Deciphering Acute Coronary Syndromes Pathobiology Through Proteomics
Abstract
1. Introduction
2. Identification of ACS Circulating Biomarkers Through Untargeted MS-Based Technologies
2.1. Proteomics of Secretory Vesicles
2.2. Proteomics of Intraluminal Thrombus
2.3. Urine Proteomics
2.4. Lipoproteomics
3. Dissecting Atherosclerotic Plaque Through Proteomics
3.1. Proteomics of Coronary Plaque Extracts
3.2. Proteomics of Plaque Secretomes
4. Conclusions
Author Contributions
Funding

Conflicts of Interest
Abbreviations
| CVD | cardiovascular disease |
| CAD | coronary artery disease |
| CHD | coronary heart disease |
| ACS | acute coronary syndrome |
| AMI | acute myocardial infarction |
| NSTEMI | non-ST-segment elevation myocardial infarction |
| STEMI | ST-segment elevation myocardial infarction |
| hs-CRP | high-sensitivity C-reactive protein |
| H-FABP | high-sensitivity C-reactive protein |
| CK-MB | creatine kinase-MB isoform |
| hs-cTn | high-sensitivity cardiac troponin |
| VSMC | vascular smooth muscle cell |
| VLDL | very low density lipoprotein |
| LDL | low-density lipoprotein |
| HDL | high-density lipoprotein |
| ECM | extracellular matrix |
| 1-DE | one-dimensional electrophoresis |
| 2-DE | two-dimensional electrophoresis |
| MS | mass spectrometry |
| LCM | laser capture microdissection |
| MALDI | matrix-assisted laser desorption ionization |
| ESI | electrospray ionization |
| SELDI | surface-enhanced laser desorption/ionization |
| TOF | time of flight |
| LC-MS/MS | liquid chromatography coupled to MS/MS |
| 2D-DIGE | two-dimensional difference gel electrophoresis |
| LTQ-FT MS | hybrid mass spectrometer consisting of a linear ion trap mass spectrometer, LTQ XL, combined with a Fourier transform ion cyclotron resonance mass spectrometer |
| Ig | immunoglobulin |
| iTRAQ | isobaric tag for relative and absolute quantitation |
| Apo | apolipoprotein |
| α1-ACT | α1-antichymotrypsin |
| SAA | serum amyloid A protein |
| CRP | C-reactive protein |
| STEACS | ST-segment elevation acute coronary syndrome |
| NSTEACS | non-ST-segment elevation acute coronary syndrome |
| Lp(a) | lipoprotein(a) |
| LC-FTICR MS | liquid chromatography/Fourier transform ion cyclotron resonance mass spectrometry |
| PTCA | percutaneous transluminal coronary angioplasty |
| CHF | chronic heart failure |
| SCX/RP | strong cation exchange reverse |
| GELS | gelsolin |
| MMP | matrix metalloproteinase |
| SDS-PAGE | sodium dodecyl-sulphate polyacrylamide gel electrophoresis |
| PTMs | post-translational modification |
| pI | isoelectric point |
| MW | molecular weight |
References
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update from the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- Fahed, A.C.; Jang, I.-K. Plaque Erosion and Acute Coronary Syndromes: Phenotype, Molecular Characteristics and Future Directions. Nat. Rev. Cardiol. 2021, 18, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Byrne, R.A.; Rossello, X.; Coughlan, J.J.; Barbato, E.; Berry, C.; Chieffo, A.; Claeys, M.J.; Dan, G.-A.; Dweck, M.R.; Galbraith, M.; et al. 2023 ESC Guidelines for the Management of Acute Coronary Syndromes. Eur. Heart J. Acute Cardiovasc. Care 2024, 13, 55–161. [Google Scholar] [CrossRef]
- Almeida, I.; Chin, J.; Santos, H.; Miranda, H.; Santos, M.; Sá, C.; Almeida, S.; Sousa, C.; Almeida, L.; National Cardiology Data Collection Center, P.S. of C.C.P.N.R. of A.C.S.I. Prognostic Value of Brain Natriuretic Peptide in ST-Elevation Myocardial Infarction Patients: A Portuguese Registry. Rev. Port. Cardiol. 2022, 41, 87–95. [Google Scholar] [CrossRef]
- Ikeda, S.; Shinohara, K.; Enzan, N.; Matsushima, S.; Tohyama, T.; Funakoshi, K.; Kishimoto, J.; Itoh, H.; Komuro, I.; Tsutsui, H. Serial Measurement of B-Type Natriuretic Peptide and Future Cardiovascular Events in Patients with Type 2 Diabetes Mellitus without Known Cardiovascular Disease. Int. J. Cardiol. 2022, 356, 98–104. [Google Scholar] [CrossRef]
- Kamath, D.Y.; Xavier, D.; Sigamani, A.; Pais, P. High Sensitivity C-Reactive Protein (HsCRP) & Cardiovascular Disease: An Indian Perspective. Indian J. Med. Res. 2015, 142, 261–268. [Google Scholar] [CrossRef]
- Polyakova, E.A.; Mikhaylov, E.N. The Prognostic Role of High-Sensitivity C-Reactive Protein in Patients with Acute Myocardial Infarction. J. Geriatr. Cardiol. 2020, 17, 379–383. [Google Scholar] [CrossRef]
- Lee, H.S.; Lee, J.-H. Early Elevation of High-Sensitivity C-Reactive Protein as a Predictor for Cardiovascular Disease Incidence and All-Cause Mortality: A Landmark Analysis. Sci. Rep. 2023, 13, 14118. [Google Scholar] [CrossRef]
- Zhang, H.-W.; Jin, J.-L.; Cao, Y.-X.; Liu, H.-H.; Zhang, Y.; Guo, Y.-L.; Wu, N.-Q.; Zhu, C.-G.; Gao, Y.; Xu, R.-X.; et al. Heart-Type Fatty Acid Binding Protein Predicts Cardiovascular Events in Patients with Stable Coronary Artery Disease: A Prospective Cohort Study. Ann. Transl. Med. 2020, 8, 1349. [Google Scholar] [CrossRef]
- Dong, T.; Zhu, W.; Yang, Z.; Matos Pires, N.M.; Lin, Q.; Jing, W.; Zhao, L.; Wei, X.; Jiang, Z. Advances in Heart Failure Monitoring: Biosensors Targeting Molecular Markers in Peripheral Bio-Fluids. Biosens. Bioelectron. 2024, 255, 116090. [Google Scholar] [CrossRef]
- Willemsen, R.T.A.; Dinant, G.J.; Glatz, J.F.C. Biomarkers of Myocardial Cell Damage: Heart-Type Fatty Acid Binding Protein (H-FABP) for the Early Evaluation of Suspected Acute Coronary Syndrome. In Biomarkers in Cardiovascular Disease; Springer: Dordrecht, The Netherlands, 2015; pp. 1–31. [Google Scholar]
- Solymoss, B.C.; Bourassa, M.G.; Fortier, A.; Théroux, P. Evaluation and Risk Stratification of Acute Coronary Syndromes Using a Low Cut-off Level of Cardiac Troponin T, Combined with CK-MB Mass Determination. Clin. Biochem. 2004, 37, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, Y.; Lewis, B.R.; Mehta, R.A.; Ola, O.; Knott, J.D.; De Michieli, L.; Akula, A.; Lobo, R.; Yang, E.H.; Gharacholou, S.M.; et al. Rapid Exclusion of Acute Myocardial Injury and Infarction with a Single High-Sensitivity Cardiac Troponin T in the Emergency Department: A Multicenter United States Evaluation. Circulation 2022, 145, 1708–1719. [Google Scholar] [CrossRef] [PubMed]
- Katsioupa, M.; Kourampi, I.; Oikonomou, E.; Tsigkou, V.; Theofilis, P.; Charalambous, G.; Marinos, G.; Gialamas, I.; Zisimos, K.; Anastasiou, A.; et al. Novel Biomarkers and Their Role in the Diagnosis and Prognosis of Acute Coronary Syndrome. Life 2023, 13, 1992. [Google Scholar] [CrossRef]
- Krychtiuk, K.A.; Newby, L.K. High-Sensitivity Cardiac Troponin Assays: Ready for Prime Time! Annu. Rev. Med. 2024, 75, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Mach, F.; Visseren, F.L.J.; Cater, N.B.; Salhi, N.; Soronen, J.; Ray, K.K.; Delgado, V.; Jukema, J.W.; Laufs, U.; Zamorano, J.-L.; et al. Addressing Residual Risk beyond Statin Therapy: New Targets in the Management of Dyslipidaemias-A Report from the European Society of Cardiology Cardiovascular Round Table. J. Clin. Lipidol. 2024, 18, e685–e700. [Google Scholar] [CrossRef]
- Welt, F.G.P. CABG versus PCI—End of the Debate? N. Engl. J. Med. 2022, 386, 185–187. [Google Scholar] [CrossRef]
- Libby, P.; Buring, J.E.; Badimon, L.; Hansson, G.K.; Deanfield, J.; Bittencourt, M.S.; Tokgözoğlu, L.; Lewis, E.F. Atherosclerosis. Nat. Rev. Dis. Primers 2019, 5, 56. [Google Scholar] [CrossRef]
- Machin, D.R.; Phuong, T.T.; Donato, A.J. The Role of the Endothelial Glycocalyx in Advanced Age and Cardiovascular Disease. Curr. Opin. Pharmacol. 2019, 45, 66–71. [Google Scholar] [CrossRef]
- Lepedda, A.J.; Nieddu, G.; Formato, M.; Baker, M.B.; Fernández-Pérez, J.; Moroni, L. Glycosaminoglycans: From Vascular Physiology to Tissue Engineering Applications. Front. Chem. 2021, 9, 680836. [Google Scholar] [CrossRef]
- Elmarasi, M.; Elmakaty, I.; Elsayed, B.; Elsayed, A.; Zein, J.A.; Boudaka, A.; Eid, A.H. Phenotypic Switching of Vascular Smooth Muscle Cells in Atherosclerosis, Hypertension, and Aortic Dissection. J. Cell Physiol. 2024, 239, e31200. [Google Scholar] [CrossRef]
- Borén, J.; Packard, C.J.; Binder, C.J. Apolipoprotein B-Containing Lipoproteins in Atherogenesis. Nat. Rev. Cardiol. 2025, 22, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Mendis, S.; Nordet, P.; Fernandez-Britto, J.E.; Sternby, N. Atherosclerosis in Children and Young Adults: An Overview of the World Health Organization and International Society and Federation of Cardiology Study on Pathobiological Determinants of Atherosclerosis in Youth Study (1985–1995). Glob. Heart 2005, 1, 3. [Google Scholar] [CrossRef]
- Hansson, G.K.; Libby, P.; Tabas, I. Inflammation and Plaque Vulnerability. J. Intern. Med. 2015, 278, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Gallone, G.; Bellettini, M.; Gatti, M.; Tore, D.; Bruno, F.; Scudeler, L.; Cusenza, V.; Lanfranchi, A.; Angelini, A.; de Filippo, O.; et al. Coronary Plaque Characteristics Associated with Major Adverse Cardiovascular Events in Atherosclerotic Patients and Lesions: A Systematic Review and Meta-Analysis. JACC Cardiovasc. Imaging 2023, 16, 1584–1604. [Google Scholar] [CrossRef]
- Libby, P.; Pasterkamp, G.; Crea, F.; Jang, I.-K. Reassessing the Mechanisms of Acute Coronary Syndromes. Circ. Res. 2019, 124, 150–160. [Google Scholar] [CrossRef]
- Chi, G.; Lee, J.J.; Kazmi, S.H.A.; Fitzgerald, C.; Memar Montazerin, S.; Kalayci, A.; Korjian, S.; Heise, M.; Deckelbaum, L.I.; Libby, P.; et al. Early and Late Recurrent Cardiovascular Events among High-Risk Patients with an Acute Coronary Syndrome: Meta-Analysis of Phase III Studies and Implications on Trial Design. Clin. Cardiol. 2022, 45, 299–307. [Google Scholar] [CrossRef]
- Formato, M.; Farina, M.; Spirito, R.; Maggioni, M.; Guarino, A.; Cherchi, G.M.; Biglioli, P.; Edelstein, C.; Scanu, A.M. Evidence for a Proinflammatory and Proteolytic Environment in Plaques from Endarterectomy Segments of Human Carotid Arteries. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 129–135. [Google Scholar] [CrossRef]
- Lepedda, A.J.; Cigliano, A.; Cherchi, G.M.; Spirito, R.; Maggioni, M.; Carta, F.; Turrini, F.; Edelstein, C.; Scanu, A.M.; Formato, M. A Proteomic Approach to Differentiate Histologically Classified Stable and Unstable Plaques from Human Carotid Arteries. Atherosclerosis 2009, 203, 112–118. [Google Scholar] [CrossRef]
- Lepedda, A.J.; Formato, M. Oxidative Modifications in Advanced Atherosclerotic Plaques: A Focus on In Situ Protein Sulfhydryl Group Oxidation. Oxidative Med. Cell. Longev. 2020, 2020, 6169825. [Google Scholar] [CrossRef]
- Lepedda, A.J.; Zinellu, A.; Nieddu, G.; Zinellu, E.; Carru, C.; Spirito, R.; Guarino, A.; De Muro, P.; Formato, M. Protein Sulfhydryl Group Oxidation and Mixed-Disulfide Modifications in Stable and Unstable Human Carotid Plaques. Oxidative Med. Cell. Longev. 2013, 2013, 403973. [Google Scholar] [CrossRef]
- Tuñón, J.; Martín-Ventura, J.L.; Blanco-Colio, L.M.; Lorenzo, O.; López, J.A.; Egido, J. Proteomic Strategies in the Search of New Biomarkers in Atherothrombosis. J. Am. Coll. Cardiol. 2010, 55, 2009–2016. [Google Scholar] [CrossRef] [PubMed]
- Herrington, D.M.; Mao, C.; Parker, S.J.; Fu, Z.; Yu, G.; Chen, L.; Venkatraman, V.; Fu, Y.; Wang, Y.; Howard, T.D.; et al. Proteomic Architecture of Human Coronary and Aortic Atherosclerosis. Circulation 2018, 137, 2741–2756. [Google Scholar] [CrossRef]
- Florens, N.; Calzada, C.; Delolme, F.; Page, A.; Guebre Egziabher, F.; Juillard, L.; Soulage, A.C.O. Proteomic Characterization of High-Density Lipoprotein Particles from Non-Diabetic Hemodialysis Patients. Toxins 2019, 11, 671. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.; Krebs, K.; Paré, G.; Milani, L. Pharmacogenomics in Stroke and Cardiovascular Disease: State of the Art. Stroke 2023, 54, 270–278. [Google Scholar] [CrossRef]
- Napoli, C.; Zullo, A.; Picascia, A.; Infante, T.; Mancini, F.P. Recent Advances in Proteomic Technologies Applied to Cardiovascular Disease. J. Cell. Biochem. 2013, 114, 7–20. [Google Scholar] [CrossRef]
- Yang, P.-Y.; Rui, Y.-C.; Yang, P.-Y.; Yu, Y.-L. Proteomic Analysis of Foam Cells. Methods Mol. Biol. 2007, 357, 297–305. [Google Scholar] [CrossRef]
- Yu, J.; Fu, J.; Zhang, X.; Cui, X.; Cheng, M. The Integration of Metabolomic and Proteomic Analyses Revealed Alterations in Inflammatory-Related Protein Metabolites in Endothelial Progenitor Cells Subjected to Oscillatory Shear Stress. Front. Physiol. 2022, 13, 825966. [Google Scholar] [CrossRef]
- Eligini, S.; Gianazza, E.; Mallia, A.; Ghilardi, S.; Banfi, C. Macrophage Phenotyping in Atherosclerosis by Proteomics. Int. J. Mol. Sci. 2023, 24, 2613. [Google Scholar] [CrossRef]
- Eslava-Alcon, S.; Extremera-García, M.J.; González-Rovira, A.; Rosal-Vela, A.; Rojas-Torres, M.; Beltran-Camacho, L.; Sanchez-Gomar, I.; Jiménez-Palomares, M.; Alonso-Piñero, J.A.; Conejero, R.; et al. Molecular Signatures of Atherosclerotic Plaques: An up-Dated Panel of Protein Related Markers. J. Proteom. 2020, 221, 103757. [Google Scholar] [CrossRef]
- de la Cuesta, F.; Alvarez-Llamas, G.; Maroto, A.S.; Barderas, M.G.; Vivanco, F. Laser Microdissection and Saturation Labeling DIGE Method for the Analysis of Human Arteries. Methods Mol. Biol. 2013, 1000, 21–32. [Google Scholar] [CrossRef]
- Rocchiccioli, S.; Pelosi, G.; Rosini, S.; Marconi, M.; Viglione, F.; Citti, L.; Ferrari, M.; Trivella, M.G.; Cecchettini, A. Secreted Proteins from Carotid Endarterectomy: An Untargeted Approach to Disclose Molecular Clues of Plaque Progression. J. Transl. Med. 2013, 11, 260. [Google Scholar] [CrossRef] [PubMed]
- Lygirou, V.; Latosinska, A.; Makridakis, M.; Mullen, W.; Delles, C.; Schanstra, J.P.; Zoidakis, J.; Pieske, B.; Mischak, H.; Vlahou, A. Plasma Proteomic Analysis Reveals Altered Protein Abundances in Cardiovascular Disease. J. Transl. Med. 2018, 16, 104. [Google Scholar] [CrossRef] [PubMed]
- Delles, C.; Diez, J.; Dominiczak, A.F. Urinary Proteomics in Cardiovascular Disease: Achievements, Limits and Hopes. Proteom. Clin. Appl. 2011, 5, 222–232. [Google Scholar] [CrossRef]
- Hoofnagle, A.N.; Heinecke, J.W. Lipoproteomics: Using Mass Spectrometry-Based Proteomics to Explore the Assembly, Structure, and Function of Lipoproteins. J. Lipid Res. 2009, 50, 1967–1975. [Google Scholar] [CrossRef]
- Shah, A.S.; Tan, L.; Long, J.L.; Davidson, W.S. Proteomic Diversity of High Density Lipoproteins: Our Emerging Understanding of Its Importance in Lipid Transport and Beyond. J. Lipid Res. 2013, 54, 2575–2585. [Google Scholar] [CrossRef]
- Mallia, A.; Gianazza, E.; Zoanni, B.; Brioschi, M.; Barbieri, S.S.; Banfi, C. Proteomics of Extracellular Vesicles: Update on Their Composition, Biological Roles and Potential Use as Diagnostic Tools in Atherosclerotic Cardiovascular Diseases. Diagnostics 2020, 10, 843. [Google Scholar] [CrossRef]
- Wagner, D.D.; Burger, P.C. Platelets in Inflammation and Thrombosis. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 2131–2137. [Google Scholar] [CrossRef]
- Thomas, M.R.; Storey, R.F. The Role of Platelets in Inflammation. Thromb. Haemost. 2015, 114, 449–458. [Google Scholar] [CrossRef]
- Gutmann, C.; Joshi, A.; Mayr, M. Platelet “-Omics” in Health and Cardiovascular Disease. Atherosclerosis 2020, 307, 87–96. [Google Scholar] [CrossRef]
- Pelosi, G.; Rocchiccioli, S.; Cecchettini, A.; Viglione, F.; Puntoni, M.; Parodi, O.; Capobianco, E.; Trivella, M.G. Inflammation Blood and Tissue Factors of Plaque Growth in an Experimental Model Evidenced by a Systems Approach. Front. Genet. 2014, 5, 70. [Google Scholar] [CrossRef]
- Rocchiccioli, S.; Cecchettini, A.; Ucciferri, N.; Terreni, M.; Viglione, F.; Trivella, M.G.; Citti, L.; Parodi, O.; Pelosi, G. Site-Specific Secretome Map Evidences VSMC-Related Markers of Coronary Atherosclerosis Grade and Extent in the Hypercholesterolemic Swine. Dis. Markers 2015, 2015, 465242. [Google Scholar] [CrossRef] [PubMed]
- Ekroos, K.; Jänis, M.; Tarasov, K.; Hurme, R.; Laaksonen, R. Lipidomics: A Tool for Studies of Atherosclerosis. Curr. Atheroscler. Rep. 2010, 12, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Stegemann, C.; Pechlaner, R.; Willeit, P.; Langley, S.R.; Mangino, M.; Mayr, U.; Menni, C.; Moayyeri, A.; Santer, P.; Rungger, G.; et al. Lipidomics Profiling and Risk of Cardiovascular Disease in the Prospective Population-Based Bruneck Study. Circulation 2014, 129, 1821–1831. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Yu, B.; Li, Q.; Guo, Y.; Koike, T.; Koike, Y.; Wu, Q.; Zhang, J.; Mao, L.; Tang, X.; et al. HDL Quality Features Revealed by Proteome–lipidome Connectivity Are Associated with Atherosclerotic Disease. J. Mol. Cell Biol. 2022, 14, mjac004. [Google Scholar] [CrossRef]
- Stegemann, C.; Drozdov, I.; Shalhoub, J.; Humphries, J.; Ladroue, C.; Didangelos, A.; Baumert, M.; Allen, M.; Davies, A.H.; Monaco, C.; et al. Comparative Lipidomics Profiling of Human Atherosclerotic Plaques. Circ. Cardiovasc. Genet. 2011, 4, 232–242. [Google Scholar] [CrossRef]
- Moerman, A.M.; Visscher, M.; Slijkhuis, N.; Van Gaalen, K.; Heijs, B.; Klein, T.; Burgers, P.C.; De Rijke, Y.B.; Van Beusekom, H.M.M.; Luider, T.M.; et al. Lipid Signature of Advanced Human Carotid Atherosclerosis Assessed by Mass Spectrometry Imaging. J. Lipid Res. 2021, 62, 100020. [Google Scholar] [CrossRef]
- Shelbaya, K.; Arthur, V.; Yang, Y.; Dorbala, P.; Buckley, L.; Claggett, B.; Skali, H.; Dufresne, L.; Yang, T.-Y.; Engert, J.C.; et al. Large-Scale Proteomics Identifies Novel Biomarkers and Circulating Risk Factors for Aortic Stenosis. J. Am. Coll. Cardiol. 2024, 83, 577–591. [Google Scholar] [CrossRef]
- Schwenk, J.M.; Omenn, G.S.; Sun, Z.; Campbell, D.S.; Baker, M.S.; Overall, C.M.; Aebersold, R.; Moritz, R.L.; Deutsch, E.W. The Human Plasma Proteome Draft of 2017: Building on the Human Plasma PeptideAtlas from Mass Spectrometry and Complementary Assays. J. Proteome Res. 2017, 16, 4299–4310. [Google Scholar] [CrossRef]
- Righetti, P.G.; Castagna, A.; Antonucci, F.; Piubelli, C.; Cecconi, D.; Campostrini, N.; Rustichelli, C.; Antonioli, P.; Zanusso, G.; Monaco, S.; et al. Proteome Analysis in the Clinical Chemistry Laboratory: Myth or Reality? Clin. Chim. Acta 2005, 357, 123–139. [Google Scholar] [CrossRef]
- Rice, S.J.; Belani, C.P. Characterization of Effective, Simple, and Low-Cost Precipitation Methods for Depleting Abundant Plasma Proteins to Enhance the Depth and Breadth of Plasma Proteomics. Proteomics 2024, 24, e2400071. [Google Scholar] [CrossRef]
- Ahsan, N.; Fornelli, L.; Najar, F.Z.; Gamagedara, S.; Hossan, M.R.; Rao, R.S.P.; Punyamurtula, U.; Bauer, A.; Yang, Z.; Foster, S.B.; et al. Proteomics Evaluation of Five Economical Commercial Abundant Protein Depletion Kits for Enrichment of Diseases-Specific Biomarkers from Blood Serum. Proteomics 2023, 23, e2300150. [Google Scholar] [CrossRef] [PubMed]
- Mateos-Cáceres, P.J.; García-Méndez, A.; López Farré, A.; Macaya, C.; Núñez, A.; Gómez, J.; Alonso-Orgaz, S.; Carrasco, C.; Burgos, M.E.; de Andrés, R.; et al. Proteomic Analysis of Plasma from Patients during an Acute Coronary Syndrome. J. Am. Coll. Cardiol. 2004, 44, 1578–1583. [Google Scholar] [CrossRef]
- Donahue, M.P.; Rose, K.; Hochstrasser, D.; Vonderscher, J.; Grass, P.; Chibout, S.-D.; Nelson, C.L.; Sinnaeve, P.; Goldschmidt-Clermont, P.J.; Granger, C.B. Discovery of Proteins Related to Coronary Artery Disease Using Industrial-Scale Proteomics Analysis of Pooled Plasma. Am. Heart J. 2006, 152, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Dardé, V.M.; de la Cuesta, F.; Dones, F.G.; Alvarez-Llamas, G.; Barderas, M.G.; Vivanco, F. Analysis of the Plasma Proteome Associated with Acute Coronary Syndrome: Does a Permanent Protein Signature Exist in the Plasma of ACS Patients? J. Proteome Res. 2010, 9, 4420–4432. [Google Scholar] [CrossRef]
- Prentice, R.L.; Paczesny, S.; Aragaki, A.; Amon, L.M.; Chen, L.; Pitteri, S.J.; McIntosh, M.; Wang, P.; Buson Busald, T.; Hsia, J.; et al. Novel Proteins Associated with Risk for Coronary Heart Disease or Stroke among Postmenopausal Women Identified by In-Depth Plasma Proteome Profiling. Genome Med. 2010, 2, 48. [Google Scholar] [CrossRef]
- Banfi, C.; Parolari, A.; Brioschi, M.; Barcella, S.; Loardi, C.; Centenaro, C.; Alamanni, F.; Mussoni, L.; Tremoli, E. Proteomic Analysis of Plasma from Patients Undergoing Coronary Artery Bypass Grafting Reveals a Protease/Antiprotease Imbalance in Favor of the Serpin α1-Antichymotrypsin. J. Proteome Res. 2010, 9, 2347–2357. [Google Scholar] [CrossRef]
- Maly, M.A.; Majek, P.; Reicheltova, Z.; Kotlin, R.; Suttnar, J.; Oravec, M.; Veselka, J.; Dyr, J.E. Proteomic Analysis of Plasma Samples from Acute Coronary Syndrome Patients—The Pilot Study. Int. J. Cardiol. 2012, 157, 126–128. [Google Scholar] [CrossRef]
- Dong, S.-Y.; Sun, X.-N.; Zeng, Q.; Xu, Y.; Sun, J.; Ma, L.-H. Proteomic Analysis of Adverse Outcomes in Patients with Acute Coronary Syndromes. Clin. Chim. Acta 2013, 416, 60–66. [Google Scholar] [CrossRef]
- Kristensen, L.P.; Larsen, M.R.; Mickley, H.; Saaby, L.; Diederichsen, A.C.P.; Lambrechtsen, J.; Rasmussen, L.M.; Overgaard, M. Plasma Proteome Profiling of Atherosclerotic Disease Manifestations Reveals Elevated Levels of the Cytoskeletal Protein Vinculin. J. Proteom. 2014, 101, 141–153. [Google Scholar] [CrossRef]
- Laborde, C.M.; Alonso-Orgaz, S.; Mourino-Alvarez, L.; Moreu, J.; Vivanco, F.; Padial, L.R.; Barderas, M.G. The Plasma Proteomic Signature as a Strategic Tool for Early Diagnosis of Acute Coronary Syndrome. Proteome Sci. 2014, 12, 43. [Google Scholar] [CrossRef]
- Xu, S.; Jiang, J.; Zhang, Y.; Chen, T.; Zhu, M.; Fang, C.; Mi, Y. Discovery of Potential Plasma Protein Biomarkers for Acute Myocardial Infarction via Proteomics. J. Thorac. Dis. 2019, 11, 3962–3972. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, M.; Chiariello, C.; Conte, E.; Castagna, A.; Robotti, E.; Gosetti, F.; Patrone, M.; Martinelli, N.; Bassi, A.; Cecconi, D.; et al. Plasma Proteome Profiles of Stable CAD Patients Stratified According to Total Apo C-III Levels. Proteom. Clin. Appl. 2019, 13, e1800023. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.; Park, S.H.; Mun, S.; Lee, J.; Kang, H.-G. Biomarker Discovery of Acute Coronary Syndrome Using Proteomic Approach. Molecules 2021, 26, 1136. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.; Mun, S.; Park, S.H.; Lee, J.; Kang, H.-G. Serum Biomarker Discovery Related to Pathogenesis in Acute Coronary Syndrome by Proteomic Approach. Biosci. Rep. 2021, 41, BSR20210344. [Google Scholar] [CrossRef]
- Núñez, E.; Fuster, V.; Gómez-Serrano, M.; Valdivielso, J.M.; Fernández-Alvira, J.M.; Martínez-López, D.; Rodríguez, J.M.; Bonzon-Kulichenko, E.; Calvo, E.; Alfayate, A.; et al. Unbiased Plasma Proteomics Discovery of Biomarkers for Improved Detection of Subclinical Atherosclerosis. EBioMedicine 2022, 76, 103874. [Google Scholar] [CrossRef]
- Pula, G.; Perera, S.; Prokopi, M.; Sidibe, A.; Boulanger, C.M.; Mayr, M. Proteomic Analysis of Secretory Proteins and Vesicles in Vascular Research. Proteom. Clin. Appl. 2008, 2, 882–891. [Google Scholar] [CrossRef]
- Boulanger, C.M.; Loyer, X.; Rautou, P.-E.; Amabile, N. Extracellular Vesicles in Coronary Artery Disease. Nat. Rev. Cardiol. 2017, 14, 259–272. [Google Scholar] [CrossRef]
- Olejarz, W.; Sadowski, K.; Radoszkiewicz, K. Extracellular Vesicles in Atherosclerosis: State of the Art. Int. J. Mol. Sci. 2023, 25, 388. [Google Scholar] [CrossRef]
- Distelmaier, K.; Adlbrecht, C.; Jakowitsch, J.; Winkler, S.; Dunkler, D.; Gerner, C.; Wagner, O.; Lang, I.M.; Kubicek, M. Local Complement Activation Triggers Neutrophil Recruitment to the Site of Thrombus Formation in Acute Myocardial Infarction. Thromb. Haemost. 2009, 102, 564–572. [Google Scholar] [CrossRef]
- Alonso-Orgaz, S.; Moreno-Luna, R.; López, J.A.; Gil-Dones, F.; Padial, L.R.; Moreu, J.; de la Cuesta, F.; Barderas, M.G. Proteomic Characterization of Human Coronary Thrombus in Patients with ST-Segment Elevation Acute Myocardial Infarction. J. Proteom. 2014, 109, 368–381. [Google Scholar] [CrossRef]
- Decramer, S.; Gonzalez de Peredo, A.; Breuil, B.; Mischak, H.; Monsarrat, B.; Bascands, J.-L.; Schanstra, J.P. Urine in Clinical Proteomics. Mol. Cell Proteom. 2008, 7, 1850–1862. [Google Scholar] [CrossRef] [PubMed]
- Thongboonkerd, V. Practical Points in Urinary Proteomics. J. Proteome Res. 2007, 6, 3881–3890. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Chen, Z.; Zheng, Y.; Wu, J. Urine Proteomics in Cardiovascular Disease: Advances in Biomarker Discovery and Clinical Applications. Expert. Rev. Proteom. 2024, 21, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Zimmerli, L.U.; Schiffer, E.; Zürbig, P.; Good, D.M.; Kellmann, M.; Mouls, L.; Pitt, A.R.; Coon, J.J.; Schmieder, R.E.; Peter, K.H.; et al. Urinary Proteomic Biomarkers in Coronary Artery Disease. Mol. Cell. Proteom. 2008, 7, 290–298. [Google Scholar] [CrossRef]
- von Zur Muhlen, C.; Schiffer, E.; Zuerbig, P.; Kellmann, M.; Brasse, M.; Meert, N.; Vanholder, R.C.; Dominiczak, A.F.; Chen, Y.C.; Mischak, H.; et al. Evaluation of Urine Proteome Pattern Analysis for Its Potential to Reflect Coronary Artery Atherosclerosis in Symptomatic Patients. J. Proteome Res. 2009, 8, 335–345. [Google Scholar] [CrossRef]
- Delles, C.; Schiffer, E.; von Zur Muhlen, C.; Peter, K.; Rossing, P.; Parving, H.-H.; Dymott, J.A.; Neisius, U.; Zimmerli, L.U.; Snell-Bergeon, J.K.; et al. Urinary Proteomic Diagnosis of Coronary Artery Disease: Identification and Clinical Validation in 623 Individuals. J. Hypertens. 2010, 28, 2316–2322. [Google Scholar] [CrossRef]
- Brown, C.E.; McCarthy, N.S.; Hughes, A.D.; Sever, P.; Stalmach, A.; Mullen, W.; Dominiczak, A.F.; Sattar, N.; Mischak, H.; Thom, S.; et al. Urinary Proteomic Biomarkers to Predict Cardiovascular Events. Proteom. Clin. Appl. 2015, 9, 610–617. [Google Scholar] [CrossRef]
- Jonas, A.; Phillips, M.C. Biochemistry of Lipids, Lipoproteins and Membranes, 5th ed.; Vance, D.E., Vance, J.E., Eds.; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar]
- Lucero, D.; Wolska, A.; Aligabi, Z.; Turecamo, S.; Remaley, A.T. Lipoprotein Assessment in the Twenty-First Century. Endocrinol. Metab. Clin. N. Am. 2022, 51, 459–481. [Google Scholar] [CrossRef]
- Fichtner, I.; Macchi, C.; Rizzuto, A.S.; Carugo, S.; Corsini, A.; Ruscica, M. Lipoprotein(a) and the Atherosclerotic Burden—Should We Wait for Clinical Trial Evidence before Taking Action? Atheroscler. Plus 2024, 58, 16–23. [Google Scholar] [CrossRef]
- Kronenberg, F.; Utermann, G. Lipoprotein(a): Resurrected by Genetics. J. Intern. Med. 2013, 273, 6–30. [Google Scholar] [CrossRef]
- Kumarapperuma, H.; Chia, Z.-J.; Malapitan, S.M.; Wight, T.N.; Little, P.J.; Kamato, D. Response to Retention Hypothesis as a Source of Targets for Arterial Wall-Directed Therapies to Prevent Atherosclerosis: A Critical Review. Atherosclerosis 2024, 397, 118552. [Google Scholar] [CrossRef] [PubMed]
- Linton, M.F.; Yancey, P.G.; Tao, H.; Davies, S.S. HDL Function and Atherosclerosis: Reactive Dicarbonyls as Promising Targets of Therapy. Circ. Res. 2023, 132, 1521–1545. [Google Scholar] [CrossRef] [PubMed]
- Kajani, S.; Curley, S.; McGillicuddy, F.C. Unravelling HDL-Looking beyond the Cholesterol Surface to the Quality Within. Int. J. Mol. Sci. 2018, 19, 1971. [Google Scholar] [CrossRef] [PubMed]
- Vaisar, T.; Pennathur, S.; Green, P.S.; Gharib, S.A.; Hoofnagle, A.N.; Cheung, M.C.; Byun, J.; Vuletic, S.; Kassim, S.; Singh, P.; et al. Shotgun Proteomics Implicates Protease Inhibition and Complement Activation in the Antiinflammatory Properties of HDL. J. Clin. Investig. 2007, 117, 746–756. [Google Scholar] [CrossRef]
- Holzer, M.; Ljubojevic-Holzer, S.; Souza Junior, D.R.; Stadler, J.T.; Rani, A.; Scharnagl, H.; Ronsein, G.E.; Marsche, G. HDL Isolated by Immunoaffinity, Ultracentrifugation, or Precipitation Is Compositionally and Functionally Distinct. J. Lipid Res. 2022, 63, 100307. [Google Scholar] [CrossRef]
- Havel, R.J.; Eder, H.A.; Bragdon, J.H. The Distribution and Chemical Composition of Ultracentrifugally Separated Lipoproteins in Human Serum. J. Clin. Investig. 1955, 34, 1345–1353. [Google Scholar] [CrossRef]
- Ståhlman, M.; Davidsson, P.; Kanmert, I.; Rosengren, B.; Borén, J.; Fagerberg, B.; Camejo, G. Proteomics and Lipids of Lipoproteins Isolated at Low Salt Concentrations in D2O/Sucrose or in KBr. J. Lipid Res. 2008, 49, 481–490. [Google Scholar] [CrossRef]
- Böttcher, A.; Schlosser, J.; Kronenberg, F.; Dieplinger, H.; Knipping, G.; Lackner, K.J.; Schmitz, G. Preparative Free-Solution Isotachophoresis for Separation of Human Plasma Lipoproteins: Apolipoprotein and Lipid Composition of HDL Subfractions. J. Lipid Res. 2000, 41, 905–915. [Google Scholar] [CrossRef]
- Collins, L.A.; Olivier, M. Quantitative Comparison of Lipoprotein Fractions Derived from Human Plasma and Serum by Liquid Chromatography-Tandem Mass Spectrometry. Proteome Sci. 2010, 8, 42. [Google Scholar] [CrossRef]
- Javeed, R.; Hussain, D.; Jabeen, F.; Saeed, A.; Fatima, B.; Ashiq, M.N.; Najam-Ul-Haq, M. Enrichment of HDL Proteome and Phospholipidome from Human Serum via IMAC/MOAC Affinity. Biomed. Chromatogr. 2020, 34, e4693. [Google Scholar] [CrossRef]
- Gordon, S.M.; Deng, J.; Lu, L.J.; Davidson, W.S. Proteomic Characterization of Human Plasma High Density Lipoprotein Fractionated by Gel Filtration Chromatography. J. Proteome Res. 2010, 9, 5239–5249. [Google Scholar] [CrossRef] [PubMed]
- Davidson, W.S.; Shah, A.S.; Sexmith, H.; Gordon, S.M. The HDL Proteome Watch: Compilation of Studies Leads to New Insights on HDL Function. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2022, 1867, 159072. [Google Scholar] [CrossRef] [PubMed]
- Finamore, F.; Nieddu, G.; Rocchiccioli, S.; Spirito, R.; Guarino, A.; Formato, M.; Lepedda, A.J. Apolipoprotein Signature of HDL and LDL from Atherosclerotic Patients in Relation with Carotid Plaque Typology: A Preliminary Report. Biomedicines 2021, 9, 1156. [Google Scholar] [CrossRef]
- Nieddu, G.; Michelucci, E.; Formato, M.; Ciampelli, C.; Obino, G.; Signore, G.; Di Giorgi, N.; Rocchiccioli, S.; Lepedda, A.J. Molecular Characterization of Plasma HDL, LDL, and VLDL Lipids Cargos from Atherosclerotic Patients with Advanced Carotid Lesions: A Preliminary Report. Int. J. Mol. Sci. 2022, 23, 12449. [Google Scholar] [CrossRef]
- Heller, M.; Stalder, D.; Schlappritzi, E.; Hayn, G.; Matter, U.; Haeberli, A. Mass Spectrometry-Based Analytical Tools for the Molecular Protein Characterization of Human Plasma Lipoproteins. Proteomics 2005, 5, 2619–2630. [Google Scholar] [CrossRef]
- Mancone, C.; Amicone, L.; Fimia, G.M.; Bravo, E.; Piacentini, M.; Tripodi, M.; Alonzi, T. Proteomic Analysis of Human Very Low-Density Lipoprotein by Two-Dimensional Gel Electrophoresis and MALDI-TOF/TOF. Proteomics 2007, 7, 143–154. [Google Scholar] [CrossRef]
- Collins, L.A.; Mirza, S.P.; Kissebah, A.H.; Olivier, M. Integrated Approach for the Comprehensive Characterization of Lipoproteins from Human Plasma Using FPLC and Nano-HPLC-Tandem Mass Spectrometry. Physiol. Genom. 2010, 40, 208–215. [Google Scholar] [CrossRef]
- Queiroz, K.C.S.; Tio, R.A.; Zeebregts, C.J.; Bijlsma, M.F.; Zijlstra, F.; Badlou, B.; de Vries, M.; Ferreira, C.V.; Spek, C.A.; Peppelenbosch, M.P.; et al. Human Plasma Very Low Density Lipoprotein Carries Indian Hedgehog. J. Proteome Res. 2010, 9, 6052–6059. [Google Scholar] [CrossRef]
- Sun, H.-Y.; Chen, S.-F.; Lai, M.-D.; Chang, T.-T.; Chen, T.-L.; Li, P.-Y.; Shieh, D.-B.; Young, K.-C. Comparative Proteomic Profiling of Plasma Very-Low-Density and Low-Density Lipoproteins. Clin. Chim. Acta 2010, 411, 336–344. [Google Scholar] [CrossRef]
- von Zychlinski, A.; Williams, M.; McCormick, S.; Kleffmann, T. Absolute Quantification of Apolipoproteins and Associated Proteins on Human Plasma Lipoproteins. J. Proteom. 2014, 106, 181–190. [Google Scholar] [CrossRef]
- Dashty, M.; Motazacker, M.M.; Levels, J.; de Vries, M.; Mahmoudi, M.; Peppelenbosch, M.P.; Rezaee, F. Proteome of Human Plasma Very Low-Density Lipoprotein and Low-Density Lipoprotein Exhibits a Link with Coagulation and Lipid Metabolism. Thromb. Haemost. 2014, 111, 518–530. [Google Scholar] [CrossRef]
- von Zychlinski, A.; Kleffmann, T.; Williams, M.J.A.; McCormick, S.P. Proteomics of Lipoprotein(a) Identifies a Protein Complement Associated with Response to Wounding. J. Proteom. 2011, 74, 2881–2891. [Google Scholar] [CrossRef] [PubMed]
- Bourgeois, R.; Girard, A.; Perrot, N.; Guertin, J.; Mitchell, P.L.; Couture, C.; Gotti, C.; Bourassa, S.; Poggio, P.; Mass, E.; et al. A Comparative Analysis of the Lipoprotein(a) and Low-Density Lipoprotein Proteomic Profiles Combining Mass Spectrometry and Mendelian Randomization. CJC Open 2021, 3, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Mueller, P.A.; Yerkes, E.; Bergstrom, P.; Rosario, S.; Hay, J.; Pamir, N. A Method for Lipoprotein (a) Isolation from a Small Volume of Plasma with Applications for Clinical Research. Sci. Rep. 2022, 12, 9138. [Google Scholar] [CrossRef]
- Green, P.S.; Vaisar, T.; Pennathur, S.; Kulstad, J.J.; Moore, A.B.; Marcovina, S.; Brunzell, J.; Knopp, R.H.; Zhao, X.-Q.; Heinecke, J.W. Combined Statin and Niacin Therapy Remodels the High-Density Lipoprotein Proteome. Circulation 2008, 118, 1259–1267. [Google Scholar] [CrossRef]
- Vaisar, T.; Mayer, P.; Nilsson, E.; Zhao, X.-Q.; Knopp, R.; Prazen, B.J. HDL in Humans with Cardiovascular Disease Exhibits a Proteomic Signature. Clin. Chim. Acta 2010, 411, 972–979. [Google Scholar] [CrossRef]
- Cubedo, J.; Padró, T.; García-Moll, X.; Pintó, X.; Cinca, J.; Badimon, L. Proteomic Signature of Apolipoprotein J in the Early Phase of New-Onset Myocardial Infarction. J. Proteome Res. 2011, 10, 211–220. [Google Scholar] [CrossRef]
- Alwaili, K.; Bailey, D.; Awan, Z.; Bailey, S.D.; Ruel, I.; Hafiane, A.; Krimbou, L.; Laboissiere, S.; Genest, J. The HDL Proteome in Acute Coronary Syndromes Shifts to an Inflammatory Profile. Biochim. Biophys. Acta 2012, 1821, 405–415. [Google Scholar] [CrossRef]
- Riwanto, M.; Rohrer, L.; Roschitzki, B.; Besler, C.; Mocharla, P.; Mueller, M.; Perisa, D.; Heinrich, K.; Altwegg, L.; von Eckardstein, A.; et al. Altered Activation of Endothelial Anti- and Proapoptotic Pathways by High-Density Lipoprotein from Patients with Coronary Artery Disease: Role of High-Density Lipoprotein-Proteome Remodeling. Circulation 2013, 127, 891–904. [Google Scholar] [CrossRef]
- Tan, Y.; Liu, T.R.; Hu, S.W.; Tian, D.; Li, C.; Zhong, J.K.; Sun, H.G.; Luo, T.T.; Lai, W.Y.; Guo, Z.-G. Acute Coronary Syndrome Remodels the Protein Cargo and Functions of High-Density Lipoprotein Subfractions. PLoS ONE 2014, 9, e94264. [Google Scholar] [CrossRef]
- Jorge, I.; Burillo, E.; Mesa, R.; Baila-Rueda, L.; Moreno, M.; Trevisan-Herraz, M.; Silla-Castro, J.C.; Camafeita, E.; Ortega-Muñoz, M.; Bonzon-Kulichenko, E.; et al. The Human HDL Proteome Displays High Inter-Individual Variability and Is Altered Dynamically in Response to Angioplasty-Induced Atheroma Plaque Rupture. J. Proteom. 2014, 106, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Huang, J.; Lee, H.; Guerrero, A.; Berglund, L.; Anuurad, E.; Lebrilla, C.B.; Zivkovic, A.M. Combined High-Density Lipoprotein Proteomic and Glycomic Profiles in Patients at Risk for Coronary Artery Disease. J. Proteome Res. 2015, 14, 5109–5118. [Google Scholar] [CrossRef] [PubMed]
- Oberbach, A.; Adams, V.; Schlichting, N.; Heinrich, M.; Kullnick, Y.; Lehmann, S.; Lehmann, S.; Feder, S.; Correia, J.C.; Mohr, F.-W.; et al. Proteome Profiles of HDL Particles of Patients with Chronic Heart Failure Are Associated with Immune Response and Also Include Bacteria Proteins. Clin. Chim. Acta 2016, 453, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.M.; Chung, J.H.; Playford, M.P.; Dey, A.K.; Sviridov, D.; Seifuddin, F.; Chen, Y.-C.; Pirooznia, M.; Chen, M.Y.; Mehta, N.N.; et al. High Density Lipoprotein Proteome Is Associated with Cardiovascular Risk Factors and Atherosclerosis Burden as Evaluated by Coronary CT Angiography. Atherosclerosis 2018, 278, 278–285. [Google Scholar] [CrossRef]
- Natarajan, P.; Collier, T.S.; Jin, Z.; Lyass, A.; Li, Y.; Ibrahim, N.E.; Mukai, R.; McCarthy, C.P.; Massaro, J.M.; D’Agostino, R.B.; et al. Association of an HDL Apolipoproteomic Score with Coronary Atherosclerosis and Cardiovascular Death. J. Am. Coll. Cardiol. 2019, 73, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Collier, T.S.; Dai, D.L.Y.; Chen, V.; Hollander, Z.; Ng, R.T.; McManus, B.M.; Balshaw, R.; Apostolidou, S.; Penn, M.S.; et al. Development and Validation of Apolipoprotein AI-Associated Lipoprotein Proteome Panel for the Prediction of Cholesterol Efflux Capacity and Coronary Artery Disease. Clin. Chem. 2019, 65, 282–290. [Google Scholar] [CrossRef]
- Camont, L.; Lhomme, M.; Rached, F.; Le Goff, W.; Nègre-Salvayre, A.; Salvayre, R.; Calzada, C.; Lagarde, M.; Chapman, M.J.; Kontush, A. Small, Dense High-Density Lipoprotein-3 Particles Are Enriched in Negatively Charged Phospholipids: Relevance to Cellular Cholesterol Efflux, Antioxidative, Antithrombotic, Anti-Inflammatory, and Antiapoptotic Functionalities. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 2715–2723. [Google Scholar] [CrossRef]
- Borodzicz, S.; Czarzasta, K.; Kuch, M.; Cudnoch-Jedrzejewska, A. Sphingolipids in Cardiovascular Diseases and Metabolic Disorders. Lipids Health Dis. 2015, 14, 55. [Google Scholar] [CrossRef]
- Dang, V.T.; Huang, A.; Zhong, L.H.; Shi, Y.; Werstuck, G.H. Comprehensive Plasma Metabolomic Analyses of Atherosclerotic Progression Reveal Alterations in Glycerophospholipid and Sphingolipid Metabolism in Apolipoprotein E-Deficient Mice. Sci. Rep. 2016, 6, 35037. [Google Scholar] [CrossRef]
- Hilvo, M.; Meikle, P.J.; Pedersen, E.R.; Tell, G.S.; Dhar, I.; Brenner, H.; Schöttker, B.; Lääperi, M.; Kauhanen, D.; Koistinen, K.M.; et al. Development and Validation of a Ceramide- and Phospholipid-Based Cardiovascular Risk Estimation Score for Coronary Artery Disease Patients. Eur. Heart J. 2020, 41, 371–380. [Google Scholar] [CrossRef]
- Mantovani, A.; Bonapace, S.; Lunardi, G.; Canali, G.; Dugo, C.; Vinco, G.; Calabria, S.; Barbieri, E.; Laaksonen, R.; Bonnet, F.; et al. Associations between Specific Plasma Ceramides and Severity of Coronary-Artery Stenosis Assessed by Coronary Angiography. Diabetes Metab. 2020, 46, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Poss, A.M.; Maschek, J.A.; Cox, J.E.; Hauner, B.J.; Hopkins, P.N.; Hunt, S.C.; Holland, W.L.; Summers, S.A.; Playdon, M.C. Machine Learning Reveals Serum Sphingolipids as Cholesterol-Independent Biomarkers of Coronary Artery Disease. J. Clin. Investig. 2020, 130, 1363–1376. [Google Scholar] [CrossRef] [PubMed]
- Di Giorgi, N.; Michelucci, E.; Smit, J.M.; Scholte, A.J.H.A.; El Mahdiui, M.; Knuuti, J.; Buechel, R.R.; Teresinska, A.; Pizzi, M.N.; Roque, A.; et al. A Specific Plasma Lipid Signature Associated with High Triglycerides and Low HDL Cholesterol Identifies Residual CAD Risk in Patients with Chronic Coronary Syndrome. Atherosclerosis 2021, 339, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Michelucci, E.; Giorgi, N.D.; Finamore, F.; Smit, J.M.; Scholte, A.J.H.A.; Signore, G.; Rocchiccioli, S. Lipid Biomarkers in Statin Users with Coronary Artery Disease Annotated by Coronary Computed Tomography Angiography. Sci. Rep. 2021, 11, 12899. [Google Scholar] [CrossRef]
- Meikle, P.J.; Wong, G.; Tsorotes, D.; Barlow, C.K.; Weir, J.M.; Christopher, M.J.; MacIntosh, G.L.; Goudey, B.; Stern, L.; Kowalczyk, A.; et al. Plasma Lipidomic Analysis of Stable and Unstable Coronary Artery Disease. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2723–2732. [Google Scholar] [CrossRef]
- Ellims, A.H.; Wong, G.; Weir, J.M.; Lew, P.; Meikle, P.J.; Taylor, A.J. Plasma Lipidomic Analysis Predicts Non-Calcified Coronary Artery Plaque in Asymptomatic Patients at Intermediate Risk of Coronary Artery Disease. Eur. Heart J. Cardiovasc. Imaging 2014, 15, 908–916. [Google Scholar] [CrossRef]
- Cheng, J.M.; Suoniemi, M.; Kardys, I.; Vihervaara, T.; de Boer, S.P.M.; Akkerhuis, K.M.; Sysi-Aho, M.; Ekroos, K.; Garcia-Garcia, H.M.; Oemrawsingh, R.M.; et al. Plasma Concentrations of Molecular Lipid Species in Relation to Coronary Plaque Characteristics and Cardiovascular Outcome: Results of the ATHEROREMO-IVUS Study. Atherosclerosis 2015, 243, 560–566. [Google Scholar] [CrossRef]
- Karjalainen, J.-P.; Mononen, N.; Hutri-Kähönen, N.; Lehtimäki, M.; Hilvo, M.; Kauhanen, D.; Juonala, M.; Viikari, J.; Kähönen, M.; Raitakari, O.; et al. New Evidence from Plasma Ceramides Links ApoE Polymorphism to Greater Risk of Coronary Artery Disease in Finnish Adults. J. Lipid Res. 2019, 60, 1622–1629. [Google Scholar] [CrossRef]
- Meikle, P.J.; Formosa, M.F.; Mellett, N.A.; Jayawardana, K.S.; Giles, C.; Bertovic, D.A.; Jennings, G.L.; Childs, W.; Reddy, M.; Carey, A.L.; et al. HDL Phospholipids, but Not Cholesterol Distinguish Acute Coronary Syndrome from Stable Coronary Artery Disease. J. Am. Heart Assoc. 2019, 8, e011792. [Google Scholar] [CrossRef]
- Ding, M.; Rexrode, K.M. A Review of Lipidomics of Cardiovascular Disease Highlights the Importance of Isolating Lipoproteins. Metabolites 2020, 10, 163. [Google Scholar] [CrossRef]
- Denimal, D.; Pais de Barros, J.-P.; Petit, J.-M.; Bouillet, B.; Vergès, B.; Duvillard, L. Significant Abnormalities of the HDL Phosphosphingolipidome in Type 1 Diabetes despite Normal HDL Cholesterol Concentration. Atherosclerosis 2015, 241, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Kostara, C.E.; Ferrannini, E.; Bairaktari, E.T.; Papathanasiou, A.; Elisaf, M.; Tsimihodimos, V. Early Signs of Atherogenic Features in the HDL Lipidomes of Normolipidemic Patients Newly Diagnosed with Type 2 Diabetes. Int. J. Mol. Sci. 2020, 21, 8835. [Google Scholar] [CrossRef] [PubMed]
- Kostara, C.E.; Karakitsou, K.S.; Florentin, M.; Bairaktari, E.T.; Tsimihodimos, V. Progressive, Qualitative, and Quantitative Alterations in HDL Lipidome from Healthy Subjects to Patients with Prediabetes and Type 2 Diabetes. Metabolites 2022, 12, 683. [Google Scholar] [CrossRef]
- Denimal, D.; Nguyen, A.; Pais de Barros, J.-P.; Bouillet, B.; Petit, J.-M.; Vergès, B.; Duvillard, L. Major Changes in the Sphingophospholipidome of HDL in Non-Diabetic Patients with Metabolic Syndrome. Atherosclerosis 2016, 246, 106–114. [Google Scholar] [CrossRef]
- Mocciaro, G.; D’Amore, S.; Jenkins, B.; Kay, R.; Murgia, A.; Herrera-Marcos, L.V.; Neun, S.; Sowton, A.P.; Hall, Z.; Palma-Duran, S.A.; et al. Lipidomic Approaches to Study HDL Metabolism in Patients with Central Obesity Diagnosed with Metabolic Syndrome. Int. J. Mol. Sci. 2022, 23, 6786. [Google Scholar] [CrossRef]
- Orsoni, A.; Thérond, P.; Tan, R.; Giral, P.; Robillard, P.; Kontush, A.; Meikle, P.J.; Chapman, M.J. Statin Action Enriches HDL3 in Polyunsaturated Phospholipids and Plasmalogens and Reduces LDL-Derived Phospholipid Hydroperoxides in Atherogenic Mixed Dyslipidemia. J. Lipid Res. 2016, 57, 2073–2087. [Google Scholar] [CrossRef]
- Kostara, C.E.; Tsimihodimos, V.; Elisaf, M.S.; Bairaktari, E.T. NMR-Based Lipid Profiling of High Density Lipoprotein Particles in Healthy Subjects with Low, Normal, and Elevated HDL-Cholesterol. J. Proteome Res. 2017, 16, 1605–1616. [Google Scholar] [CrossRef]
- Chen, Y.; Wen, S.; Jiang, M.; Zhu, Y.; Ding, L.; Shi, H.; Dong, P.; Yang, J.; Yang, Y. Atherosclerotic Dyslipidemia Revealed by Plasma Lipidomics on ApoE-/- Mice Fed a High-Fat Diet. Atherosclerosis 2017, 262, 78–86. [Google Scholar] [CrossRef]
- Takeda, H.; Izumi, Y.; Tamura, S.; Koike, T.; Koike, Y.; Shiomi, M.; Bamba, T. Lipid Profiling of Serum and Lipoprotein Fractions in Response to Pitavastatin Using an Animal Model of Familial Hypercholesterolemia. J. Proteome Res. 2020, 19, 1100–1108. [Google Scholar] [CrossRef]
- Chapman, M.J.; Orsoni, A.; Tan, R.; Mellett, N.A.; Nguyen, A.; Robillard, P.; Giral, P.; Thérond, P.; Meikle, P.J. LDL Subclass Lipidomics in Atherogenic Dyslipidemia: Effect of Statin Therapy on Bioactive Lipids and Dense LDL. J. Lipid Res. 2020, 61, 911–932. [Google Scholar] [CrossRef]
- Padro, T.; Vilahur, G.; Sánchez-Hernández, J.; Hernández, M.; Antonijoan, R.M.; Perez, A.; Badimon, L. Lipidomic Changes of LDL in Overweight and Moderately Hypercholesterolemic Subjects Taking Phytosterol- and Omega-3-Supplemented Milk. J. Lipid Res. 2015, 56, 1043–1056. [Google Scholar] [CrossRef] [PubMed]
- Sutter, I.; Velagapudi, S.; Othman, A.; Riwanto, M.; Manz, J.; Rohrer, L.; Rentsch, K.; Hornemann, T.; Landmesser, U.; von Eckardstein, A. Plasmalogens of High-Density Lipoproteins (HDL) Are Associated with Coronary Artery Disease and Anti-Apoptotic Activity of HDL. Atherosclerosis 2015, 241, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yang, J.S.; Lee, S.-H.; Moon, M.H. Analysis of Lipoprotein-Specific Lipids in Patients with Acute Coronary Syndrome by Asymmetrical Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography-Tandem Mass Spectrometry. J. Chromatogr. B Analyt Technol. Biomed. Life Sci. 2018, 1099, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Avela, H.F.; Sirén, H. Advances in Lipidomics. Clin. Chim. Acta 2020, 510, 123–141. [Google Scholar] [CrossRef]
- Ponnaiah, M.; Zakiev, E.; Lhomme, M.; Rached, F.; Camont, L.; Serrano, C.V.; Santos, R.D.; Chapman, M.J.; Orekhov, A.; Kontush, A. Acute Myocardial Infarction Preferentially Alters Low-Abundant, Long-Chain Unsaturated Phospholipid and Sphingolipid Species in Plasma High-Density Lipoprotein Subpopulations. Atheroscler. Plus 2024, 55, 21–30. [Google Scholar] [CrossRef]
- Hancock-Cerutti, W.; Lhomme, M.; Dauteuille, C.; Lecocq, S.; Chapman, M.J.; Rader, D.J.; Kontush, A.; Cuchel, M. Paradoxical Coronary Artery Disease in Humans with Hyperalphalipoproteinemia Is Associated with Distinct Differences in the High-Density Lipoprotein Phosphosphingolipidome. J. Clin. Lipidol. 2017, 11, 1192–1200.e3. [Google Scholar] [CrossRef]
- Giammanco, A.; Noto, D.; Barbagallo, C.M.; Nardi, E.; Caldarella, R.; Ciaccio, M.; Averna, M.R.; Cefalù, A.B. Hyperalphalipoproteinemia and Beyond: The Role of HDL in Cardiovascular Diseases. Life 2021, 11, 581. [Google Scholar] [CrossRef]
- Badimon, L.; Vilahur, G. Thrombosis Formation on Atherosclerotic Lesions and Plaque Rupture. J. Intern. Med. 2014, 276, 618–632. [Google Scholar] [CrossRef]
- Stary, H.C. Natural History and Histological Classification of Atherosclerotic Lesions: An Update. Arterioscler. Thromb. Vasc. Biol. 2000, 20, 1177–1178. [Google Scholar] [CrossRef]
- Virmani, R.; Kolodgie, F.D.; Burke, A.P.; Farb, A.; Schwartz, S.M. Lessons from Sudden Coronary Death: A Comprehensive Morphological Classification Scheme for Atherosclerotic Lesions. Arterioscler. Thromb. Vasc. Biol. 2000, 20, 1262–1275. [Google Scholar] [CrossRef]
- Nieddu, G.; Formato, M.; Lepedda, A.J. Searching for Atherosclerosis Biomarkers by Proteomics: A Focus on Lesion Pathogenesis and Vulnerability. Int. J. Mol. Sci. 2023, 24, 15175. [Google Scholar] [CrossRef] [PubMed]
- You, S.-A.; Archacki, S.R.; Angheloiu, G.; Moravec, C.S.; Rao, S.; Kinter, M.; Topol, E.J.; Wang, Q. Proteomic Approach to Coronary Atherosclerosis Shows Ferritin Light Chain as a Significant Marker: Evidence Consistent with Iron Hypothesis in Atherosclerosis. Physiol. Genom. 2003, 13, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Bagnato, C.; Thumar, J.; Mayya, V.; Hwang, S.-I.; Zebroski, H.; Claffey, K.P.; Haudenschild, C.; Eng, J.K.; Lundgren, D.H.; Han, D.K. Proteomics Analysis of Human Coronary Atherosclerotic Plaque: A Feasibility Study of Direct Tissue Proteomics by Liquid Chromatography and Tandem Mass Spectrometry. Mol. Cell Proteom. 2007, 6, 1088–1102. [Google Scholar] [CrossRef] [PubMed]
- de la Cuesta, F.; Alvarez-Llamas, G.; Maroto, A.S.; Donado, A.; Zubiri, I.; Posada, M.; Padial, L.R.; Pinto, A.G.; Barderas, M.G.; Vivanco, F. A Proteomic Focus on the Alterations Occurring at the Human Atherosclerotic Coronary Intima. Mol. Cell. Proteom. 2011, 10, M110.003517. [Google Scholar] [CrossRef]
- Lee, R.; Fischer, R.; Charles, P.D.; Adlam, D.; Valli, A.; Di Gleria, K.; Kharbanda, R.K.; Choudhury, R.P.; Antoniades, C.; Kessler, B.M.; et al. A Novel Workflow Combining Plaque Imaging, Plaque and Plasma Proteomics Identifies Biomarkers of Human Coronary Atherosclerotic Plaque Disruption. Clin. Proteom. 2017, 14, 22. [Google Scholar] [CrossRef]
- Stakhneva, E.M.; Meshcheryakova, I.A.; Demidov, E.A.; Starostin, K.V.; Sadovski, E.V.; Peltek, S.E.; Voevoda, M.I.; Chernyavskii, A.M.; Volkov, A.M.; Ragino, Y.I. A Proteomic Study of Atherosclerotic Plaques in Men with Coronary Atherosclerosis. Diagnostics 2019, 9, 177. [Google Scholar] [CrossRef]
- Hwang, S.-I.; Thumar, J.; Lundgren, D.H.; Rezaul, K.; Mayya, V.; Wu, L.; Eng, J.; Wright, M.E.; Han, D.K. Direct Cancer Tissue Proteomics: A Method to Identify Candidate Cancer Biomarkers from Formalin-Fixed Paraffin-Embedded Archival Tissues. Oncogene 2007, 26, 65–76. [Google Scholar] [CrossRef]
- Duran, M.C.; Mas, S.; Martin-Ventura, J.L.; Meilhac, O.; Michel, J.B.; Gallego-Delgado, J.; Lázaro, A.; Tuñon, J.; Egido, J.; Vivanco, F. Proteomic Analysis of Human Vessels: Application to Atherosclerotic Plaques. Proteomics 2003, 3, 973–978. [Google Scholar] [CrossRef]
- Martin-Ventura, J.L.; Duran, M.C.; Blanco-Colio, L.M.; Meilhac, O.; Leclercq, A.; Michel, J.-B.; Jensen, O.N.; Hernandez-Merida, S.; Tuñón, J.; Vivanco, F.; et al. Identification by a Differential Proteomic Approach of Heat Shock Protein 27 as a Potential Marker of Atherosclerosis. Circulation 2004, 110, 2216–2219. [Google Scholar] [CrossRef]
- Durán, M.C.; Martín-Ventura, J.L.; Mohammed, S.; Barderas, M.G.; Blanco-Colio, L.M.; Mas, S.; Moral, V.; Ortega, L.; Tuñón, J.; Jensen, O.N.; et al. Atorvastatin Modulates the Profile of Proteins Released by Human Atherosclerotic Plaques. Eur. J. Pharmacol. 2007, 562, 119–129. [Google Scholar] [CrossRef]
- de la Cuesta, F.; Barderas, M.G.; Calvo, E.; Zubiri, I.; Maroto, A.S.; Darde, V.M.; Martin-Rojas, T.; Gil-Dones, F.; Posada-Ayala, M.; Tejerina, T.; et al. Secretome Analysis of Atherosclerotic and Non-Atherosclerotic Arteries Reveals Dynamic Extracellular Remodeling during Pathogenesis. J. Proteom. 2012, 75, 2960–2971. [Google Scholar] [CrossRef]

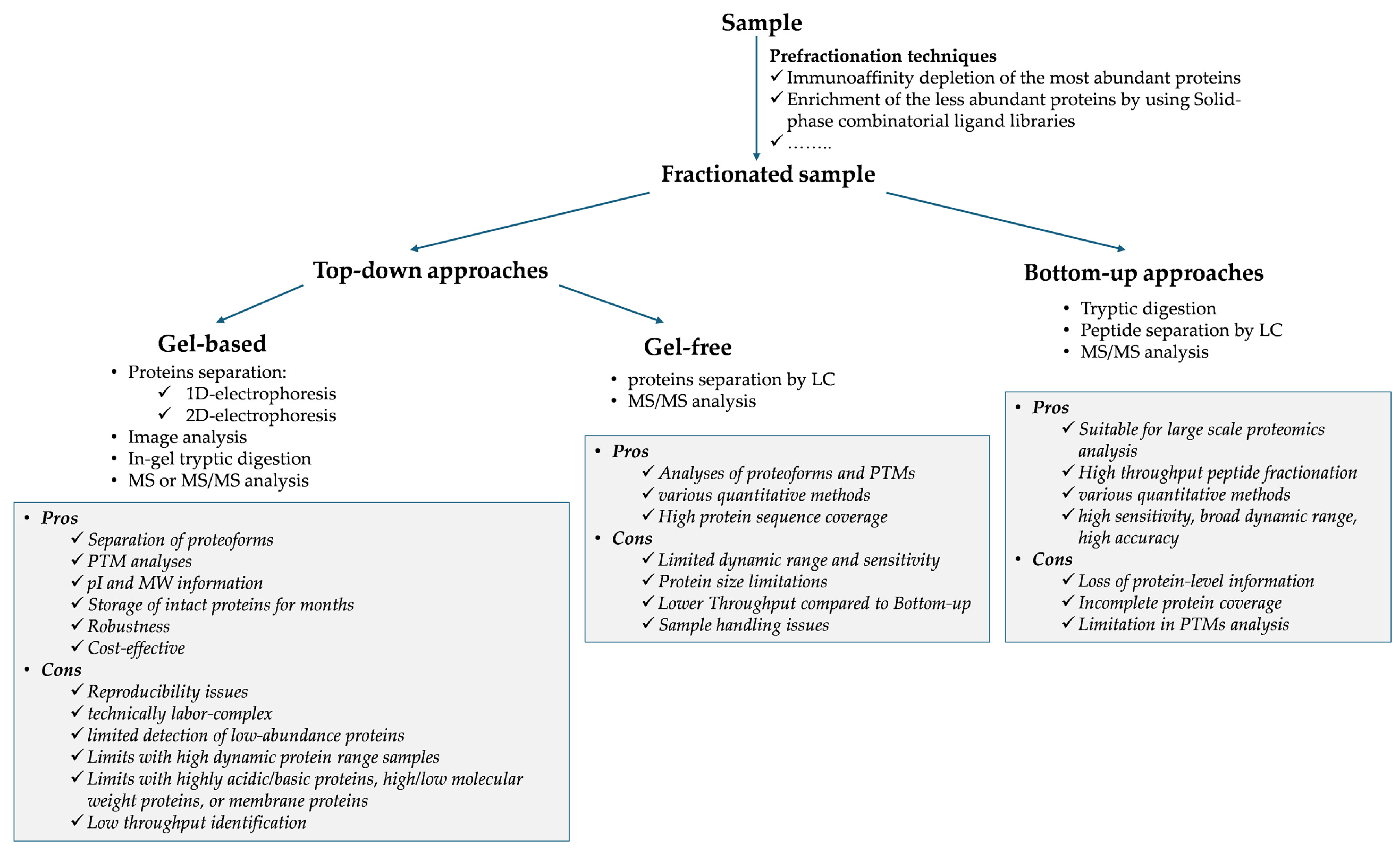
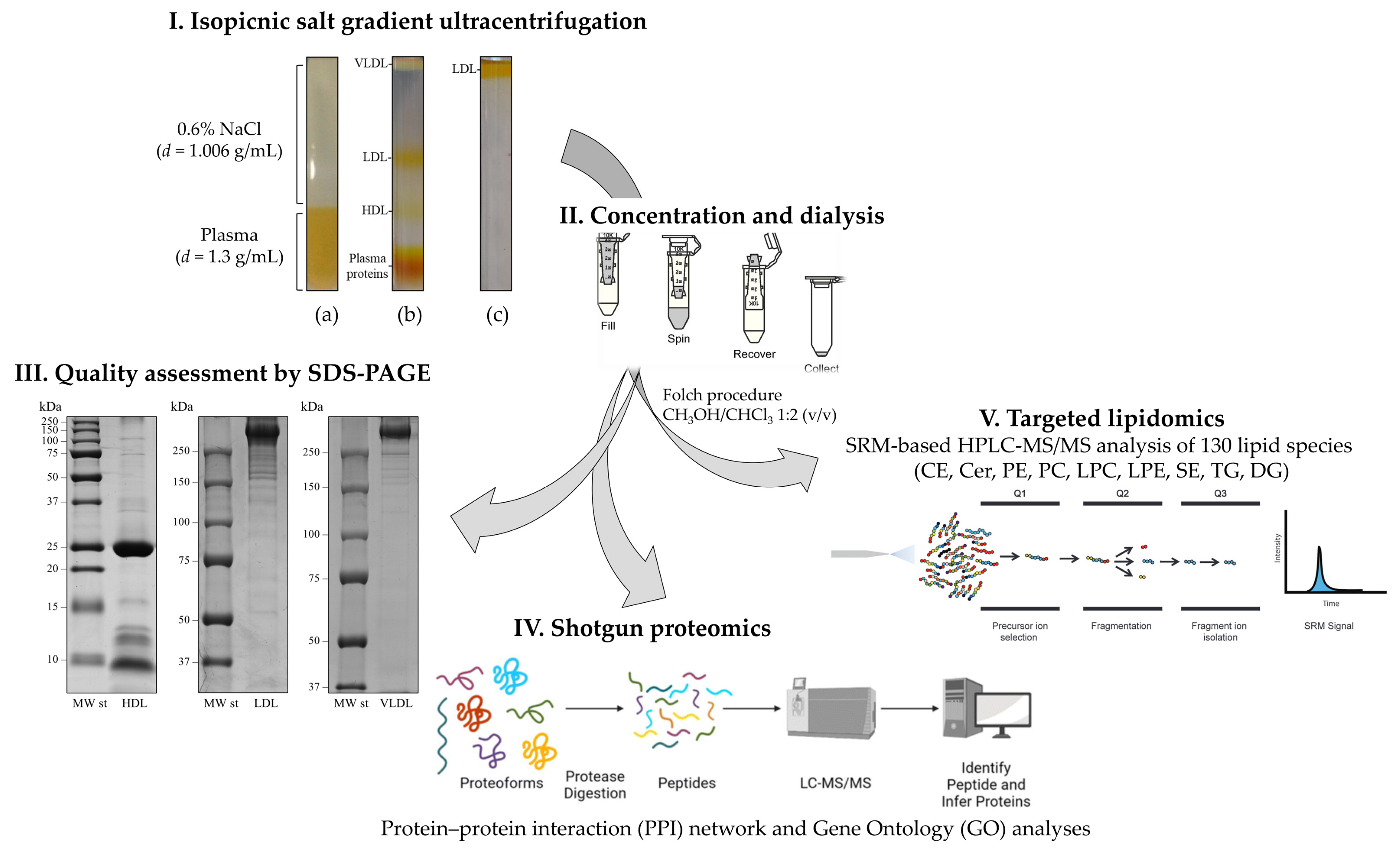
| Plasma Samples | Proteomics Approach | Main Findings | References |
|---|---|---|---|
| 11 AMI vs. 8 unstable angina vs. 9 healthy controls | 2-DE coupled to MALDI-TOF MS | ↓ alpha1-antitrypsin, ↑ Fibrinogen gamma chain, ↓ apolipoprotein A-I, ↑ ɣ-immunoglobulin heavy chains | [63] |
| 53 pooled CAD vs. 53 pooled controls | albumin and immunoglobulin removal, low-molecular weight proteins enrichment, cation exchange, and reverse phase chromatography fractionation, LC-MS/MS | 95 differentially expressed proteins, including inflammatory mediators, defense mechanism proteins, coagulation system proteins, and growth factors | [64] |
| 48 non-ST elevation ACS vs. 10 CAD vs. 20 controls (6 months follow-up) | six most abundant proteins depletion, 2-DE and 2D-DIGE, MALDI-MS/MS | 33 differentially expressed proteins belonging to the following functional groups: coagulation proteins, metabolism and/or lipid transport, inflammation and immune response, metabolite transport, cytoskeleton, miscellaneous proteins | [65] |
| 800 pooled CAD vs. 800 pooled stroke vs. 1600 pooled controls (postmenopausal women) | six most abundant proteins depletion, protein labeling with C13 acrylamide, anion exchange, and reverse phase chromatography fractionation, LTQ-FT MS | beta-2 microglobulin and insulin-like growth factor binding protein 4 as risk markers for CHD and stroke, respectively; a number of candidate markers of disease risk and candidate mediators of hormone therapy effects on CHD and stroke | [66] |
| 5 coronary bypass surgery patients (30-day follow-up) | 2-DE coupled to LC-ESI-MS/MS | ↑ α1-ACT and cathepsin G levels, which reflect a post-surgery pro-inflammatory and prothrombotic state due to neutrophil activation | [67] |
| 22 ACS vs. 23 patients with normal coronary angiogram | 2-DE coupled to nano LC-MS/MS | ↑ serum amyloid A protein, ↑ alpha-1-antitrypsin, and fibrinogen A and B chains | [68] |
| 409 ACS (3-year follow-up) | SELDI-TOF MS (SELDI protein chip technology) | m/z 4174.39 peak is associated with an increased incidence of 3-year events | [69] |
| 30 pooled ACS vs. 30 pooled stable atherosclerotic disease vs. 30 pooled subclinical CVD vs. 30 pooled controls | albumin, IgG, IgA, haptoglobin, transferrin, and antitrypsin depletion; 4-plex iTRAQ followed by LC–MS/MS | increased expression of serum amyloid protein A (SAA), C-reactive protein (CRP), apolipoprotein(a), and vinculin, from controls to ACS | [70] |
| 30 STEACS vs. 30 NSTEACS vs. 30 controls | 14 most abundant plasma proteins depletion, 2D-DIGE, MALDI-TOF/TOF | differential expression of proteins involved in metabolism (~3.2%), lipid metabolism and transport (~29.3%), inflammation and immune response (~25.8%), blood homeostasis and coagulation (~16.2%), proteases and protease inhibitors (~9.7%), others (~6.4%) and unknown (~6.4%) | [71] |
| 10 AMI vs. 5 controls | albumin, IgG, IgA, transferrin, haptoglobin, and antitrypsin depletion, iTRAQ followed by LC–MS/MS | a panel of 12 higher and 19 lower expressed proteins in AMI; the top 3 biological process terms were regulation of cell proliferation, response to wounding, and wound healing | [72] |
| 20 Apo CIII < 10 mg/dL CAD vs. 32 Apo CIII > 10 mg/dL CAD | solid phase extraction followed by LC–MS/MS | ↑ Apo C-II, ↑ Apo E, ↑ retinol-binding protein 4, and ↑ vitronectin, ↓ alpha-1 antitrypsin | [73] |
| 20 ACS vs. 20 healthy controls | six most abundant proteins depletion, LC-ESI-MS/MS | ↑ hemopexin, ↑ leucine-rich-2-glycoprotein, ↑ vitronectin, ↓ fibronectin | [74,75] |
| 50 ACS vs. 50 healthy controls | albumin, antitrypsin, haptoglobin, IgA, IgG, and transferrin depletion, LC–MS/MS | ↑ α-1-acid glycoprotein 1, complement C5, leucine-rich α-2-glycoprotein, vitronectin, ↓ GELS | [75] |
| Lipoprotein Fractions | Purification | Proteomics Approach | Main Findings | References |
|---|---|---|---|---|
| Group 1: HDL from 20 healthy volunteers; Group 2: HDL3 from 6 controls and 7 CAD patients; Group 3: HDL3 from 32 controls and 32 CAD; HDL from atherosclerotic tissue | Ultracentrifugation/immunoaffinity chromatography | LC-ESI-MS/MS analysis | 48 proteins identified in HDL involved in lipid metabolism, complement regulation, proteinase inhibition, and acute-phase response. ↑ Apo C-IV, ↑ paraoxonase-1, ↑ Complement C3, ↑ apo A-IV, ↑ apo E (confirmed by WB on HDL3 fraction from 64 subjects) in HDL3 from CAD. More than 100 proteins identified in HDL from atherosclerotic plaque. | [96] |
| Group 1: HDL3 from 6 CAD, 6 CAD + Statin/Niacin, 6 healthy controls; Group 2: HDL3 from 18 CAD, 18 CAD + Statin/Niacin, 12 months therapy | Sequential density ultracentrifugation | LC-FTICR MS | After 12 months of therapy, ↓ apo E, ↑ apo F, and ↑ phospholipid transfer protein; combined therapy may revert CAD-associated changes in HDL3 protein composition. | [117] |
| HDL2 from 18 CAD vs. 20 controls | Sequential density ultracentrifugation | MALDI-TOF-TOF MS and pattern recognition analysis; LC-MALDI-TOF/TOF MS | HDL2 from CAD contains methionine sulfoxide residues in apolipoprotein A-I and elevated levels of apolipoprotein C-III; specific proteomic signatures of HDL2 accurately classify CAD and control subjects. | [118] |
| HDL from 39 new-onset AMI-patients vs. 60 healthy individuals | Salt gradient ultracentrifugation | 2-DE coupled with MALDI-TOF MS analysis | Altered glycosylation pattern in AMI-patients within the first 6 h after the onset of the event. | [119] |
| HDL from 10 healthy controls vs. 10 CAD vs. 10 ACS | Sequential ultracentrifugation | 1-DE coupled with LC-ESI-MS/MS analysis | 67 HDL-associated proteins identified; ↓ apo A-IV, ↑ SAA, ↑ Complement C3 in ACS vs. both CAD and controls. | [120] |
| HDL from CAD, ACS, and healthy controls | Sequential ultracentrifugation or gel filtration chromatography | LC-ESI-MS/MS analysis | ↓ clusterin, ↑ apolipoprotein C-III in both CAD and ACS; HDL-proteome remodeling plays an important role in the altered functional properties of HDL (stimulation of proapoptotic pathways). | [121] |
| HDL2 and HDL3 from 40 ACS vs. 40 controls | Two-step discontinuous density-gradient ultracentrifugation | 2-DE and 2D-DIGE analysis coupled with MALDI-TOF/TOF MS | 17 differentially expressed HDL-associated proteins identified, shifting to dysfunctional HDL subfractions. | [122] |
| HDL from 21 CAD pre- and post-percutaneous transluminal coronary angioplasty (PTCA) | Immunoaffinity chromatography (anti-apo A-I antibodies) | Quantitative 16O/18O analysis/iTRAQ technology coupled with LC–MS/MS/system biology analysis | 225 identified proteins; high protein variability in HDL composition between individuals; post-PTCA increase in two protein clusters that included several apolipoproteins, fibrinogen-like protein 1, and other intracellular proteins, and a decrease in antithrombin-III, annexin A1, and several immunoglobulins. | [123] |
| HDL from 20 subjects at risk for CAD: 10 patients had CAD and 10 did not | Sequential ultracentrifugation | Shotgun proteomic, glycomic, and ganglioside analyses using LC-MS | Combined HDL proteomic and glycomic profiles distinguished at-risk subjects with atherosclerosis from those without; CAD patients had ↓ apo A-I, ↓ apo A-II, ↓ apo E, ↓ SAA2, ↓ SAA4 (p = 0.007), ↑ sialylated glycans. | [124] |
| HDL from 5 chronic heart failure (CHF) patients vs. 5 controls | Sequential ultracentrifugation | SCX/RP LC–MS/MS | 494 proteins identified; 23 newly identified HDL-associated proteins; 223 bacterial peptides were found in both CHF and controls. | [125] |
| HDL from 126 subjects with clinical indication for a coronary computed tomography angiography | High-resolution size exclusion chromatography followed by phospholipid-associated proteins capture (calcium silicate hydrate) | LC-ESI-MS/MS | 72 HDL-associated proteins detected in at least 75% of subjects; 13 proteins significantly associated with calcified plaque burden including cathelicidin antimicrobial peptide, GELS, kininogen-1, and paraoxonase-1 (inverse relationships), apo A-IV, vitamin D binding protein, alpha-2- macroglobulin, and apo C-II (positive relationships); 15 proteins significantly associated with non-calcified plaque burden including apo A-I, apo F, antithrombin III, and apo C-I (inverse relationships), serum amyloid A1, immunoglobulin heavy constant alpha 1, complement factor B, complement C2, complement C3, complement bC1s subcomponent (positive relationships); among the evaluated risk factors, body mass index has the greatest overall impact on the protein composition of HDL. | [126] |
| HDL from 943 participants without prevalent myocardial infarction referred for coronary angiography in the CASABLANCA study | 15NHis6Apo A-I was added to human serum, incubated, diluted, and then purified using PhyTips (Phynexus, San Jose, CA, USA), packed with Ni-NTA HisBind Superflow stationary phase | Targeted proteomics (apolipoprotein A-I, C-1, C-2, C-3, and C-4): LC-ESI-MS/MS analysis | An HDL apolipoproteomic score is associated with CAD, independent of circulating apo A-I and apo B concentrations and other conventional cardiovascular risk factors. Among individuals with CAD, this score may be independently associated with cardiovascular death. | [127] |
| Apolipoprotein AI-associated lipoproteins from 231 healthy individuals and patients with CAD | Metal chelate affinity chromatography | Targeted proteomics (21 proteins): LC-MS/MS analysis | A multiplexed proteomic assay useful for the estimation of cholesterol efflux and CAD risk in the clinical laboratory. | [128] |
| Samples | Proteomics Approach | Main Findings | References |
|---|---|---|---|
| 10 coronary arteries from CAD vs. 7 coronary arteries from normal individuals | 2-DE coupled with LC-MS/MS | Increased expression of the ferritin light chain in CAD (1.41 vs. 0.75; p = 0.01). | [164] |
| 35 human coronary atherosclerotic specimens (frozen, embedded, LCM specimens) | LC-MS/MS | 806 proteins identified; first large-scale proteomics map of human coronary atherosclerotic plaques. | [165] |
| Intimal proteome from the human atherosclerotic coronary artery (LCM) vs. preatherosclerotic coronary vs. radial arteries | 2D DIGE coupled with MALDI TOF/TOF MS | 13 proteins were differentially expressed (7 upregulated and 6 downregulated), and are implicated in the migrative capacity of vascular SMCs, ECM composition, coagulation, apoptosis, heat shock response, and intraplaque hemorrhage deposition. | [166] |
| 23 lipid-rich plaques vs. 13 non-lipid-rich plaques | LC MS/MS | Library of 423 proteins that are present in coronary plaque debris; MMP-9 and another 5 plaque-enriched proteins (lipopolysaccharide binding protein, Annexin A5, eukaryotic translocation initiation factor, syntaxin 11, cytochrome B5 reductase 3) significantly enriched in plaque and in plasma after plaque disruption. | [167] |
| Human coronary arteries (n = 100) and aortas (n = 100) | LC MS/MS | Identified hundreds of proteins (n = 1925) and numerous networks and pathways that are associated with early atherosclerosis. | [33] |
| Coronary arteries at different stages of development, from 15 patients | 2-DE followed by MALDI-TOF MS | The amounts of the following proteins were increased in stable atherosclerotic plaques at the stage of lipidosis and fibrosis: vimentin, tropomyosin β-chain, actin, keratin, tubulin β-chain, microfibril-associated glycoprotein 4, serum amyloid P-component, and annexin 5. In plaques at the stage of fibrosis and calcification, the amounts of mimecan and fibrinogen were increased. In the unstable atherosclerotic plaque of the necrotic–dystrophic type, the amounts of human serum albumin, mimecan, fibrinogen, serum amyloid P-component, and annexin were increased. | [168] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nieddu, G.; Formato, M.; Lepedda, A.J. Deciphering Acute Coronary Syndromes Pathobiology Through Proteomics. J. Cardiovasc. Dev. Dis. 2025, 12, 188. https://doi.org/10.3390/jcdd12050188
Nieddu G, Formato M, Lepedda AJ. Deciphering Acute Coronary Syndromes Pathobiology Through Proteomics. Journal of Cardiovascular Development and Disease. 2025; 12(5):188. https://doi.org/10.3390/jcdd12050188
Chicago/Turabian StyleNieddu, Gabriele, Marilena Formato, and Antonio Junior Lepedda. 2025. "Deciphering Acute Coronary Syndromes Pathobiology Through Proteomics" Journal of Cardiovascular Development and Disease 12, no. 5: 188. https://doi.org/10.3390/jcdd12050188
APA StyleNieddu, G., Formato, M., & Lepedda, A. J. (2025). Deciphering Acute Coronary Syndromes Pathobiology Through Proteomics. Journal of Cardiovascular Development and Disease, 12(5), 188. https://doi.org/10.3390/jcdd12050188








