scholar] [CrossRef]
- Dittiger, L.D.; Chaudhary, S.; Furch, A.C.U.; Mithöfer, A.; Schirawski, J. Plant Responses of Maize to Two formae speciales of Sporisorium reilianum Support Recent Fungal Host Jump. Int. J. Mol. Sci. 2023, 24, 15604. [Google Scholar] [CrossRef]
- Zuther, K.; Kahnt, J.; Utermark, J.; Imkampe, J.; Uhse, S.; Schirawski, J. Host specificity of Sporisorium reilianum is tightly linked to generation of the phytoalexin luteolinidin by Sorghum bicolor. Mol. Plant Microbe Interact. 2012, 25, 1230–1237. [Google Scholar] [CrossRef]
- Rauwane, M.E.; Ogugua, U.V.; Kalu, C.M.; Ledwaba, L.K.; Woldesemayat, A.A.; Ntushelo, K. Pathogenicity and Virulence Factors of Fusarium graminearum Including Factors Discovered Using Next Generation Sequencing Technologies and Proteomics. Microorganisms 2020, 8, 305. [Google Scholar] [CrossRef]
- Frederiksen, R.A. Head smuts of corn and sorghum. In Proceedings of the 32nd Corn and Sorghum Research Conference, Chicago, IL, USA, 6–8 December 1977; pp. 89–105. [Google Scholar]
- Bai, C.; Liu, Y.; Lu, X.; Tao, C. Progress in sorghum head smut research. In Proceedings of the International Conference on Civil, Structure, Environmental Engineering (13CSEE 2016), Guangzhou, China, 12–13 March 2016; pp. 317–320. [Google Scholar]
- Wilson, J.M.; Frederiksen, R.A. Histopathology of the interaction of Sorghum bicolor and Sphacelotheca reiliana. Phytopathology 1970, 60, 828–832. [Google Scholar] [CrossRef]
- Prom, L.K.; Montes-Garcia, N.; Odvody, G.N. Influence of Planting Depths on the Incidence of Sorghum Head Smut, caused by Sporisorium reilianum. Trop. Subtrop. Agroecosystems 2014, 17, 33–38. [Google Scholar]
- Torres-Montalvo, H.; McDonald, B.A.; Magill, C.W.; Frederiksen, R.A. Structure of Sporisorium reilianum populations from Mexico, USA, and Niger. Int. Sorghum Millets Newsl. 1998, 39, 110–113. [Google Scholar]
- Prom, L.K.; Perumal, R.; Isakeit, T.; Erattaimuthu, S.; Magill, C. Response of sorghum accessions against newly documented pathotypes 5 and 6 of head smut pathogen, Sporisorium reilianum. Am. J. Plant Sci. 2021, 12, 432–443. [Google Scholar] [CrossRef]
- Indira, S.; Xu, X.; Iamsupasit, N.; Shetty, H.S.; Vasanthi, N.S.; Singh, S.D.; Bandyopadhyay, R. Diseases of sorghum and pearl millet in Asia. In Sorghum and Millets Diseases; Leslie, J.F., Ed.; Iowa State Press, A Blackwell Publishing Company: Ames, IA, USA, 2002; pp. 393–402. ISBN 978-0-8138-0389-0. [Google Scholar]
- Frowd, J.A. A World Review of Sorghum Smut. In Sorghum Diseases, A World Review, Proceedings of the International Workshop on Sorghum Diseases, Hyderabad, India, 11–15 December 1978; Bengtson, G.D., Ed.; ICRISAT: Patancheruvu, India, 1980; pp. 331–348. [Google Scholar]
- Frederiksen, R.A. Head Smut. In Compendium of Sorghum Diseases; Frederiksen, R.A., Odvody, G.N., Eds.; The American Phytopathological Society: St. Paul, MN, USA, 2000; pp. 18–20. [Google Scholar]
- Pecina-Quintero, V.; Williams-Alanís, H.; Montes-García, N.; Rodríguez-Herrera, R.; Rosales-Robles, E.; Vidal-Martínez, V.A. Incidence of head smut Sporisorium reilianum (Kühn) Langdon and Fullerton in sorghum [Sorghum bicolor (L.) Moench.] hybrids with A1 and A2 cytoplasms. Rev. Mex. Fitopatol. 2004, 22, 315–319. [Google Scholar]
- Torres-Montalvo, J.H. The Population Genetics of Sporisorium reilianum, the Head Smut Pathogen of Sorghum and Maize. Ph.D. Thesis, Texas A&M University, College Station, TX, USA, 1998; p. 90. [Google Scholar]
- Sánchez Maya, H.E.; Mercado-Flores, Y.; Téllez-Jurado, A.; Pérez-Camarillo, J.P.; Mejía, O.; Anducho-Reyes, M.A. Molecular Variation of the Phytopathogenic Fungus Sporisorium reilianum in Valle del Mezquital, Hidalgo. Front. Ecol. Evol. 2020, 8, 36. [Google Scholar] [CrossRef]
- Frederiksen, R.A.; Rosenow, D.T.; Reyes, L. Races of Sphacelotheca reiliana on sorghum in Texas. Plant Dis. Rep. 1975, 59, 549–551. [Google Scholar]
- Dodman, R.L.; Obst, N.R.; Henzell, R.G. Races of sorghum head smut (Sporisorium reilianum) in South-East Queensland. Australas. Plant Pathol. 1985, 14, 45. [Google Scholar] [CrossRef]
- Sajnani, M.R.; Bhatt, V.D.; Joshi, C.G. Comparing SNPS Identification by CLC and Seqman from transcriptome sequencing data. Intern. J. Comput. Biol. 2013, 4, 56–60. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- FAO. How to Feed the World in 2050. Available online: https://www.fao.org/fileadmin/templates/wsfs/docs/expert_paper/How_to_Feed_the_World_in_2050.pdf (accessed on 27 March 2023).
- Kangama, C.O. Importance of Sorghum bicolor in African’s cultures. J. Agric. Environ. Sci. 2017, 6, 134–137. [Google Scholar] [CrossRef][Green Version]
- Pereira, L.M.; Hawkes, C. Leveraging the potential of sorghum as a healthy food and resilient crop in the South African food system. Front. Sustain. Food Syst. 2022, 6, 786151. [Google Scholar] [CrossRef]
- Radwan, G.L.; Prom, L.K.; Odvody, G.; Magill, C. Mating type a locus alleles and genomic polymorphism in Sporisorium reilianum: Comparison of sorghum isolates to those from maize. Australas. Plant Pathol. 2019, 48, 119–129. [Google Scholar] [CrossRef]
- Drees, K.P.; Lorch, J.M.; Puechmaille, S.J.; Parise, K.L.; Wibbelt, G.; Hoyt, J.R.; Sun, K.; Jargalsaikhan, A.; Dalannast, M.; Palmer, J.M.; et al. Phylogenetics of a fungal invasion: Origins and widespread dispersal of white-nose syndrome. mBio 2017, 8, e01941-17. [Google Scholar] [CrossRef] [PubMed]
- Derbyshire, M.C.; Denton-Giles, M.; Hane, J.K.; Chang, S.; Mousavi-Derazmahalleh, M.; Raffaele, S.; Buchwaldt, L.; Kamphuis, L.G. A whole genome scan of SNP data suggests a lack of abundant hard selective sweeps in the genome of the broad host range plant pathogenic fungus Sclerotinia sclerotiorum. PLoS ONE 2019, 14, e0214201. [Google Scholar] [CrossRef]
- Kulik, T.; Molcan, T.; Fiedorowicz, G.; van Diepeningen, A.; Stakheev, A.; Treder, K.; Olszewski, J.; Bilska, K.; Beyer, M.; Pasquali, M.; et al. Whole-genome single nucleotide polymorphism analysis for typing the pandemic pathogen Fusarium graminearum sensu stricto. Front. Microbiol. 2022, 13, 885978. [Google Scholar] [CrossRef]

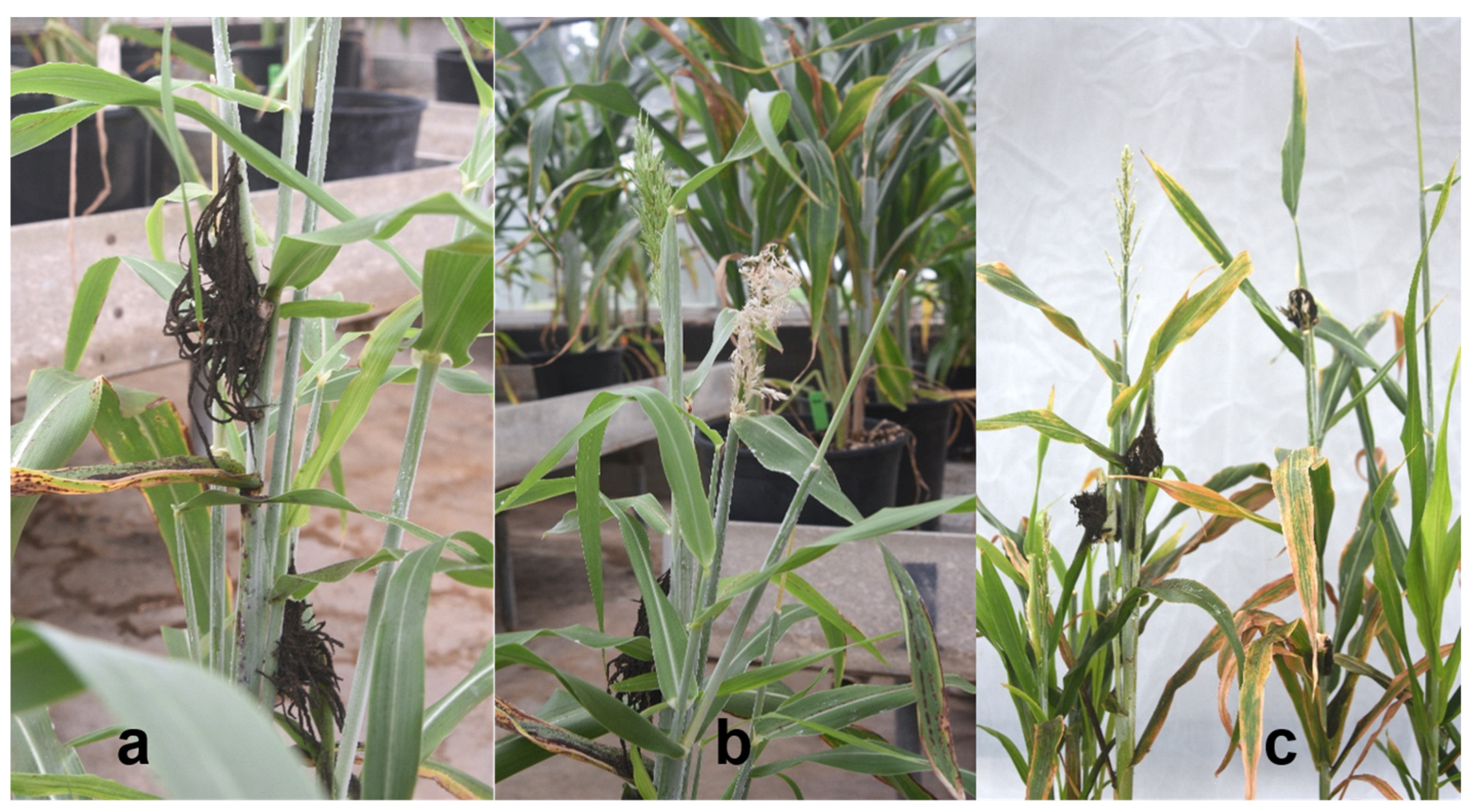
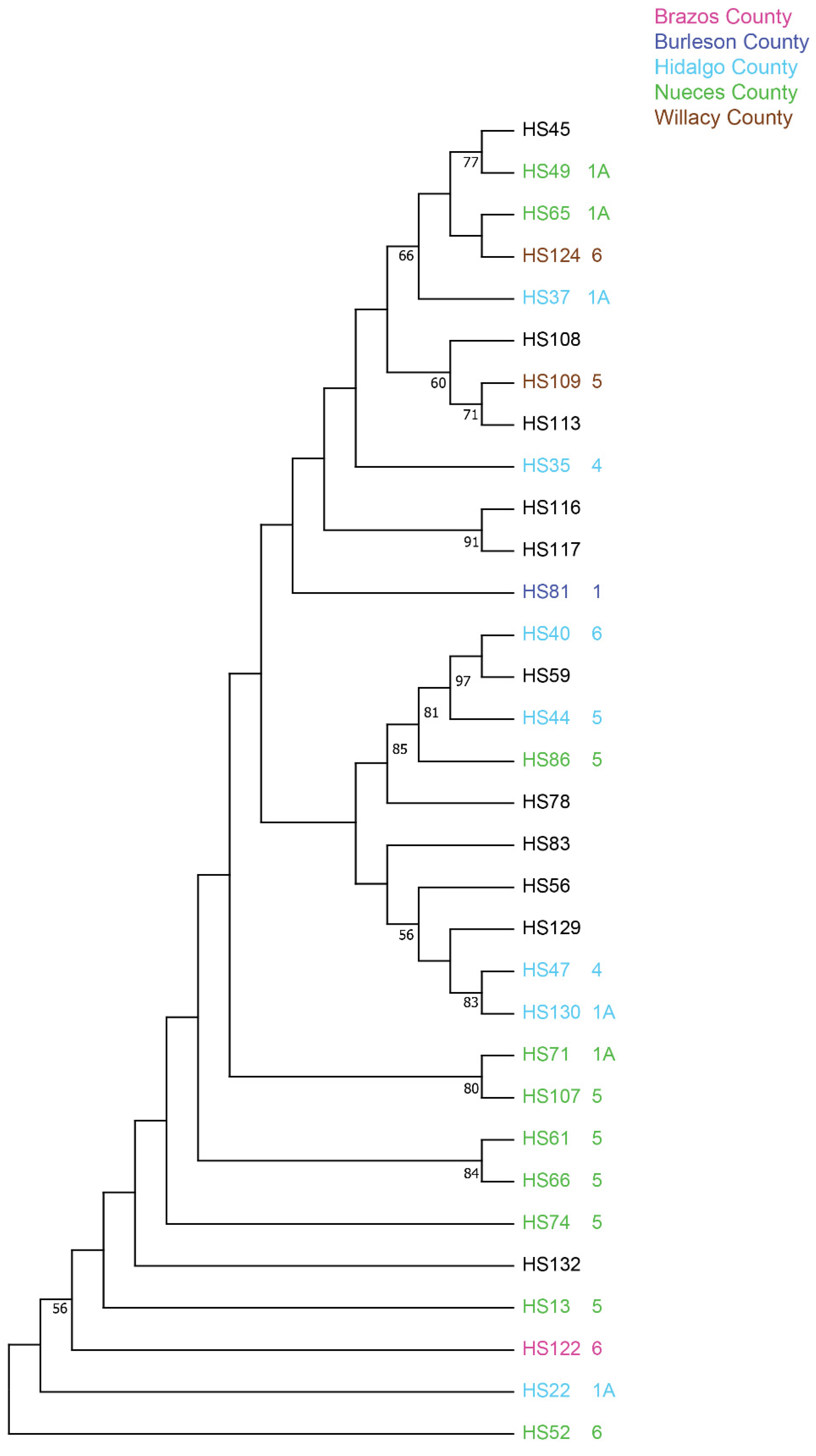
| Texas—County | Isolate | TX7078 | SA281 (Early Hegari) | TX414 | SC170-6-17 (TAM2571) | Btx635 | BTx643 | Pathotype |
|---|---|---|---|---|---|---|---|---|
| Nueces | HS13 | S | R | R | S | R | S | 5 |
| Burleson | HS22 | R | R | R | S | R | S | 1A |
| Hidalgo | HS35 | S | R | S | S | R | S | 4 |
| Hidalgo | HS37 | R | R | R | S | R | S | 1A |
| Hidalgo | HS40 | R | R | S | S | R | S | 6 |
| Hidalgo | HS44 | S | R | R | S | R | S | 5 |
| Hidalgo | HS47 | S | R | S | S | R | S | 4 |
| Nueces | HS49 | R | R | R | S | R | S | 1A |
| Nueces | HS52 | R | R | S | S | R | S | 6 |
| Nueces | HS61 | S | R | R | S | R | S | 5 |
| Nueces | HS65 | R | R | R | S | R | S | 1A |
| Nueces | HS66 | S | R | R | S | R | S | 5 |
| Nueces | HS71 | R | R | R | S | R | S | 1A |
| Nueces | HS74 | S | R | R | S | R | S | 5 |
| Burleson | HS81 | S | R | R | R | R | S | 1 |
| Nueces | HS86 | S | R | R | S | R | S | 5 |
| Nueces | HS107 | S | R | R | S | R | S | 5 |
| Willacy | HS109 | S | R | R | S | R | S | 5 |
| Brazos | HS122 | R | R | S | S | R | S | 6 |
| Willacy | HS124 | R | R | S | S | R | S | 6 |
| Hidalgo | HS130 | R | R | R | S | R | S | 1A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prom, L.K.; Ahn, E.J.S.; Perumal, R.; Isakeit, T.S.; Odvody, G.N.; Magill, C.W. Genetic and Pathogenic Variability among Isolates of Sporisorium reilianum Causing Sorghum Head Smut. J. Fungi 2024, 10, 62. https://doi.org/10.3390/jof10010062
Prom LK, Ahn EJS, Perumal R, Isakeit TS, Odvody GN, Magill CW. Genetic and Pathogenic Variability among Isolates of Sporisorium reilianum Causing Sorghum Head Smut. Journal of Fungi. 2024; 10(1):62. https://doi.org/10.3390/jof10010062
Chicago/Turabian StyleProm, Louis K., Ezekiel Jin Sung Ahn, Ramasamy Perumal, Thomas S. Isakeit, Gary N. Odvody, and Clint W. Magill. 2024. "Genetic and Pathogenic Variability among Isolates of Sporisorium reilianum Causing Sorghum Head Smut" Journal of Fungi 10, no. 1: 62. https://doi.org/10.3390/jof10010062
APA StyleProm, L. K., Ahn, E. J. S., Perumal, R., Isakeit, T. S., Odvody, G. N., & Magill, C. W. (2024). Genetic and Pathogenic Variability among Isolates of Sporisorium reilianum Causing Sorghum Head Smut. Journal of Fungi, 10(1), 62. https://doi.org/10.3390/jof10010062






