Abstract
Ganoderma is a genus of biomedical fungus that is used in the development of numerous health products throughout the world. The Lower Volta River Basin of Ghana is an undulating land surface covered by extensive vegetation and water bodies and is rich in polypore mushrooms resembling various members of the Ganoderma genus. Despite the extensive biopharmaceutical benefits of Ganoderma spp., the isolates from the Lower Volta River Basin have not been properly characterized, thus limiting their use in the development of biotechnological products. In this study, Ganoderma spp. collected from the Lower Volta River Basin were genetically analyzed using the nuclear ribosomal sequences, the internal transcribed spacer 2 (ITS 2), the complete internal transcribed spacer (ITS), and the nuclear large subunit (nLSU). Blastn search and sequence analysis revealed that the sample we coded as Ganoderma LVRB-2 belongs to G. mbrekobenum, whereas Ganoderma LVRB-1, Ganoderma LVRB-14, and Ganoderma LVRB-16 belong to the species G. enigmaticum. Our analysis further demonstrates that Ganoderma LVRB-17 belongs to the species G. resinaceum. Thus, the five samples collected in the present study were positioned in three different distinct groups, namely G. mbrekobenum, G. enigmaticum, and G. resinaceum. The current data may serve as reference points for future studies.
1. Introduction
Members of the genus Ganoderma are economically and ecologically important white-rot fungi, with an extensive and impressive range of applications. The worldwide usage of Ganoderma spp. for biomedical and alternative health purposes is significant, and an ever-increasing number of products that incorporate ganoderma as an active ingredient are commercially available. They include extracts and isolated constituents in various formulations, which are marketed all over the world in the form of capsules, creams, hair tonics, and syrups [1]. The genus Ganoderma, comprising more than 250 species [2,3], has gained acceptance as a biomedical material for the development of many types of health products in China, Japan, and the USA [4]. In China, for example, extracts from various Ganoderma species have been used for adjuvant anticancer clinical therapy [5]. Several natural bioactive compounds, including polysaccharides, triterpenoids, nucleosides, sterols, alkaloids, polypeptides, fatty acids, and steroids, have been isolated from the mycelia, fruiting body, and spores of ganoderma. These compounds have been reported to demonstrate numerous biologically relevant properties, but perhaps the most significant are their immunomodulatory and antitumor activities [6,7,8].
Given the growing importance of ganoderma as a health-promoting biomedical fungus, the species used in formulating various health products must be properly identified. Until recently, the identification of different Ganoderma spp. was based mainly on morphological and cultural characteristics, but this method is known to be affected by pleomorphic and environmentally influenced characteristics, imposing serious limitations on their identification [9]. Furthermore, the identification of Ganoderma spp. based on their morphological and cultural characteristics is an experience-based science; however, most mycologists lack the necessary expertise to accurately differentiate the various species. The genetic approach, involving DNA sequence analysis, is one of the most reliable methods for the identification of Ganoderma spp. [10,11]; it is convenient, rapid, and accurate, and it only requires a small quantity of sample [12].
Recent biogeographic studies suggest that Africa harbors the highest diversity of Ganoderma spp. [13]. Unfortunately, many species have not been properly characterized to establish their phylogenetic positions in relation to Ganoderma spp. from other parts of the world. In Ghana, their identification is mainly performed by physical examination of the phenotypic characteristics of the fruiting bodies using the naked eye. As a result, most identified Ganoderma spp. from Ghana lack the supporting molecular data [14], thus making their molecular phylogenetic positions dubious. Recently, the Food Research Institute of Ghana, in collaboration with the University of Minnesota, conducted molecular phylogenetic studies on Ganoderma spp. collected from the Brong Ahafo and Upper West regions of Ghana. This collaborative research led to the identification and naming of three new Ganoderma spp.: G. wiiroense [15], G. mbrekobenum, and G. enigmaticum [16]. Our goal, as presented in this communication, was to characterize the various ganoderma samples collected from the Lower Volta River Basin (LVRB) of Ghana through Bayesian phylogenetic analysis, a statistical inference approach for estimating species phylogeny and divergence.
2. Materials and Methods
2.1. Chemicals and Reagents
Antibiotic malt extract agar (AMEA) and malt extract agar (MEA) were from Fungi Perfecti, LLC (Olympia, WA, USA); UltraClean Plant DNA Isolation Kits were from MoBio Laboratories, Inc. (Carlsbad, CA, USA); GeneClean Spin kits were from Qbiogene, Inc. (Santa Ana, CA, USA); BigDye Terminator sequencing enzyme v.3.1 was from Applied Biosystems (Foster City, CA, USA); and the PCR master mix was from Promega Corp. (Madison, WI, USA).
2.2. Origin and Sampling of Isolates of Ganoderma Species
Fresh mushroom fruiting bodies resembling Ganoderma spp. were collected during mycological surveys from May to June 2015. Samples were collected from different locations (Agortigagorme, Azaglo Torkor, Lukunu, and Degorme) in Mepe and the surrounding communities, all in the Lower Volta River Basin of Ghana (Figure 1). The fresh mushroom fruiting bodies were cleaned and aseptically transferred in paper bags for transport to the Science Laboratory Technology Department of Accra Technical University for hyphae pseudoparenchyma fragment isolation.
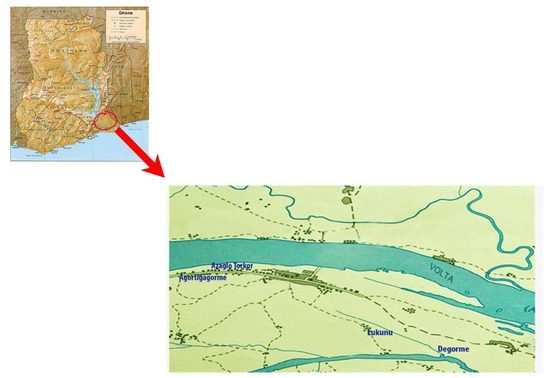
Figure 1.
Locational map of sample collection sites in the Lower Volta River Basin of Ghana Subsection.
2.3. Hyphae Pseudoparenchyma Fragment Isolation
The fruiting bodies of the respective isolates (Ganoderma LVRB-1, Ganoderma LVRB-2, Ganoderma LVRB-14, Ganoderma LVRB-16, and Ganoderma LVRB-17) were surface sterilized with 70% alcohol, cut longitudinally with a sterilized scalpel, and then a small piece of this sample was taken aseptically from the inner core of each fruiting body. The isolated samples were placed on AMEA in a Petri dish and incubated in the dark at 28 °C for 10 days. The resulting clean and genetically pure mycelium was transferred to MEA, consisting of 2% w/v malt extract and 1.5% w/v agar without antibiotics, and cultured for another 10 days to obtain pure mycelium from the fungal culture.
2.4. DNA Extraction
Genomic DNA extraction was performed following a slightly modified protocol outlined in Aime and Phillips-Mora [17]. In brief, approximately 2–4 mm3 of mycelium was aseptically excised from the actively growing edge of the colony and extracted using the UltraClean Plant DNA Isolation Kit (UltraClean Plant DNA Isolation Kits were from MoBio Laboratories, Inc. (Carlsbad, CA, USA) per the manufacturer’s instructions. DNA extractions with evidence of co-extracted fungal pigments were cleaned with the GeneClean Spin kit according to the manufacturer’s protocol.
2.5. Polymerase Chain Reactions (PCR)
The ITS2 region was amplified with the primer pairs ITS3 and ITS4 as previously described and with a slightly modified protocol [18]. Briefly, PCR was performed in 25 µL reaction mixtures, containing 12.5 µL of 2× PCR buffer, 1 µL of each primer (2.5 µM), and 2 µL of DNA extract, and the total volume was adjusted to 25 µL with sterile, deionized water. PCR amplification was conducted using the following procedure: 94 °C for 5 min, 40 cycles of 94 °C for 30 s, 50 °C for 30 s, 72 °C for 1 min, and a final extension at 72 °C for 10 min [19].
The primers ITS1F [20] and ITS4 [18] were used to amplify the internal transcribed spacer (ITS) region. The PCR amplification of the ITS ribosomal region was performed following the protocol outlined in [17], with a slight modification. Briefly, 12.5 µL of PCR master mix, 1.25 µL each of 10 mM primers (upstream and downstream), and 10 µL of diluted (10- to 100-fold) DNA template were mixed, yielding a 25 µL reaction volume [15]. Amplification was achieved with a 2 min denaturation step at 94 °C, then 35 cycles of 94 °C for 39 s, 50 °C for 15 s, and a final extension of 45 s at 60 °C [17].
The nLSU ribosomal region was amplified using the primer pairs LROR and LR6 as previously described [21], and the reaction was performed in 25 µL volumes, as described above for the ITS. LSU amplification was carried out under the same cycling program for the ITS, except the cyclic extension step was increased to 4 min. All the PCR products were cleaned using MontageTM PCR Centrifugal Filter Devices, from Millipore Corp., Burlington, MA, USA [17].
2.6. Cycle Sequencing
Sequencing reactions were cleaned using ethanol precipitation and sequenced with BigDye Terminator sequencing enzyme v.3.1 following a modified protocol described in [17]. The sequencing was performed as follows: 2 µL of diluted BigDye in 1:3 dilutions of BigDye dilution buffer (400 mM Tris pH 8.0, 10 mM MgCl2); 0.3 µL of 10 mM primer; 10–20 ng of cleaned PCR template; and H2O to 5 µL total reaction volume. For ITS2 and ITS, the sequencing primers were the same as those used for PCR, but for LSU, LR0R, LR3R, LR5, and LR16, they were used as internal sequencing primers [22].
2.7. DNA Sequence Comparisons and Data Sets
The identification of the isolates based on DNA sequence data was conducted by a similarity search of the nuclear ribosomal ITS2, ITS, and LSU sequences using NCBI BLASTn (release 2.12.0) search against Ganoderma spp. in GenBank of the National Center for Biotechnology Information (NCBI). DNA sequences sharing at least 99% nucleotide identity with those from the Lower Volta River Basin were downloaded and used for comparison.
2.8. Phylogenetic Analyses
Phylogenetic trees were generated based on Bayesian inference (BI). The sequence data set generated was aligned with Clustal Omega in Geneious Prime Version 2022.01 and refinements to the alignment were executed manually. BI analysis was conducted for the ITS2, ITS, and nLSU using Mr. Bayes in Geneious Prime Version 2022.01 with the GTR model and chain length set to 1,100,000 generations.
3. Results
3.1. Origin and Sampling of Ganoderma Species
Five isolates designated Ganoderma LVRB-1, Ganoderma LVRB-2, Ganoderma LVRB-14, Ganoderma LVRB-16, and Ganoderma LVRB-17 were opportunistically collected from different locations (Agortigagorme, Azaglo Torkor, Kizito Campus, Lukunu, and Degorme) in the Lower Volta River Basin of Ghana (Figure 1) during the rainy season of May–June 2015.
Of the five collections, two isolates coded Ganoderma LVRB-2 (Figure 2B) and Ganoderma LVRB-17 (Figure 2E) were found growing on a dead Acacia spp. tree, whereas the isolate coded Ganoderma LVRB-16 (Figure 2D) was found growing on Mangifera indica. The collections designated Ganoderma LVRB-1 (Figure 2A) were found growing on Azadirachta indica, while the last collection, designated Ganoderma LVRB-14 (Figure 2C), was found growing on Baphia nitida.
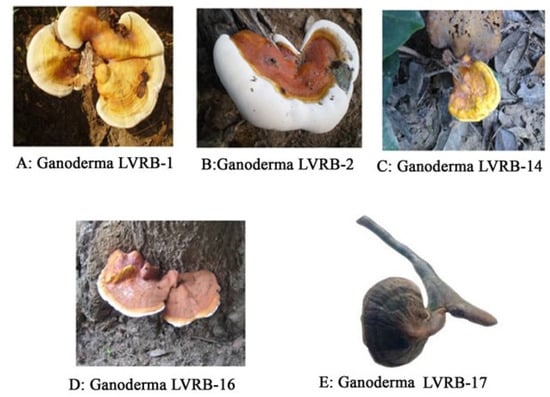
Figure 2.
(A) Ganoderma isolate LVRB-1 growing on dead Azadirachta indica collected from a house in Agortigagorme. (B) Ganoderma isolate LVRB-2 growing on dead Acacia spp. collected from Degorme. (C) Ganoderma isolate LVRB-14 growing on dead Baphia nitida collected from a farm in Lukunu. (D) Ganoderma isolate LVRB-16 growing at the base of Mangifera indica collected from Lukunu. (E) Ganoderma isolate LVRB-17 growing on dead Acacia spp. collected from Azaglo Torkor.
The fruiting bodies of the collections from M. indica and Acacia spp. were reddish brown in color, while collections from A. indica and B. nitida were yellowish brown. The sample collection site, along the Lower Volta River Basin, was found to have extensive bodies of water (Volta River, Aklakpa, and Aklamador) and mostly undulating land covered by luxuriant vegetation (Figure 1).
3.2. Sequence Generation
Amplification and sequencing of the ITS2 region were carried out for all five Ganoderma isolates. The analysis, however, revealed that the ITS2 sequence was insufficient for resolving the phylogenetic position of Ganoderma LVRB-2 and Ganoderma LVRB-17. As a result, the complete ITS region was amplified and sequenced for these two isolates. Similarly, the nLSU region was amplified and sequenced for all five isolates. The ITS2, ITS, and nLSU sequences of the isolates are presented in Supplementary Table S1A, S1B and S1C, respectively.
3.3. DNA Sequence Comparisons
Blastn comparisons of the ITS2 sequence data revealed that Ganoderma LVRB-1, Ganoderma LVRB-14, and Ganoderma LVRB-16 showed the highest similarity (99.49%, 99.74%, and 99.74%), respectively, with G. enigmaticum (Supplementary Table S1A). As indicated in Supplementary Table S1A, Ganoderma LVRB-2 showed the highest similarity (100%) with G. mbrekobenum, while Ganoderma LVRB-17 also showed the highest similarity (99.48%) with G. mbrekobenum. Likewise, in the ITS Blastn search, Ganoderma LVRB-2 displayed the highest similarity (99.47%) with G. mbrekobenum, while Ganoderma LVRB-17 showed the highest similarity (99.4%) with G. resinaceum in the ITS Blastn search (Supplementary Table S1B). BLASTn search analysis of the nLSU sequence, however, revealed that Ganoderma LVRB-1, Ganoderma LVRB-14, and Ganoderma LVRB-16, like the ITS2, showed the highest similarity, 99.90%, 100.00%, and 99.71%, respectively, with G. enigmaticum (Supplementary Table S1C). As summarized in Supplementary Table S1C, Ganoderma LVRB-2 showed the highest similarity (100%) with G. mbrekobenum. Finally, Ganoderma LVRB-17 showed the highest similarity (100%) with G. resinaceum (Supplementary Table S1C).
3.4. ITS2 Phylogenetic Analysis
Bayesian posterior probability (BPP) was conducted for all the sequenced Ganoderma spp. collections (n = 5) using MrBayes in Geneious Prime Version 2022.01, and the resulting tree is presented in Figure 3. As described in Figure 3, Ganoderma LVRB-1, Ganoderma LVRB-14, and Ganoderma LVRB-16 formed a well-supported clade (BPP = 0.984) with Ganoderma enigmaticum. As illustrated in Figure 3, Ganoderma LVRB-2 and Ganoderma LVRB-17 statistically clustered strongly with G. mbrekobenum (BPP = 1.00). The ITS2 Bayesian phylogenetic analysis revealed the five isolates are separated into two distinct clades, namely G. enigmaticum and G. mbrekobenum (Figure 3).
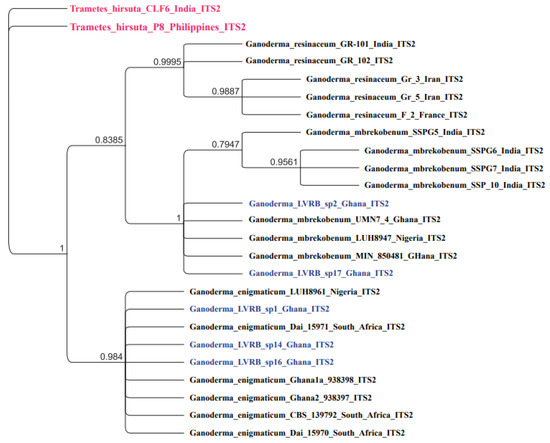
Figure 3.
Bayesian phylogenetic tree showing the phylogenetic position of Ganoderma spp. collections from the Lower Volta River Basin of Ghana compared with available ITS2 rDNA sequence data of the Ganoderma genus in GenBank. Trametes hirsuta CLF6 and Trametes hirsuta P8 were used as outgroups. Numbers at the branch nodes represent BPP values.
3.5. ITS Phylogenetic Analysis
The ITS Bayesian phylogenetic analysis of Ganoderma LVRB-2 and Ganoderma LVRB-17 is presented in Figure 4. As illustrated in Figure 4, Ganoderma LVRB-17 strongly clustered with G. resinaceum (BPP = 1.00).
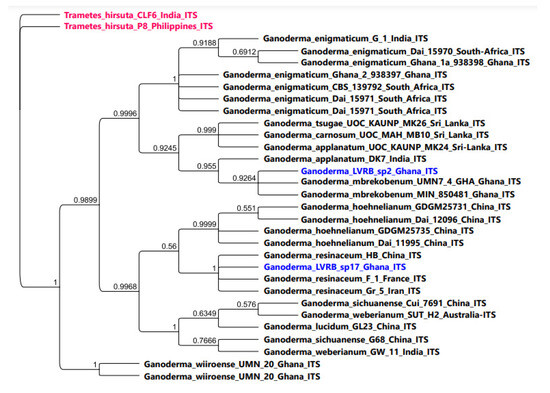
Figure 4.
Bayesian phylogenetic tree showing the phylogenetic position of Ganoderma spp. collections from the Lower Volta River Basin of Ghana compared with available ITS rDNA sequence data of the genus Ganoderma in GenBank. Trametes hirsuta CLF6 and Trametes hirsuta P8 were used as outgroups. Numbers at the branch nodes represent BPP values.
As shown in Figure 4, Ganoderma LVRB-2 strongly clustered with two isolates of G. mbrekonenum from Ghana (BPP = 0.9264), thus providing further evidence that Ganoderma LVRB-2 belongs to G. mbrekobenum. The ITS Bayesian phylogenetic analysis demonstrated that Ganoderma LVRB-2 and Ganoderma LVRB-17 were separated into two clades, namely G. mbrekobenum and G. resinaceum, respectively (Figure 4).
3.6. nLSU Phylogenetic Analysis
The nLSU was sequenced for the five isolates, and the resultant phylogenetic tree is presented in Figure 5.
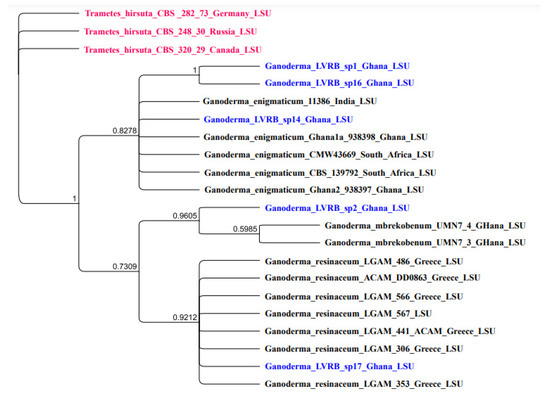
Figure 5.
Bayesian posterior probability (BPP) tree showing the phylogenetic position of Ganoderma spp. collections from the Lower Volta River Basin of Ghana compared with available nLSU sequence data of the genus Ganoderma in GenBank. Trametes hirsuta CBS 282, Trametes hirsuta CBS 320, and Trametes hirsuta CBS 248 were used as outgroups. Numbers at the branch nodes represent BPP values.
As illustrated in Figure 5, Ganoderma LVRB-2 strongly clustered with G. mbrekobenum (BPP = 0.9605), further confirming that Ganoderma LVRB-2 belongs to the species G. mbrekobenum. Ganoderma LVRB-17, on the other hand, clustered with G. resinaceum (BPP = 0.9212). The finding of nLSU phylogenetic analysis was consistent with the observation made in the ITS phylogenetic analysis, further suggesting that Ganoderma LVRB-17 belongs to the species G. resinaceum. The LSU phylogenetic tree generated from the Bayesian analysis of the LSU in this study also revealed that Ganoderma LVRB-1, Ganoderma LVRB-14, and Ganoderma LVRB-16 formed a clade with G. enigmaticum with high support (BPP = 0.8272), as observed in the ITS2 phylogenetic analysis.
4. Discussion
Numerous studies have established that medicinal mushrooms have great therapeutic potential in human health as they possess many significant medicinal properties, including anti-diabetic, anticancer, anti-obesity, immunomodulatory, hypocholesteremia, hepatoprotective, and anti-aging [23]. Indeed, Ganoderma is one of the most extensively studied genera of medicinal mushrooms, and it has proven to be a panacea in alternative medicine and a significant reservoir of medically important bioactive compounds [24]. It is also worth mentioning that medicinal mushrooms represent an important source of functional enzymes with biocatalytic potential [25,26]. In the present study, we conducted a mycological survey and collected five novel Ganoderma isolates from different locations along the banks of the Lower Volta River Basin of Ghana, in West Africa. Previously, molecular phylogenetic analysis of the internal transcribed spacer 2 (ITS2) [19], complete internal transcribed spacer (ITS) [25], and nuclear large subunit (nLSU) [26,27,28] were used to identify Ganoderma spp. from different geographical regions of the world. Recently, G. mbrekobenum, a wood-rotting fungus originally isolated from Brong Ahafo and Greater Accra, was characterized using ITS and LSU genes and named after the Ghanaian Twi word ‘mbrekoben’, which translates to reddish brown mushroom [16]. In 2021, Parihar and coworkers identified G. mbrekobenum in India [29], while Ofodile et al., just one year ago, isolated G. mbrekobenum from Nigeria [30] based on ITS Blastn search and phylogenetic analysis. In the present study, we targeted three nuclear ribosomal sequences to characterize Ghanaian Ganoderma spp. using the Bayesian method instead of neighbor-joining (distance-based) and parsimony methods. The choice for the Bayesian method is because neighbor-joining (distance-based) and parsimony are known not to perform well with molecular sequences that are evolving at different rates [31]. Besides the above explanation, the Bayesian method is also known to provide a faster measure of clade support and differs from the maximum likelihood method in that it allows for complex models of nucleotide or amino acid evolution to be implemented [31].
The results of the ITS2, ITS, and nLSU Blastn search and molecular phylogenetic analysis revealed that the isolate designated Ganoderma LVRB-2 belongs to the species G. mbrekobenum originally isolated from the Brong Ahafo and Greater Accra regions of Ghana. Aside from Brong Ahafo and Greater Accra, G. mbrekobenum has not been reported in any other region of Ghana, making the current isolation from the Volta River Basin a very important observation. Since G. mbrekobenum from Ghana is not well studied, there is a need to comprehensively study Ghanaian G. mbrekobenum to help reveal its potential utilization in comparison with Ganoderma species from other geographical regions of the world. Recently, in search of medicinally active compounds in mushrooms of the genus Ganoderma, eleven undescribed lanostane triterpenoids were isolated from artificially cultivated fruiting bodies of G. mbrekobenum [32]. Interestingly, two of the undescribed lanostane triterpenoids exhibited moderate antimalarial activity. This finding suggests that Ganoderma LVRB-2, which formed a well-supported clade with G. mbrekobenum in this present study, may also have antimalarial activity. In another recent study, extracts of the G. mbrekobenum fruiting body collected from Tanzania effectively inhibited the proliferation of the human liver cancer cell line HepG2, the human breast cancer cell line MDA-MB-231, and the human brain cancer cell line U87 [33]. These findings suggest that G. mbrekobenum may prove to be a natural source of biological precursors for the development of antimalarial and anticancer drugs.
Similar to G. mbrekobenum, several medicinally active compounds could be present in the other Ganoderma species from Ghana. Coetzee et al., (2015) studied the Ganoderma species, including a new taxon associated with root rot of the iconic Jacaranda mimosifolia in Pretoria, South Africa. The phylogenetic trees, which were generated using DNA sequences obtained from the ITS and LSU regions of the ribosomal RNA operon, confirmed G. enigmaticum as a new species [34]. Recently, G. enigmaticum was reported in Nigeria using internal transcribed spacer sequences [32]. In the present study, ITS2 and nLSU Blastn search and molecular phylogenetic analysis demonstrated that Ganoderma LVRB-1, Ganoderma LVRB-14, and Ganoderma LVRB-16 belong to the G. enigmaticum described previously from the Pretoria Province of South Africa [34]. The second ever G. enigmaticum isolation reported in the literature was from the Upper West region of Ghana [16]. This current isolation of G. enigmaticum from the Lower Volta River Basin is very useful information and represents the second new record of G. enigmaticum in Ghana, although the third-ever isolation was made from Nigeria [30]. Recently, four lanostane triterpenoids were identified in the artificially cultivated mycelial biomass of G. enigmaticum from Ghana [35]. The identified lanostane compounds include ganoderic acid C6, ganoderenic acid D, ganoderenic acid A, and ganoderic acid G, together with ganoderenic acid K and ganoderenic acid AM1 as annotated compounds.
G. resinaceum is used in ethnomedicine for immune regulation and treating hyperglycemia and liver disease [36,37]. Recently, El-Fallal et al., (2015) confirmed the status of Ganoderma species collected in the Northeast Nile Delta, Egyptm as G. resinaceum by analyzing the ribosomal 5.8S rRNA gene and the flanking internal transcribed spacers (ITS) [38]. In the current study, ITS and nLSU molecular phylogenetic analyses revealed that the Ganoderma sample LVRB-17 clustered with the species G. resinaceum. The isolation of G. resinaceum in the present study represents the first molecular evidence of the occurrence of G. resinaceum in Ghana. The results of the ITS2 phylogenetic analysis, on the other hand, showed that Ganoderma LVRB-2 and Ganoderma LVRB-17 belong to the same species, suggesting ITS2 was not able to resolve the identity of Ganoderma LVRB-17 in the current study, contrary to the observations of some researchers [19,39] that suggest that ITS2 was suitable for identification of Ganoderma species. In the present study, ITS1 was not used because ITS1 and ITS2 have been reported to yield, to a large extent, similar results when used as DNA metabarcodes for fungi [40]. Since Ganoderma species from different geographic regions have different bioactive compounds and biological activities, the isolates identified in the present molecular phylogenetic analysis need to be properly characterized using metabolomics approaches and their bioactivities investigated to provide insight into their biopharmaceutical potentials, thereby unlocking their potential nutraceutical and biopharmaceutical applications.
5. Conclusions
The present study confirmed that the Ganoderma species collected from the Lower Volta River Basin in Ghana belong to three distinct species, namely G. mbrekobenum, G. enigmaticum, and G. resinaceum. The molecular phylogenetic analysis of Ganoderma species from the Lower Volta Basin in this present study was generally consistent with the earlier findings by the Food Research Institute of Ghana and the University of Minnesota, in the USA, in which the occurrence of G. mbrekobenum and G. enigmaticum was reported in Ghana. To the best of our knowledge, this is the first phylogenetic study that provides molecular evidence for the occurrence of G. resinaceum in Ghana. The current study has provided information that will be useful for future studies regarding the molecular evolution, biomedical implications, and phytopathogenic significance of Ganoderma species in Ghana.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/jof10010006/s1, Table S1A: ITS2 sequence matching results of isolates Ganoderma LVRB-1, Ganoderma LVRB-2, Ganoderma LVRB-14, Ganoderma LVRB-16 and Ganoderma LVRB-17 from the Lower Volta River Basin of Ghana. Table S1B: ITS sequence matching results of isolates of isolate Ganoderma LVRB-2 and Ganoderma LVRB-17 from the Lower Volta River Basin of GhanaITS2 sequence. Table S1C: nLSU nucleotide sequence matching results of isolates Ganoderma LVRB-1, Ganoderma LVRB-2, Ganoderma LVRB-14, Ganoderma LVRB-16 and Ganoderma LVRB-17 from the Lower Volta River Basin of Ghana.
Author Contributions
Conceptualization, G.A., R.N.A., V.C.L., A.K.A., L.K.N.O. and W.S.K.G.; methodology, G.A., R.N.A., P.Y., J.C.H. and P.O.; software, G.A., A.Q., R.N.A. and V.C.L.; validation, G.A., R.N.A. and V.C.L.; investigation, G.A., R.N.A., A.Q., A.K.A. and V.C.L.; resources, G.A., A.Q., R.N.A., P.Y. and V.C.L.; writing—original draft preparation, G.A., R.N.A., P.O., A.Q., V.C.L. and P.Y.; writing—review and editing, G.A., R.N.A., V.C.L., A.Q., J.C.H., P.O. and A.K.A.; supervision, A.K.A., A.Q., L.K.N.O., W.S.K.G. and J.C.H.; funding acquisition, R.N.A., G.A., A.K.A. and A.Q. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available upon request.
Acknowledgments
We appreciate the kind support given by Kofi Bedzra of Ghana Atomic Energy Commission (GAEC). We also thank Kwasi Asigbe, Kwadzo Obani Adotey, Vicent Amaglo Gakpetor, and Prosper Akumani for their invaluable assistance in taking the team around Ghana’s Lower Volta River Basin.
Conflicts of Interest
J.C.H. is the founder of Aloha Medicinals Inc., a medicinal mushroom biotechnology company. No other conflict exist.
References
- Benzie, I.F.F.; Wachtel-Galor, S. (Eds.) Herbal Medicine: Biomolecular and Clinical Aspects, 2nd ed.; CRC Press/Taylor & Francis: Boca Raton, FL, USA, 2011. [Google Scholar]
- Ryvarden, L.; Iturriaga, T. Studies in neotropical polypores 10. New polypores from Venezuela. Mycologia 2003, 95, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, G.C. Ainsworth & Bisby’s Dictionary of the Fungi, 10th ed.; CABI Publishing: Wallingford, UK, 2008; Volume 4, p. 43. [Google Scholar]
- Wasser, S.P.; Coates, P.; Blackman, M.; Cragg, G.; Levine, M.; Moss, J.; White, J. Encyclopedia of Dietary Supplements; Marcel Dekker: New York, NY, USA, 2005. [Google Scholar]
- Zhao, R.-L.; He, Y.-M. Network pharmacology analysis of the anti-cancer pharmacological mechanisms of Ganoderma lucidum extract with experimental support using Hepa1-6-bearing C57 BL/6 mice. J. Ethnopharmacol. 2018, 210, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Batra, P.; Sharma, A.K.; Khajuria, R. Probing Lingzhi or Reishi medicinal mushroom Ganoderma lucidum (Higher Basidiomycetes): A bitter mushroom with amazing health benefits. Int. J. Med. Mushrooms 2013, 15, 127–143. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Xu, J.; Liu, C.; Zhu, Y.; Nelson, D.R.; Zhou, S.; Li, C.; Wang, L.; Guo, X.; Sun, Y.; et al. Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat. Commun. 2012, 3, 913. [Google Scholar] [CrossRef] [PubMed]
- Shiao, M.-S. Natural products of the medicinal fungus Ganoderma lucidum: Occurrence, biological activities, and pharmacological functions. Chem. Rec. 2003, 3, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Fryssouli, V.; Zervakis, G.; Polemis, E.; Typas, M.A. A global meta-analysis of ITS rDNA sequences from material belonging to the genus Ganoderma (Basidiomycota, Polyporales) including new data from selected taxa. MycoKeys 2020, 75, 71–143. [Google Scholar] [CrossRef] [PubMed]
- Richter, C.; Wittstein, K.; Kirk, P.M.; Stadler, M. An assessment of the taxonomy and chemotaxonomy of Ganoderma. Fungal Divers. 2015, 71, 1–15. [Google Scholar] [CrossRef]
- Welti, S.; Moreau, P.; Decock, C.; Danel, C.; Duhal, N.; Favel, A.; Courtecuisse, R. Oxygenated lanostane-type triterpenes profiling in laccate Ganoderma chemotaxonomy. Mycol. Prog. 2015, 14, 45. [Google Scholar] [CrossRef]
- Singh, M.; Bandana; Ahuja, P.S. Isolation and PCR Amplification of Genomic DNA from Market Samples of Dry Tea. Plant Mol. Biol. Rep. 1999, 17, 171–178. [Google Scholar] [CrossRef]
- Moncalvo, J.-M. Molecular Systematics of Ganoderma: What Is Reishi? Int. J. Med. Mushrooms 2005, 7, 353–354. [Google Scholar] [CrossRef]
- Obodai, M.; Mensah, D.L.N.; Fernandes, A.; Kortei, N.K.; Dzomeku, M.; Teegarden, M.; Schwartz, S.J.; Barros, L.; Prempeh, J.; Takli, R.K.; et al. Chemical Characterization and Antioxidant Potential of Wild Ganoderma Species from Ghana. Molecules 2017, 22, 196. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.; Wingfield, M.; Le Roux, J.; Richardson, D.; Strasberg, D.; Shivas, R.; Alvarado, P.; Edwards, J.; Moreno, G.; Sharma, R.; et al. Fungal Planet description sheets: 371–399. Persoonia Mol. Phylogeny Evol. Fungi 2015, 35, 264–327. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Wingfield, M.J.; Richardson, D.M.; Le Roux, J.J.; Strasberg, D.; Edwards, J.; Roets, F.; Hubka, V.; Taylor, P.W.; Heykoop, M.; et al. Fungal Planet description sheets: 400–468. Persoonia 2016, 36, 316–458. [Google Scholar] [CrossRef] [PubMed]
- Aime, M.; Phillips-Mora, W. The causal agents of witches’ broom and frosty pod rot of cacao (chocolate, Theobroma cacao) form a new lineage of Marasmiaceae. Mycologia 2005, 97, 1012–1022. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.D.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Eds.; Academic Press: London, UK, 1990; pp. 315–322. [Google Scholar]
- Liao, B.; Chen, X.; Han, J.; Dan, Y.; Wang, L.; Jiao, W.; Song, J.; Chen, S. Identification of commercial Ganoderma (Lingzhi) species by ITS2 sequences. Chin. Med. 2015, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Hopple, J.S.; Vilgalys, R. Phylogenetic relationships among coprinoid txa and allies based on data from restriction site mapping of nuclear rDNA. Mycologia 1994, 86, 96–107. [Google Scholar] [CrossRef]
- Moncalvo, J.; Flood, J.; Bridge, P.D.; Holderness, M. Systematics of Ganoderma. In Ganoderma Diseases of Perennial Crops; CABI Publishing: Wallingford, UK, 2000; pp. 23–45. [Google Scholar]
- Chaturvedi, V.K.; Agarwal, S.; Gupta, K.K.; Ramteke, P.W.; Singh, M.P. Medicinal mushroom: Boon for therapeutic applications. 3 Biotech 2018, 8, 334. [Google Scholar] [CrossRef]
- Chen, X.; Veena, R.K.; Ramya, H.; Janardhanan, K.K.; George, V. Gano oil: A novel antinociceptive agent extracted from Ganoderma lucidum inhibits paw oedema and relieves pain by hypnotic and analgesic actions of fatty acid amides. J. Ethnopharmacol. 2020, 263, 113144. [Google Scholar] [CrossRef]
- Tan, H.; Tang, J.; Li, X.; Liu, T.; Miao, R.; Huang, Z.; Wang, Y.; Gan, B.; Peng, W. Biochemical Characterization of a Psychrophilic Phytase from an Artificially Cultivable Morel Morchella importuna. J. Microbiol. Biotechnol. 2017, 27, 2180–2189. [Google Scholar] [CrossRef]
- Zhang, Q.; Miao, R.; Liu, T.; Huang, Z.; Peng, W.; Gan, B.; Zhang, X.; Tan, H. Biochemical characterization of a key laccase-like multicopper oxidase of artificially cultivable Morchella importuna provides insights into plant-litter decomposition. 3 Biotech 2019, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Gunnels, T.; Creswell, M.; McFerrin, J.; Whittall, J.B. The ITS region provides a reliable DNA barcode for identifying reishi/lingzhi (Ganoderma) from herbal supplements. PLoS ONE 2020, 15, e0236774. [Google Scholar] [CrossRef] [PubMed]
- Luangharn, T.; Karunarathna, S.C.; Mortimer, P.E.; Hyde, K.D.; Xu, J. Additions to the knowledge of Ganoderma in Thailand: Ganoderma casuarinicola, a new record; and Ganoderma thailandicum sp. nov. MycoKeys 2019, 59, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Parihar, S.S.; Sahu, S.; Gupta, G.; Prakash, A. Ganoderma mbrekobenum: A pharmacologically Important Mushroom Naturally Growing in Raisen, India. Curr. Trends Biotechnol. Pharm. 2021, 15, 383–389. [Google Scholar] [CrossRef]
- Ofodile, L.N.; Isikhuemhen, O.S.; Anike, F.N.; Adekunle, A.A. The Domestication and Cultivation of Ganoderma (Agaricomycetes) Medicinal Mushroom Species from Nigeria. Int. J. Med. Mushrooms 2022, 24, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal Identification Using Molecular Tools: A Primer for the Natural Products Research Community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef] [PubMed]
- Yangchum, A.; Fujii, R.; Choowong, W.; Rachtawee, P.; Pobkwamsuk, M.; Boonpratuang, T.; Mori, S.; Isaka, M. Lanostane triterpenoids from cultivated fruiting bodies of basidiomycete Ganoderma mbrekobenum. Phytochemistry 2022, 196, 113075. [Google Scholar] [CrossRef]
- Huiping, H.; Yuanchao, L.; Xiaowei, L.; Xiangmin, L.; Weipeng, M.; Yizhen, X.; Zhi, Z.; Qingping, W. Artificial Cultivation Anti-tumor Activity of Ganoderma mbrekobenum. Sains Malays. 2021, 50, 723–733. [Google Scholar] [CrossRef]
- Coetzee, M.P.A.; Marincowitz, S.; Muthelo, V.G.; Wingfield, M.J. Ganoderma species, including new taxa associated with root rot of the iconic Jacaranda mimosifolia in Pretoria, South Africa. IMA Fungus 2015, 6, 249–256. [Google Scholar] [CrossRef]
- Adotey, G.; Alolga, R.N.; Quarcoo, A.; Gedel, M.A.; Anang, A.K.; Holliday, J.C. Ultra Performance Liquid Chromatography-Quadrupole Time-of-Flight Mass Spectrometry (UPLC-Q-TOF-MS)-based metabolomic analysis of mycelial biomass of three Ganoderma isolates from the Lower Volta River Basin of Ghana. J. Pharm. Biomed. Anal. 2021, 205, 114355. [Google Scholar] [CrossRef]
- Peng, X.-R.; Liu, J.-Q.; Han, Z.-H.; Yuan, X.-X.; Luo, H.-R.; Qiu, M.-H. Protective effects of triterpenoids from Ganoderma resinaceum on H2O2-induced toxicity in HepG2 cells. Food Chem. 2013, 141, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Oyetayo, O.V. Medicinal uses of mushrooms in Nigeria: Towards full and sustainable exploitation. Afr. J. Tradit. Complement. Altern. Med. 2011, 8, 267–274. [Google Scholar] [CrossRef] [PubMed]
- El-Fallal, A. First record of two Ganoderma species from North East Nile DeltaEgypt. Mycosphere 2015, 6, 248–259. [Google Scholar] [CrossRef]
- Han, J.; Zhu, Y.; Chen, X.; Liao, B.; Yao, H.; Song, J.; Chen, S.; Meng, F. The Short ITS2 Sequence Serves as an Efficient Taxonomic Sequence Tag in Comparison with the Full-Length ITS. BioMed Res. Int. 2013, 2013, 741476. [Google Scholar] [CrossRef]
- Blaalid, R.; Kumar, S.; Nilsson, R.H.; Abarenkov, K.; Kirk, P.M.; Kauserud, H. ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol. Ecol. Resour. 2013, 13, 218–224. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).