Masking the Pathogen: Evolutionary Strategies of Fungi and Their Bacterial Counterparts
Abstract
:1. Introduction
2. Sugar-Coated Killers: Capsular Structures of Bacteria and a Pathogenic Fungus
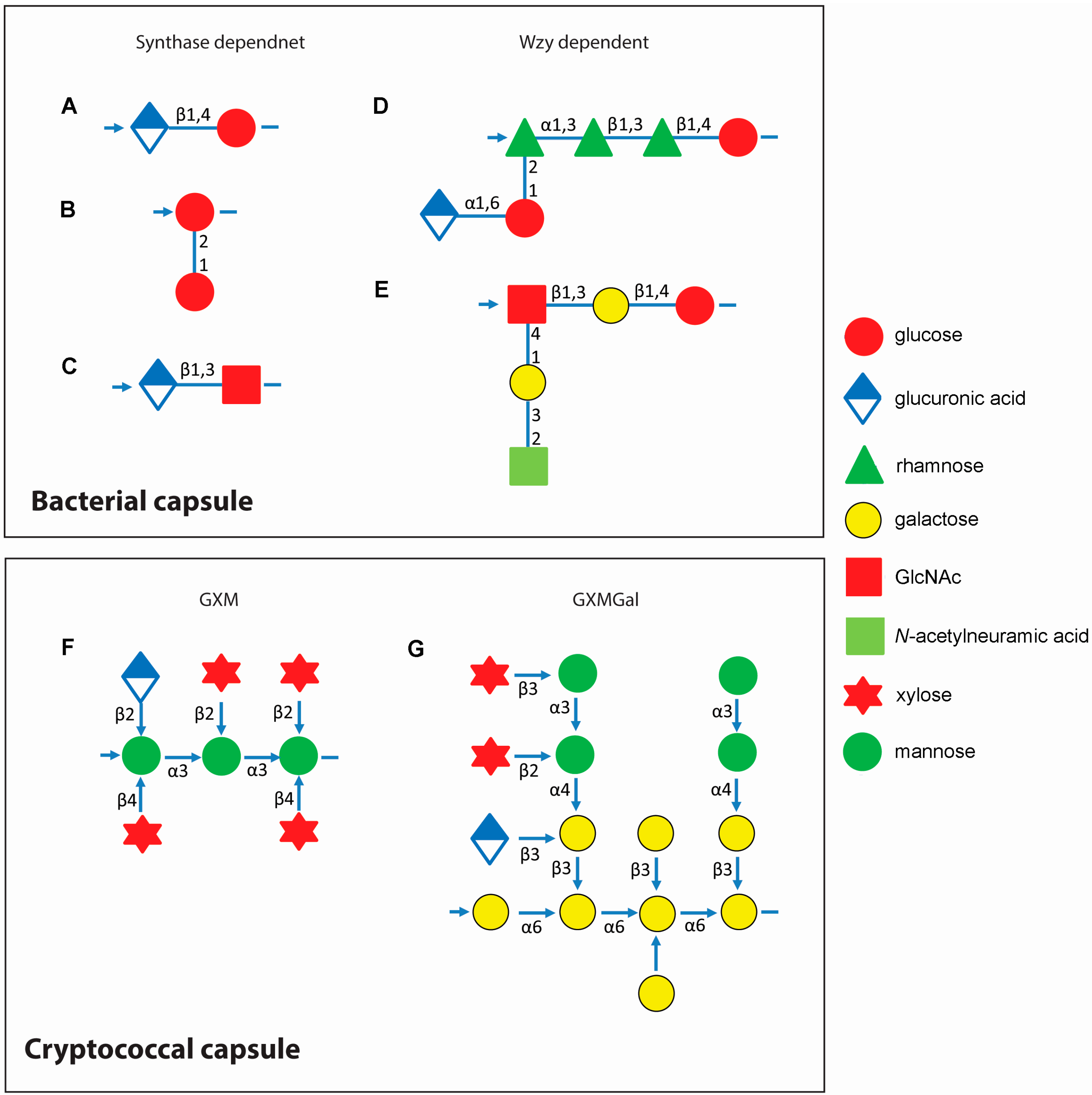
3. Assembly of Capsular Components Show Evolution in Complexity
3.1. Bacterial Capsule Synthesis
3.2. Cryptococcal Capsule Synthesis
3.3. Non-Pathogenic Fungal Capsules are Less Protective
4. Function Follows Form: Capsule Alters Host Immune Responses
4.1. Bacterial Capsules and Host Defense
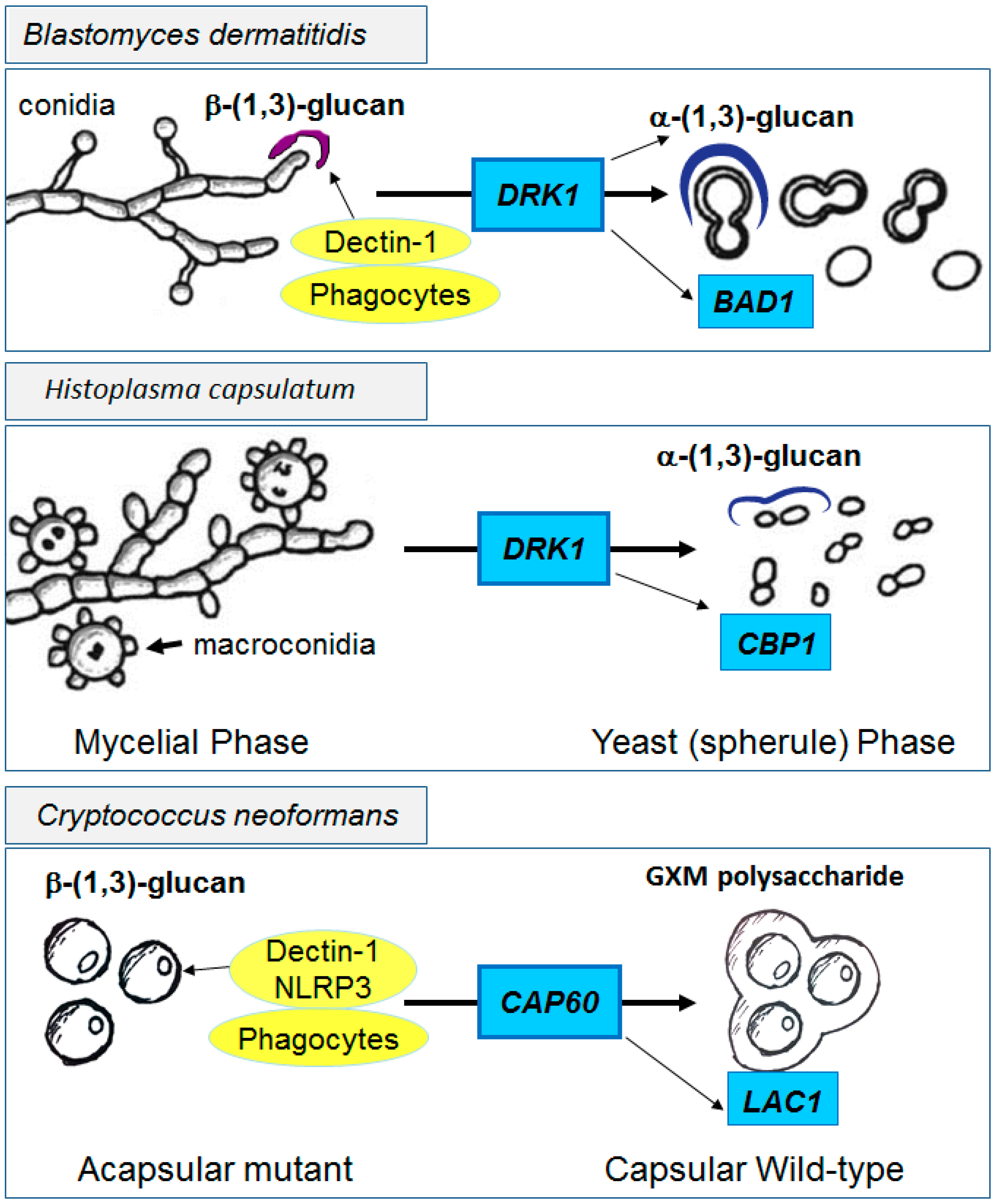
4.2. Innate Shielding of Fungi
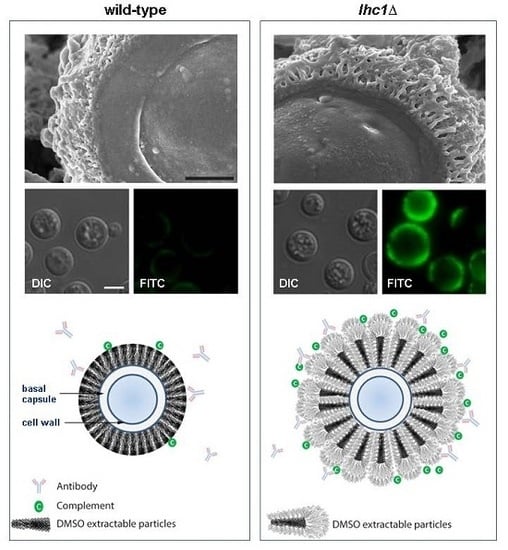
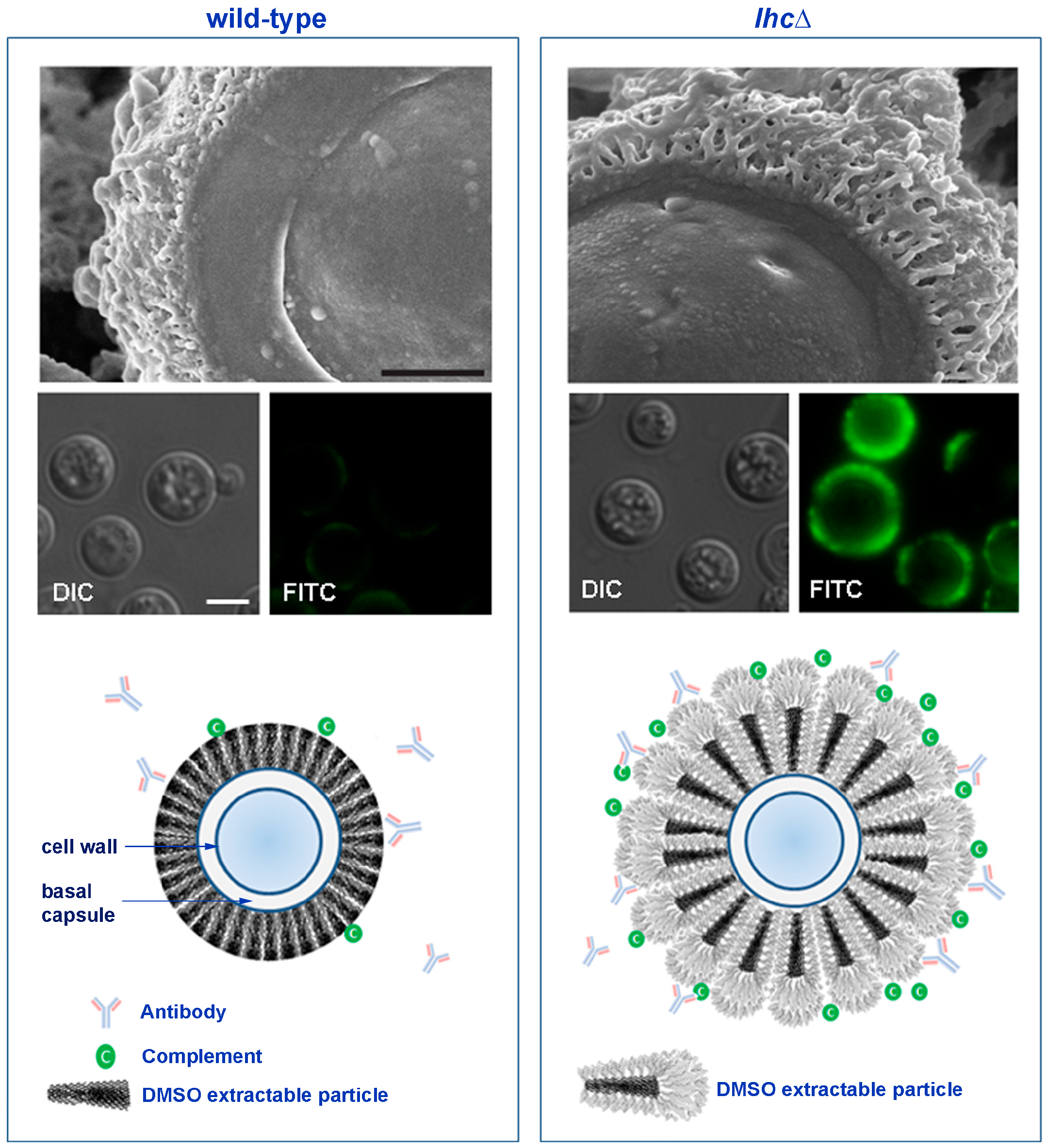
4.3. The Cryptococcal Immune Shield
5. Evolutionary Pressures in the Host and Environment: The Red Queen Paradigm
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Doering, T.L. How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans. Annu. Rev. Microbiol. 2009, 63, 223–247. [Google Scholar] [CrossRef] [PubMed]
- Roberts, I.S.; Saunders, F.K.; Boulnois, G.J. Bacterial capsules and interactions with complement and phagocytes. Biochem. Soc. Trans. 1989, 17, 462–464. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, M.A.; Silverstein, S.C. Influence of the Escherichia coli capsule on complement fixation and on phagocytosis and killing by human phagocytes. J. Clin. Investig. 1980, 65, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.C.; Exley, R.M.; Ram, S.; Sim, R.B.; Tang, C.M. Interactions between Neisseria meningitidis and the complement system. Trends Microbiol. 2007, 15, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Vecchiarelli, A. Immunoregulation by capsular components of Cryptococcus neoformans. Med. Mycol. 2000, 38, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Ravichandra, N.G. Fundamentals of Plant Pathology; Rekha Printers Private Limited: New Delhi, India, 2013; p. 204. [Google Scholar]
- Kroncke, K.D.; Golecki, J.R.; Jann, K. Further electron microscopic studies on the expression of Escherichia coli group II capsules. J. Bacteriol. 1990, 172, 3469–3472. [Google Scholar] [PubMed]
- Heritage, J.; Evans, E.G.V.; Killington, R.A. Introductory Microbiology; Cambridge University Press: Cambridge, UK, 1996; p. 41. [Google Scholar]
- Salomons, B.; Sigmond, J.; Terpstra, M. Immunoassay: A Survey of Patents, Patent Applications and Other Literature 1980–1991; Taylor and Francis: London, UK, 1992. [Google Scholar]
- Whitfield, C.; Roberts, I.S. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 1999, 31, 1307–1319. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Matsumoto, T.; Tateda, K.; Uchida, K.; Tsujimoto, S.; Yamaguchi, K. Role of bacterial capsule in local and systemic inflammatory responses of mice during pulmonary infection with Klebsiella pneumoniae. J. Med. Microbiol. 2000, 49, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Schouls, L.; van der Heide, H.; Witteveen, S.; Zomer, B.; van der Ende, A.; Burger, M.; Schot, C. Two variants among Haemophilus influenzae serotype b strains with distinct bcs4, hcsa and hcsb genes display differences in expression of the polysaccharide capsule. BMC Microbiol. 2008, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Getin, E.T.; Toreci, K.; Ang, O. Encapsulated Pseudomonas aeruginosa. J. Bacteriol. 1965, 89, 1432–1433. [Google Scholar]
- Gates, M.A.; Thorkildson, P.; Kozel, T.R. Molecular architecture of the Cryptococcus neoformans capsule. Mol. Microbiol. 2004, 52, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Cieslewicz, M.J.; Chaffin, D.; Glusman, G.; Kasper, D.; Madan, A.; Rodrigues, S.; Fahey, J.; Wessels, M.R.; Rubens, C.E. Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect. Immun. 2005, 73, 3096–3103. [Google Scholar] [CrossRef] [PubMed]
- Pelkonen, S.; Hayrinen, J.; Finne, J. Polyacrylamide gel electrophoresis of the capsular polysaccharides of Escherichia coli K1 and other bacteria. J. Bacteriol. 1988, 170, 2646–2653. [Google Scholar] [PubMed]
- Yother, J. Capsules of Streptococcus pneumoniae and other bacteria: Paradigms for polysaccharide biosynthesis and regulation. Annu. Rev. Microbiol. 2011, 65, 563–581. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Aanensen, D.M.; Mavroidi, A.; Saunders, D.; Rabbinowitsch, E.; Collins, M.; Donohoe, K.; Harris, D.; Murphy, L.; Quail, M.A.; et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2006, 2, e31. [Google Scholar] [CrossRef] [PubMed]
- Van Dam, J.E.; Fleer, A.; Snippe, H. Immunogenicity and immunochemistry of Streptococcus pneumoniae capsular polysaccharides. Antonie Van Leeuwenhoek 1990, 58, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Cherniak, R.; Valafar, H.; Morris, L.C.; Valafar, F. Cryptococcus neoformans chemotyping by quantitative analysis of 1H nuclear magnetic resonance spectra of glucuronoxylomannans with a computer-simulated artificial neural network. Clin. Diagn. Lab. Immunol. 1998, 5, 146–159. [Google Scholar] [PubMed]
- Roberts, I.S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 1996, 50, 285–315. [Google Scholar] [CrossRef] [PubMed]
- Walsh, R.L.; Camilli, A. Streptococcus pneumoniae is desiccation tolerant and infectious upon rehydration. mBio 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Vishniac, H.S. Simulated in situ competitive ability and survival of a representative soil yeast, Cryptococcus albidus. Microb. Ecol. 1995, 30, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Aksenov, S.I.; Babyeva, I.P.; Golubev, V.I. On the mechanism of adaptation of micro-organisms to conditions of extreme low humidity. Life Sci. Space Res. 1973, 11, 55–61. [Google Scholar] [PubMed]
- Golubev, V. Capsules; Academic Press: New York, NY, USA, 1991. [Google Scholar]
- Zaragoza, O.; Rodrigues, M.L.; de Jesus, M.; Frases, S.; Dadachova, E.; Casadevall, A. The capsule of the fungal pathogen Cryptococcus neoformans. Adv. Appl. Microbiol. 2009, 68, 133–216. [Google Scholar] [PubMed]
- De Jesus, M.; Park, C.G.; Su, Y.; Goldman, D.L.; Steinman, R.M.; Casadevall, A. Spleen deposition of Cryptococcus neoformans capsular glucuronoxylomannan in rodents occurs in red pulp macrophages and not marginal zone macrophages expressing the C-type lectin SIGN-R1. Med. Mycol. 2008, 46, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.S.; Kim, J.Y.; Bruening, S.A.; Pack, M.; Charalambous, A.; Pritsker, A.; Moran, T.M.; Loeffler, J.M.; Steinman, R.M.; Park, C.G. The C-type lectin SIGN-R1 mediates uptake of the capsular polysaccharide of Streptococcus pneumoniae in the marginal zone of mouse spleen. Proc. Natl. Acad. Sci. USA 2004, 101, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Maitta, R.W.; Datta, K.; Lees, A.; Belouski, S.S.; Pirofski, L.A. Immunogenicity and efficacy of Cryptococcus neoformans capsular polysaccharide glucuronoxylomannan peptide mimotope-protein conjugates in human immunoglobulin transgenic mice. Infect. Immun. 2004, 72, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Pirofski, L.A.; Casadevall, A. Cryptococcus neoformans: Paradigm for the role of antibody immunity against fungi? Zentralbl. Bakteriol. 1996, 284, 475–495. [Google Scholar] [CrossRef]
- Martinez, L.R.; Casadevall, A. Biofilm formation by Cryptococcus neoformans. Microbiol. Spectr. 2015, 3, 1–2. [Google Scholar]
- Kumar, P.; Yang, M.; Haynes, B.C.; Skowyra, M.L.; Doering, T.L. Emerging themes in cryptococcal capsule synthesis. Curr. Opin. Struct. Biol. 2011, 21, 597–602. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, T.R.; Alspaugh, J.A. The Cryptococcus neoformans capsule: A sword and a shield. Clin. Microbiol. Rev. 2012, 25, 387–408. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, A.K.; Bennett, J.E.; Glaudemans, C.P. Capsular polysaccharides of Cryptococcus neoformans. Rev. Infect. Dis. 1984, 6, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Cherniak, R.; Sundstrom, J.B. Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect. Immun. 1994, 62, 1507–1512. [Google Scholar] [PubMed]
- Kozel, T.R. Antigenic structure of Cryptococcus neoformans capsular polysaccharides. Immunol. Ser. 1989, 47, 63–86. [Google Scholar] [PubMed]
- Janbon, G.; Himmelreich, U.; Moyrand, F.; Improvisi, L.; Dromer, F. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol. Microbiol. 2001, 42, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Nosanchuk, J.D.; Casadevall, A. Cellular charge of Cryptococcus neoformans: Contributions from the capsular polysaccharide, melanin, and monoclonal antibody binding. Infect. Immun. 1997, 65, 1836–1841. [Google Scholar] [PubMed]
- Vaishnav, V.V.; Bacon, B.E.; O’Neill, M.; Cherniak, R. Structural characterization of the galactoxylomannan of Cryptococcus neoformans Cap67. Carbohydr. Res. 1998, 306, 315–330. [Google Scholar] [CrossRef]
- Oyston, P.C. Francisella tularensis: Unravelling the secrets of an intracellular pathogen. J. Med. Microbiol. 2008, 57, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Dan, J.M.; Wang, J.P.; Lee, C.K.; Levitz, S.M. Cooperative stimulation of dendritic cells by Cryptococcus neoformans mannoproteins and CpG oligodeoxynucleotides. PLoS ONE 2008, 3, e2046. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed]
- Valvano, M.A. Export of O-specific lipopolysaccharide. Front. Biosci. 2003, 8, s452–s471. [Google Scholar] [CrossRef] [PubMed]
- Cuthbertson, L.; Kos, V.; Whitfield, C. ABC transporters involved in export of cell surface glycoconjugates. Microbiol. Mol. Biol. Rev. 2010, 74, 341–362. [Google Scholar] [CrossRef] [PubMed]
- Cuthbertson, L.; Mainprize, I.L.; Naismith, J.H.; Whitfield, C. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol. Mol. Biol. Rev. 2009, 73, 155–177. [Google Scholar] [CrossRef] [PubMed]
- Willis, L.M.; Whitfield, C. Structure, biosynthesis, and function of bacterial capsular polysaccharides synthesized by abc transporter-dependent pathways. Carbohydr. Res. 2013, 378, 35–44. [Google Scholar] [CrossRef] [PubMed]
- DeAngelis, P.L. Molecular directionality of polysaccharide polymerization by the pasteurella multocida hyaluronan synthase. J. Biol. Chem. 1999, 274, 26557–26562. [Google Scholar] [CrossRef] [PubMed]
- Kundig, J.D.; Aminoff, D.; Roseman, S. The sialic acids. XII. Synthesis of colominic acid by a sialyltransferase from Escherichia coli K-235. J. Biol. Chem. 1971, 246, 2543–2550. [Google Scholar] [PubMed]
- Rohr, T.E.; Troy, F.A. Structure and biosynthesis of surface polymers containing polysialic acid in Escherichia coli. J. Biol. Chem. 1980, 255, 2332–2342. [Google Scholar] [PubMed]
- Wilson, J.W.; Schurr, M.J.; LeBlanc, C.L.; Ramamurthy, R.; Buchanan, K.L.; Nickerson, C.A. Mechanisms of bacterial pathogenicity. Postgrad. Med. J. 2002, 78, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Orskov, I.; Orskov, F. Serotyping of Klebsiella. Methods Microbiol. 1984, 14, 143–164. [Google Scholar]
- Bhattacharjee, A.K.; Bennett, J.E.; Bundle, D.R.; Glaudemans, C.P. Anticryptococcal type D antibodies raised in rabbits. Mol. Immunol. 1983, 20, 351–359. [Google Scholar] [CrossRef]
- Panackal, A.A.; Dekker, J.P.; Proschan, M.; Beri, A.; Williamson, P.R. Enzyme immunoassay versus latex agglutination cryptococcal antigen assays in adults with non-HIV-related cryptococcosis. J. Clin. Microbiol. 2014, 52, 4356–4358. [Google Scholar] [CrossRef] [PubMed]
- Kozel, T.R.; Bauman, S.K. Crag lateral flow assay for cryptococcosis. Expert Opin. Med. Diagn. 2012, 6, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Gates-Hollingsworth, M.A.; Kozel, T.R. Serotype sensitivity of a lateral flow immunoassay for cryptococcal antigen. Clin. Vaccine Immunol. 2013, 20, 634–635. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chang, Y.C.; Kwon-Chung, K.J. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 1994, 14, 4912–4919. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Penoyer, L.A.; Kwon-Chung, K.J. The second capsule gene of Cryptococcus neoformans, Cap64, is essential for virulence. Infect. Immun. 1996, 64, 1977–1983. [Google Scholar] [PubMed]
- Chang, Y.C.; Kwon-Chung, K.J. Isolation of the third capsule-associated gene, Cap60, required for virulence in Cryptococcus neoformans. Infect. Immun. 1998, 66, 2230–2236. [Google Scholar] [PubMed]
- Chang, Y.C.; Kwon-Chung, K.J. Isolation, characterization, and localization of a capsule-associated gene, Cap10, of Cryptococcus neoformans. J. Bacteriol. 1999, 181, 5636–5643. [Google Scholar] [PubMed]
- Feldmesser, M.; Kress, Y.; Casadevall, A. Dynamic changes in the morphology of Cryptococcus neoformans during murine pulmonary infection. Microbiology 2001, 147, 2355–2365. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rivera, J.; Chang, Y.C.; Kwon-Chung, K.J.; Casadevall, A. Cryptococcus neoformans Cap59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot. Cell 2004, 3, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Yoneda, A.; Doering, T.L. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol. Biol. Cell 2006, 17, 5131–5140. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.L.; Nimrichter, L.; Oliveira, D.L.; Frases, S.; Miranda, K.; Zaragoza, O.; Alvarez, M.; Nakouzi, A.; Feldmesser, M.; Casadevall, A. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell 2007, 6, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Panepinto, J.; Komperda, K.; Frases, S.; Park, Y.; Djordjevic, J.; Casadevall, A.; Williamson, P. Sec6-dependent sorting of fungal extracellular exosomes and laccase of Cryptococcus neoformans. Mol. Microbiol. 2009, 71, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Wills, E.A.; Roberts, I.S.; del Poeta, M.; Rivera, J.; Casadevall, A.; Cox, G.M.; Perfect, J.R. Identification and characterization of the Cryptococcus neoformans phosphomannose isomerase-encoding gene, MAN1, and its impact on pathogenicity. Mol. Microbiol. 2001, 40, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Bar-Peled, M.; Griffith, C.L.; Ory, J.J.; Doering, T.L. Biosynthesis of UDP-GlcA, a key metabolite for capsular polysaccharide synthesis in the pathogenic fungus Cryptococcus neoformans. Biochem. J. 2004, 381, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Moyrand, F.; Janbon, G. UGD1, encoding the Cryptococcus neoformans UDP-glucose dehydrogenase, is essential for growth at 37 °C and for capsule biosynthesis. Eukaryot. Cell 2004, 3, 1601–1608. [Google Scholar] [CrossRef] [PubMed]
- Griffith, C.L.; Klutts, J.S.; Zhang, L.; Levery, S.B.; Doering, T.L. UDP-glucose dehydrogenase plays multiple roles in the biology of the pathogenic fungus Cryptococcus neoformans. J. Biol. Chem. 2004, 279, 51669–51676. [Google Scholar] [CrossRef] [PubMed]
- Coyne, M.J.; Fletcher, C.M.; Reinap, B.; Comstock, L.E. UDP-glucuronic acid decarboxylases of bacteroides fragilis and their prevalence in bacteria. J. Bacteriol. 2011, 193, 5252–5259. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza, O.; Casadevall, A. Experimental modulation of capsule size in Cryptococcus neoformans. Biol. Proced. Online 2004, 6, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Rivera, J.; Feldmesser, M.; Cammer, M.; Casadevall, A. Organ-dependent variation of capsule thickness in Cryptococcus neoformans during experimental murine infection. Infect. Immun. 1998, 66, 5027–5030. [Google Scholar] [PubMed]
- Pukkila-Worley, R.; Gerrald, Q.D.; Kraus, P.R.; Boily, M.J.; Davis, M.J.; Giles, S.S.; Cox, G.M.; Heitman, J.; Alspaugh, J.A. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot. Cell 2005, 4, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.; Wang, Y.-L.; Whittington, A.; Li, L.; Wang, P. The RGS protein Crg2 regulates pheromone and cyclic AMP signaling in Cryptococcus neoformans. Eukaryot. Cell 2008, 7, 1540–1548. [Google Scholar] [CrossRef] [PubMed]
- Alspaugh, J.A.; Pukkila-Worley, R.; Harashima, T.; Cavallo, L.M.; Funnell, D.; Cox, G.M.; Perfect, J.R.; Kronstad, J.W.; Heitman, J. Adenylyl cyclase functions downstream of the Gα protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 2002, 1, 75–84. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, C.A.; Alspaugh, J.A.; Yue, C.; Harashima, T.; Cox, G.M.; Perfect, J.R.; Heitman, J. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 2001, 21, 3179–3191. [Google Scholar] [CrossRef] [PubMed]
- Alspaugh, J.A.; Perfect, J.R.; Heitman, J. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit Gpa1 and cAMP. Genes Dev. 1997, 11, 3206–3217. [Google Scholar] [CrossRef] [PubMed]
- Bryan, R.A.; Zaragoza, O.; Zhang, T.; Ortiz, G.; Casadevall, A.; Dadachova, E. Radiological studies reveal radial differences in the architecture of the polysaccharide capsule of Cryptococcus neoformans. Eukaryot. Cell 2005, 4, 465–475. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cordero, R.J.; Frases, S.; Guimaraes, A.J.; Rivera, J.; Casadevall, A. Evidence for branching in cryptococcal capsular polysaccharides and consequences on its biological activity. Mol. Microbiol. 2011, 79, 1101–1117. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-D.; Shin, S.; Panepinto, J.; Ramos, J.; Qiu, J.; Frases, S.; Albuquerque, P.; Cordero, R.J.B.; Zhang, N.; Himmelreich, U.; et al. A role for LHC1 in higher order structure and complement binding of the Cryptococcus neoformans capsule. PLoS Pathog. 2014, 10, e1004037. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza, O.; Telzak, A.; Bryan, R.A.; Dadachova, E.; Casadevall, A. The polysaccharide capsule of the pathogenic fungus Cryptococcus neoformans enlarges by distal growth and is rearranged during budding. Mol. Microbiol. 2006, 59, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Perfect, J. Cryptococcus Neoformans; ASM Press: Washington, DC, USA, 1998. [Google Scholar]
- Burnik, C.; Altintas, N.D.; Ozkaya, G.; Serter, T.; Selcuk, Z.T.; Firat, P.; Arikan, S.; Cuenca-Estrella, M.; Topeli, A. Acute respiratory distress syndrome due to Cryptococcus albidus pneumonia: Case report and review of the literature. Med. Mycol. 2007, 45, 469–473. [Google Scholar] [CrossRef] [PubMed]
- De Castro, L.E.; Sarraf, O.A.; Lally, J.M.; Sandoval, H.P.; Solomon, K.D.; Vroman, D.T. Cryptococcus albidus keratitis after corneal transplantation. Cornea 2005, 24, 882–883. [Google Scholar] [CrossRef] [PubMed]
- Garelick, J.M.; Khodabakhsh, A.J.; Lopez, Y.; Bamji, M.; Lister, M. Scleral ulceration caused by Cryptococcus albidus in a patient with acquired immune deficiency syndrome. Cornea 2004, 23, 730–731. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.B.; Bradley, S.F.; Kauffman, C.A. Fungaemia due to Cryptococcus laurentii and a review of non-neoformans cryptococcaemia. Mycoses 1998, 41, 277–280. [Google Scholar] [CrossRef] [PubMed]
- De Baets, S.; Du Laing, S.; Francois, C.; Vandamme, E.J. Optimization of exopolysaccharide production by tremella mesenterica NRRL Y-6158 through implementation of fed-batch fermentation. J. Ind. Microbiol. Biotechnol. 2002, 29, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Mittag, H. Fine structural investigation of malassezia furfur. II. The envelope of the yeast cells. Mycoses 1995, 38, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Thianprasit, M.; Thagerngpol, K. Rhinosporidiosis. Curr. Top. Med. Mycol. 1989, 3, 64–85. [Google Scholar] [PubMed]
- Melcher, G.P.; Reed, K.D.; Rinaldi, M.G.; Lee, J.W.; Pizzo, P.A.; Walsh, T.J. Demonstration of a cell wall antigen cross-reacting with cryptococcal polysaccharide in experimental disseminated trichosporonosis. J. Clin. Microbiol. 1991, 29, 192–196. [Google Scholar] [PubMed]
- Matsumoto, Y.; Yamada, M.; Yoshida, Y. Light-microscopical appearance and ultrastructure of blastocystis hominis, an intestinal parasite of man. Zentralbl. Bakteriol. Mikrobiol. Hyg. A. 1987, 264, 379–385. [Google Scholar] [CrossRef]
- Garrison, R.G.; Mirikitani, F.K. Electron cytochemical demonstration of the capsule of yeast-like sporothrix schenckii. Sabouraudia 1983, 21, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Araujo Gde, S.; Fonseca, F.L.; Pontes, B.; Torres, A.; Cordero, R.J.; Zancope-Oliveira, R.M.; Casadevall, A.; Viana, N.B.; Nimrichter, L.; Rodrigues, M.L.; et al. Capsules from pathogenic and non-pathogenic Cryptococcus spp. Manifest significant differences in structure and ability to protect against phagocytic cells. PLoS ONE 2012, 7, e29561. [Google Scholar] [CrossRef] [PubMed]
- Frases, S.; Nimrichter, L.; Viana, N.B.; Nakouzi, A.; Casadevall, A. Cryptococcus neoformans capsular polysaccharide and exopolysaccharide fractions manifest physical, chemical, and antigenic differences. Eukaryot. Cell 2008, 7, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Daffé, M.; Etienne, G. The capsule of mycobacterium tuberculosis and its implications for pathogenicity. Tuber. Lung Dis. 1999, 79, 153–169. [Google Scholar] [CrossRef] [PubMed]
- Robbins, J.B.; Schneerson, R. Planning for a second (23 valent) generation pneumococcal vaccine. With special reference to new developments in our understanding of the structure and biology of polysaccharides. Bull. Eur Physiopathol. Respir. 1983, 19, 215–226. [Google Scholar] [PubMed]
- Chu, C.; Schneerson, R.; Robbins, J.B.; Rastogi, S.C. Further studies on the immunogenicity of Haemophilus influenzae type b and pneumococcal type 6A polysaccharide-protein conjugates. Infect. Immun. 1983, 40, 245–256. [Google Scholar] [PubMed]
- Aliberti, S.; Mantero, M.; Mirsaeidi, M.; Blasi, F. The role of vaccination in preventing pneumococcal disease in adults. Clin. Microbiol. Infect. 2014, 20, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Pichichero, M.E. Protein carriers of conjugate vaccines: Characteristics, development, and clinical trials. Hum. Vaccines Immunother. 2013, 9, 2505–2523. [Google Scholar] [CrossRef] [PubMed]
- McClane, B.A.; Mietzner, T.A. Microbial pathogenesis; Fence Creek Publishing: Madison, CT, USA, 1999. [Google Scholar]
- Bottomley, M.J.; Serruto, D.; Safadi, M.A.; Klugman, K.P. Future challenges in the elimination of bacterial meningitis. Vaccine 2012, 30, B78–B86. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, P.B.; O’Brien, K.L.; Greenwood, B.; van de Beek, D. Effect of vaccines on bacterial meningitis worldwide. Lancet 2012, 380, 1703–1711. [Google Scholar] [CrossRef]
- Goodridge, H.S.; Reyes, C.N.; Becker, C.A.; Katsumoto, T.R.; Ma, J.; Wolf, A.J.; Bose, N.; Chan, A.S.; Magee, A.S.; Danielson, M.E.; et al. Activation of the innate immune receptor Dectin-1 upon formation of a “phagocytic synapse”. Nature 2011, 472, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Moalli, F.; Doni, A.; Deban, L.; Zelante, T.; Zagarella, S.; Bottazzi, B.; Romani, L.; Mantovani, A.; Garlanda, C. Role of complement and Fcγ receptors in the protective activity of the long pentraxin PTX3 against aspergillus fumigatus. Blood 2010, 116, 5170–5180. [Google Scholar] [CrossRef] [PubMed]
- Tomalka, J.; Ganesan, S.; Azodi, E.; Patel, K.; Majmudar, P.; Hall, B.A.; Fitzgerald, K.A.; Hise, A.G. A novel role for the NLRC4 inflammasome in mucosal defenses against the fungal pathogen Candida albicans. PLoS Pathog. 2011, 7, e1002379. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Z.; Tao, L.L.; Zhang, J.; Zhang, H.J.; Qu, J.M. Role of NOD2 in regulating the immune response to aspergillus fumigatus. Inflam. Res. 2012, 61, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Van der Meer, J.W.; van de Veerdonk, F.L.; Joosten, L.A.; Kullberg, B.J.; Netea, M.G. Severe Candida spp. Infections: New insights into natural immunity. Int. J. Antimicrob. Agents 2010, 36, S58–S62. [Google Scholar] [CrossRef] [PubMed]
- Wey, S.; Mori, M.; Pfaller, M.; Woolson, R.; Wenzel, R. Risk factors for hospital-acquired candidemia. A matched case-control study. Arch. Intern. Med. 1989, 149, 2349–2353. [Google Scholar]
- Cunha, C.; di Ianni, M.; Bozza, S.; Giovannini, G.; Zagarella, S.; Zelante, T.; D’Angelo, C.; Pierini, A.; Pitzurra, L.; Falzetti, F.; et al. Dectin-1 Y238X polymorphism associates with susceptibility to invasive aspergillosis in hematopoietic transplantation through impairment of both recipient- and donor-dependent mechanisms of antifungal immunity. Blood 2010, 116, 5394–5402. [Google Scholar] [CrossRef] [PubMed]
- Plantinga, T.S.; Johnson, M.D.; Scott, W.K.; van de Vosse, E.; Velez Edwards, D.R.; Smith, P.B.; Alexander, B.D.; Yang, J.C.; Kremer, D.; Laird, G.M.; et al. Toll-like receptor 1 polymorphisms increase susceptibility to candidemia. J. Infect. Dis. 2012, 205, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.; de Luca, A.; Bozza, S.; Cunha, C.; D’Angelo, C.; Moretti, S.; Perruccio, K.; Iannitti, R.G.; Fallarino, F.; Pierini, A.; et al. TLR3 essentially promotes protective class I-restricted memory CD8+ T-cell responses to Aspergillus fumigatus in hematopoietic transplanted patients. Blood 2012, 119, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Glocker, E.O.; Hennigs, A.; Nabavi, M.; Schaffer, A.A.; Woellner, C.; Salzer, U.; Pfeifer, D.; Veelken, H.; Warnatz, K.; Tahami, F.; et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N. Engl. J. Med. 2009, 361, 1727–1735. [Google Scholar] [CrossRef] [PubMed]
- Rambach, G.; Speth, C. Complement in Candida albicans infections. Front. Biosci. 2009, 1, 1–12. [Google Scholar]
- Gavino, C.; Cotter, A.; Lichtenstein, D.; Lejtenyi, D.; Fortin, C.; Legault, C.; Alirezaie, N.; Majewski, J.; Sheppard, D.C.; Behr, M.A.; et al. CARD9 deficiency and spontaneous central nervous system candidiasis: Complete clinical remission with GM-CSF therapy. Clin. Infect. Dis. 2014, 59, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Nemecek, J.C.; Wuthrich, M.; Klein, B.S. Global control of dimorphism and virulence in fungi. Science 2006, 312, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Klein, B.S.; Tebbets, B. Dimorphism and virulence in fungi. Curr. Opin. Microbiol. 2007, 10, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Finkel-Jimenez, B.; Wuthrich, M.; Brandhorst, T.; Klein, B.S. The WI-1 adhesin blocks phagocyte TNF-α production, imparting pathogenicity on Blastomyces dermatitidis. J. Immunol. 2001, 166, 2665–2673. [Google Scholar] [CrossRef] [PubMed]
- Batanghari, J.W.; Deepe, G.S., Jr.; di Cera, E.; Goldman, W.E. Histoplasma acquisition of calcium and expression of CBP1 during intracellular parasitism. Mol. Microbiol. 1998, 27, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Cross, C.E.; Bancroft, G.J. Ingestion of acapsular Cryptococcus neoformans occurs via mannose and β-glucan receptors, resulting in cytokine production and increased phagocytosis of the encapsulated form. Infect. Immun. 1995, 63, 2604–2611. [Google Scholar] [PubMed]
- Guo, C.; Chen, M.; Fa, Z.; Lu, A.; Fang, W.; Sun, B.; Chen, C.; Liao, W.; Meng, G. Acapsular Cryptococcus neoformans activates the NLRP3 inflammasome. Microb. Infect. 2014, 16, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Kozel, T.R. Non-encapsulated variant of Cryptococcus neoformans. II. Surface receptors for cryptococcal polysaccharide and their role in inhibition of phagocytosis by polysaccharide. Infect. Immun. 1977, 16, 99–106. [Google Scholar] [PubMed]
- Cross, C.E.; Collins, H.L.; Bancroft, G.J. CR3-dependent phagocytosis by murine macrophages: Different cytokines regulate ingestion of a defined CR3 ligand and complement-opsonized Cryptococcus Neoformans. Immunology 1997, 91, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Kozel, T.R.; Gotschlich, E.C. The capsule of Cryptococcus neoformans passively inhibits phagocytosis of the yeast by macrophages. J. Immunol. 1982, 129, 1675–1680. [Google Scholar] [PubMed]
- Granger, D.L.; Perfect, J.R.; Durack, D.T. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J. Clin. Investig. 1985, 76, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Stano, P.; Williams, V.; Villani, M.; Cymbalyuk, E.S.; Qureshi, A.; Huang, Y.; Morace, G.; Luberto, C.; Tomlinson, S.; del Poeta, M. App1: An antiphagocytic protein that binds to complement receptors 3 and 2. J. Immunol. 2009, 182, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Williamson, P. Lessons from cryptococcal laccase: From environmental saprophyte to pathogen. Curr. Fungal Infect. Rep. 2011, 5, 233–244. [Google Scholar] [CrossRef]
- Nakamura, K.; Miyazato, A.; Xiao, G.; Hatta, M.; Inden, K.; Aoyagi, T.; Shiratori, K.; Takeda, K.; Akira, S.; Saijo, S.; et al. Deoxynucleic acids from Cryptococcus neoformans activate myeloid dendritic cells via a TLR9-dependent pathway. J. Immunol. 2008, 180, 4067–4074. [Google Scholar] [CrossRef] [PubMed]
- Biondo, C.; Midiri, A.; Messina, L.; Tomasello, F.; Garufi, G.; Catania, M.R.; Bombaci, M.; Beninati, C.; Teti, G.; Mancuso, G. Myd88 and TLR2, but not TLR4, are required for host defense against Cryptococcus neoformans. Eur. J. Immunol. 2005, 35, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Kinjo, T.; Saijo, S.; Miyazato, A.; Adachi, Y.; Ohno, N.; Fujita, J.; Kaku, M.; Iwakura, Y.; Kawakami, K. Dectin-1 is not required for the host defense to Cryptococcus neoformans. Microbiol. Immunol. 2007, 51, 1115–1119. [Google Scholar] [CrossRef] [PubMed]
- Mershon-Shier, K.L.; Vasuthasawat, A.; Takahashi, K.; Morrison, S.L.; Beenhouwer, D.O. In vitro C3 deposition on Cryptococcus capsule occurs via multiple complement activation pathways. Mol. Immunol. 2011, 48, 2009–2018. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza, O.; Taborda, C.; Casadevall, A. The efficacy of complement-mediated phagocytosis of Cryptococcus neoformans is dependent on the location of C3 in the polysaccharide capsule and involves both direct and indirect C3-mediated interactions. Eur. J. Immunol. 2003, 33, 1957–1967. [Google Scholar] [CrossRef] [PubMed]
- Rohatgi, S.; Pirofski, L.A. Host immunity to Cryptococcus neoformans. Future Microbiol. 2015, 10, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.M.; Murphy, J.W. Intravascular cryptococcal culture filtrate (CneF) and its major component, glucuronoxylomannan, are potent inhibitors of leukocyte accumulation. Infect. Immun. 1995, 63, 770–778. [Google Scholar] [PubMed]
- Powderly, W.G. Fungal infections in patients infected with HIV. Mo. Med. 1990, 87, 348–350. [Google Scholar] [PubMed]
- Pyrgos, V.; Seitz, A.E.; Steiner, C.A.; Prevots, D.R.; Williamson, P.R. Epidemiology of Cryptococcal meningitis in the us: 1997–2009. PLoS ONE 2013, 8, e56269. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L.; Singh, N. Cryptococcus neoformans infection. Liv. Transpl. 2002, 8, 846–847. [Google Scholar] [CrossRef] [PubMed]
- Jowitt, S.N.; Love, E.M.; Yin, J.A.; Pumphrey, R.S. CD4 lymphocytopenia without HIV in patient with cryptococcal infection. Lancet 1991, 337, 500–501. [Google Scholar] [CrossRef]
- Vecchiarelli, A.; Pericolini, E.; Gabrielli, E.; Kenno, S.; Perito, S.; Cenci, E.; Monari, C. Elucidating the immunological function of the Cryptococcus neoformans capsule. Future Microbiol. 2013, 8, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Monari, C.; Pericolini, E.; Bistoni, G.; Casadevall, A.; Kozel, T.R.; Vecchiarelli, A. Cryptococcus neoformans capsular glucuronoxylomannan induces expression of Fas ligand in macrophages. J. Immunol. 2005, 174, 3461–3468. [Google Scholar] [CrossRef] [PubMed]
- Villena, S.N.; Pinheiro, R.O.; Pinheiro, C.S.; Nunes, M.P.; Takiya, C.M.; DosReis, G.A.; Previato, J.O.; Mendonca-Previato, L.; Freire-de-Lima, C.G. Capsular polysaccharides galactoxylomannan and glucuronoxylomannan from Cryptococcus neoformans induce macrophage apoptosis mediated by Fas ligand. Cell. Microbiol. 2008, 10, 1274–1285. [Google Scholar] [CrossRef] [PubMed]
- Piccioni, M.; Monari, C.; Bevilacqua, S.; Perito, S.; Bistoni, F.; Kozel, T.R.; Vecchiarelli, A. A critical role for FcγIIb in up-regulation of Fas ligand induced by a microbial polysaccharide. Clin. Exp. Immunol. 2011, 165, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Vecchiarelli, A.; Retini, C.; Monari, C.; Tascini, C.; Bistoni, F.; Kozel, T.R. Purified capsular polysaccharide of Cryptococcus neoformans induces interleukin-10 secretion by human monocytes. Infect. Immun. 1996, 64, 2846–2849. [Google Scholar] [PubMed]
- Chiapello, L.S.; Baronetti, J.L.; Garro, A.P.; Spesso, M.F.; Masih, D.T. Cryptococcus neoformans glucuronoxylomannan induces macrophage apoptosis mediated by nitric oxide in a caspase-independent pathway. Int. Immunol. 2008, 20, 1527–1541. [Google Scholar] [CrossRef] [PubMed]
- Rosen, L.B.; Freeman, A.F.; Yang, L.M.; Jutivorakool, K.; Olivier, K.N.; Angkasekwinai, N.; Suputtamongkol, Y.; Bennett, J.E.; Pyrgos, V.; Williamson, P.R.; et al. Anti-GM-CSF autoantibodies in patients with cryptococcal meningitis. J. Immunol. 2013, 190, 3959–3966. [Google Scholar] [CrossRef] [PubMed]
- Saijo, T.; Chen, J.; Chen, S.C.; Rosen, L.B.; Yi, J.; Sorrell, T.C.; Bennett, J.E.; Holland, S.M.; Browne, S.K.; Kwon-Chung, K.J. Anti-granulocyte-macrophage colony-stimulating factor autoantibodies are a risk factor for central nervous system infection by Cryptococcus gattii in otherwise immunocompetent patients. MBio 2014, 5, e00912–e00914. [Google Scholar] [CrossRef] [PubMed]
- Antachopoulos, C.; Walsh, T.J.; Roilides, E. Fungal infections in primary immunodeficiencies. Euro. J. Pediatr. 2007, 166, 1099–1117. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, K.; Metzger, B.; Hanau, L.H.; Guh, A.; Rucker, L.; Badri, S.; Pirofski, L.A. Igm+ memory b cell expression predicts HIV-associated cryptococcosis status. J. Infect. Dis. 2009, 200, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.M.; Murphy, J.W. Cryptococcal polysaccharides induce l-selectin shedding and tumor necrosis factor receptor loss from the surface of human neutrophils. J. Clin. Investig. 1996, 97, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, H.; Kagaya, K.; Fukazawa, Y. Anti-chemotactic activity of capsular polysaccharide of Cryptococcus neoformans in vitro. Microbiol. Immunol. 1997, 41, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Ellerbroek, P.M.; Hoepelman, A.I.; Wolbers, F.; Zwaginga, J.J.; Coenjaerts, F.E. Cryptococcal glucuronoxylomannan inhibits adhesion of neutrophils to stimulated endothelium in vitro by affecting both neutrophils and endothelial cells. Infect. Immun. 2002, 70, 4762–4771. [Google Scholar] [CrossRef] [PubMed]
- Vinh, D.C.; Patel, S.Y.; Uzel, G.; Anderson, V.L.; Freeman, A.F.; Olivier, K.N.; Spalding, C.; Hughes, S.; Pittaluga, S.; Raffeld, M.; et al. Autosomal dominant and sporadic monocytopenia with susceptibility to mycobacteria, fungi, papillomaviruses, and myelodysplasia. Blood 2010, 115, 1519–1529. [Google Scholar] [CrossRef] [PubMed]
- Van der Poll, T.; Opal, S.M. Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet 2009, 374, 1543–1556. [Google Scholar] [CrossRef]
- Carroll, L. The Penguin Complete Lewis Carroll; Penguin: London, UK, 1982. [Google Scholar]
- Jack, R.S. Evolution of immunity and pathogens. Results Probl. Cell Differ. 2015, 57, 1–20. [Google Scholar] [PubMed]
- Li, Y.; Gierahn, T.; Thompson, C.M.; Trzcinski, K.; Ford, C.B.; Croucher, N.; Gouveia, P.; Flechtner, J.B.; Malley, R.; Lipsitch, M. Distinct effects on diversifying selection by two mechanisms of immunity against Streptococcus pneumoniae. PLoS Pathog. 2012, 8, e1002989. [Google Scholar] [CrossRef] [PubMed]
- Andam, C.P.; Hanage, W.P. Mechanisms of genome evolution of Streptococcus. Infect. Geneti. Evolut. 2015, 33, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Croucher, N.J.; Hanage, W.P.; Harris, S.R.; McGee, L.; van der Linden, M.; de Lencastre, H.; Sa-Leao, R.; Song, J.H.; Ko, K.S.; Beall, B.; et al. Variable recombination dynamics during the emergence, transmission and ‘disarming’ of a multidrug-resistant pneumococcal clone. BMC Biol. 2014, 12, 49. [Google Scholar] [CrossRef] [PubMed]
- Croucher, N.J.; Harris, S.R.; Fraser, C.; Quail, M.A.; Burton, J.; van der Linden, M.; McGee, L.; von Gottberg, A.; Song, J.H.; Ko, K.S.; et al. Rapid pneumococcal evolution in response to clinical interventions. Science 2011, 331, 430–434. [Google Scholar] [CrossRef] [PubMed]
- Struve, C.; Roe, C.C.; Stegger, M.; Stahlhut, S.G.; Hansen, D.S.; Engelthaler, D.M.; Andersen, P.S.; Driebe, E.M.; Keim, P.; Krogfelt, K.A. Mapping the evolution of hypervirulent Klebsiella pneumoniae. MBio 2015, 6, e00630. [Google Scholar] [CrossRef] [PubMed]
- Ajello, L. Occurrence of Cryptococcus neoformans in soils. Am. J. Hyg. 1958, 67, 72–77. [Google Scholar] [PubMed]
- Warpeha, K.M.; Park, Y.D.; Williamson, P.R. Susceptibility of intact germinating arabidopsis thaliana to the human fungal pathogen Cryptococcus. Appl. Environ .Microbiol. 2013, 79, 2979–2988. [Google Scholar] [CrossRef] [PubMed]
- Balankura, P. Isolation of Cryptococcus neoformans from soil contaminated with pigeon droppings in bangkok. J. Med. Assoc. Thai. 1974, 57, 158–159. [Google Scholar] [PubMed]
- Gales, N.; Wallace, G.; Dickson, J. Pulmonary cryptococcosis in a striped dolphin (Stenella coeruleoalba). J. Wildl Dis. 1985, 21, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.G.; Padhye, A.A.; van Bonn, W.; Jensen, E.; Brandt, M.E.; Ridgway, S.H. Cryptococcosis in a bottlenose dolphin (Tursiops truncatus) caused by Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 2002, 40, 721–724. [Google Scholar] [CrossRef] [PubMed]
- Perfect, J.R. The impact of the host on fungal infections. Am. J. Med. 2012, 125, S39–S51. [Google Scholar] [CrossRef] [PubMed]
- Steenbergen, J.N.; Shuman, H.A.; Casadevall, A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. USA 2001, 98, 15245–15250. [Google Scholar] [CrossRef] [PubMed]
- Steenbergen, J.N.; Nosanchuk, J.D.; Malliaris, S.D.; Casadevall, A. Interaction of Blastomyces dermatitidis, sporothrix schenckii, and Histoplasma capsulatum with Acanthamoeba castellanii. Infect. Immun. 2004, 72, 3478–3488. [Google Scholar] [CrossRef] [PubMed]
- Valdez, P.A.; Vithayathil, P.J.; Janelsins, B.M.; Shaffer, A.L.; Williamson, P.R.; Datta, S.K. Prostaglandin E2 suppresses antifungal immunity by inhibiting interferon regulatory factor 4 function and interleukin-17 expression in T cells. Immunity 2012, 36, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.; Boyle, C. The endophytic continuum. Mycol. Res. 2005, 109, 661–686. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Chen, S.H.; Qiu, J.; Bennett, J.E.; Myers, T.G.; Williamson, P.R. Microevolution during serial mouse passage demonstrates FRE3 as a virulence adaptation gene in Cryptococcus neoformans. MBio 2014, 5, e00941–e00914. [Google Scholar] [CrossRef] [PubMed]
- Janbon, G.; Ormerod, K.L.; Paulet, D.; Byrnes, E.J., 3rd; Yadav, V.; Chatterjee, G.; Mullapudi, N.; Hon, C.C.; Billmyre, R.B.; Brunel, F.; et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex rna expression and microevolution leading to virulence attenuation. PLoS Genet. 2014, 10, e1004261. [Google Scholar] [CrossRef] [PubMed]
- Billmyre, R.B.; Croll, D.; Li, W.; Mieczkowski, P.; Carter, D.A.; Cuomo, C.A.; Kronstad, J.W.; Heitman, J. Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution. MBio 2014, 5, e01494–e01414. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, Y.-D.; Williamson, P.R. Masking the Pathogen: Evolutionary Strategies of Fungi and Their Bacterial Counterparts. J. Fungi 2015, 1, 397-421. https://doi.org/10.3390/jof1030397
Park Y-D, Williamson PR. Masking the Pathogen: Evolutionary Strategies of Fungi and Their Bacterial Counterparts. Journal of Fungi. 2015; 1(3):397-421. https://doi.org/10.3390/jof1030397
Chicago/Turabian StylePark, Yoon-Dong, and Peter R. Williamson. 2015. "Masking the Pathogen: Evolutionary Strategies of Fungi and Their Bacterial Counterparts" Journal of Fungi 1, no. 3: 397-421. https://doi.org/10.3390/jof1030397
APA StylePark, Y.-D., & Williamson, P. R. (2015). Masking the Pathogen: Evolutionary Strategies of Fungi and Their Bacterial Counterparts. Journal of Fungi, 1(3), 397-421. https://doi.org/10.3390/jof1030397