Rtf1 Transcriptionally Regulates Neonatal and Adult Cardiomyocyte Biology
Abstract
:1. Introduction
2. Materials and Methods
2.1. NRVM Culture
2.2. NRVM Immunostaining
2.3. RNA Isolation
2.4. Real-Time Quantitative PCR
2.5. RNA-seq and Analysis
2.6. Mice
2.7. Histology Staining
2.8. Western Blotting
2.9. Echocardiography
2.10. Immunostaining of Mouse Heart Sections
2.11. Statistics
3. Results
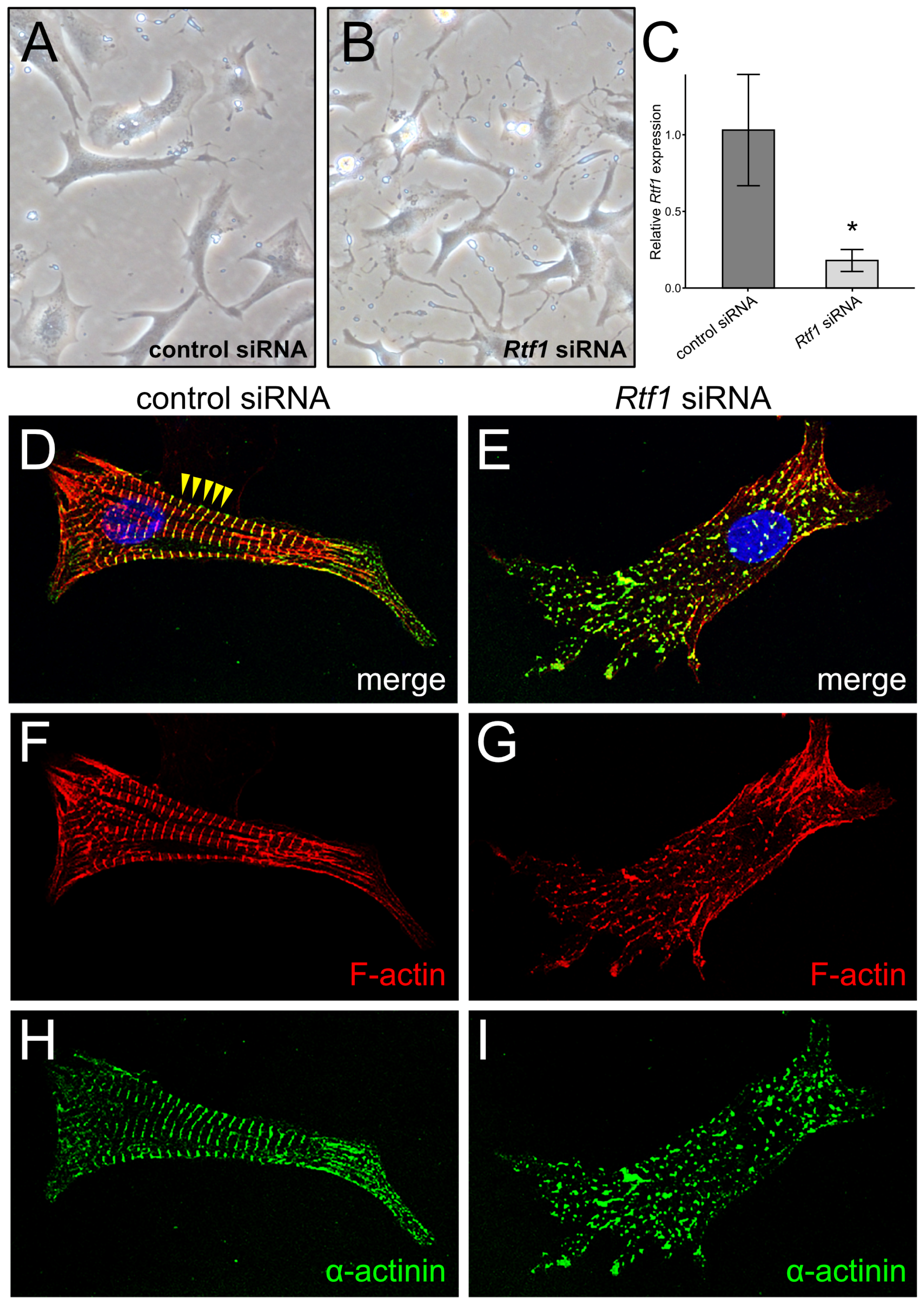
3.1. Rtf1 Regulates Neonatal Cardiomyocyte Morphology
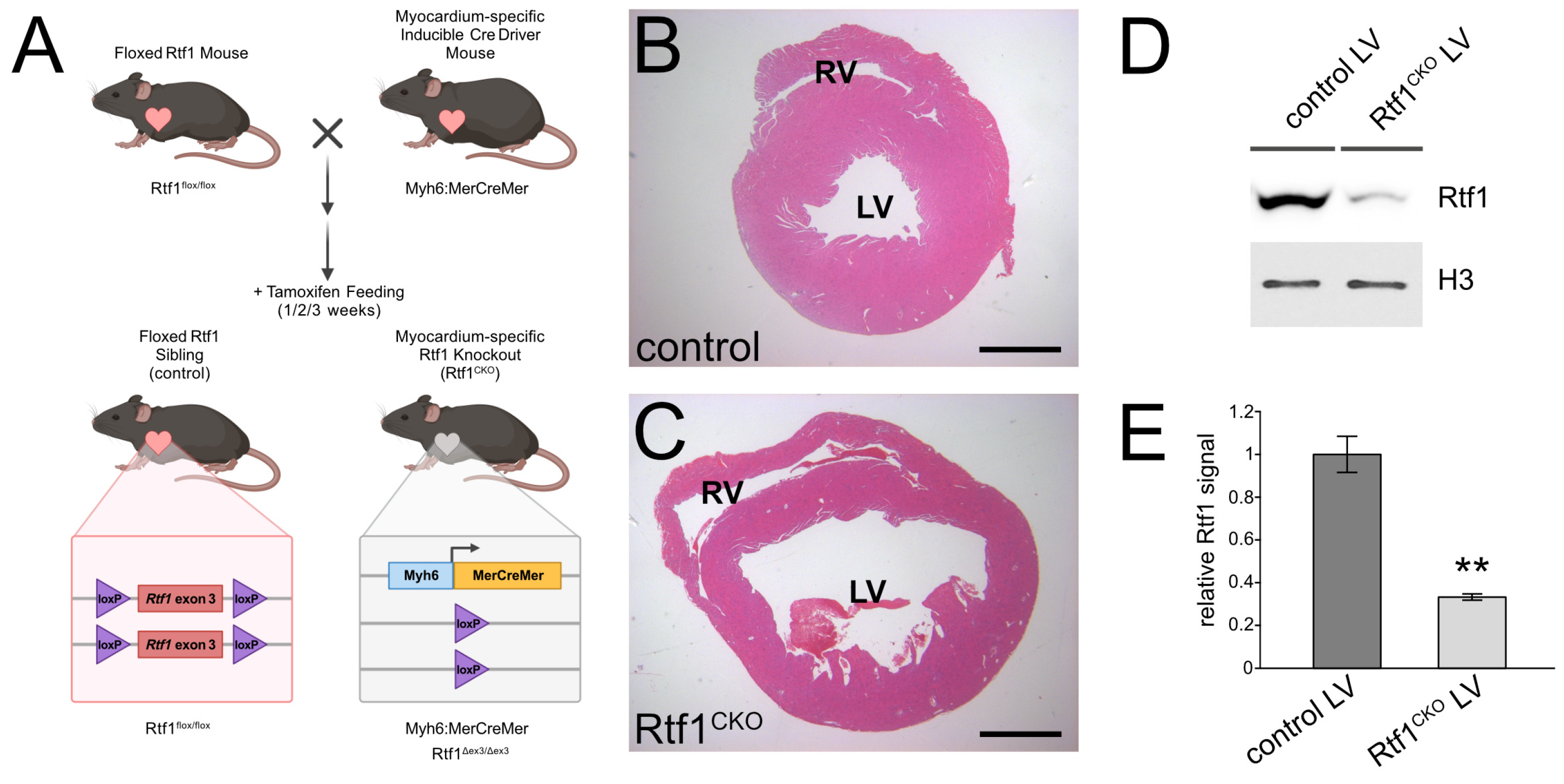
3.2. Inducible Knockout of Rtf1 in the Adult Mouse Myocardium
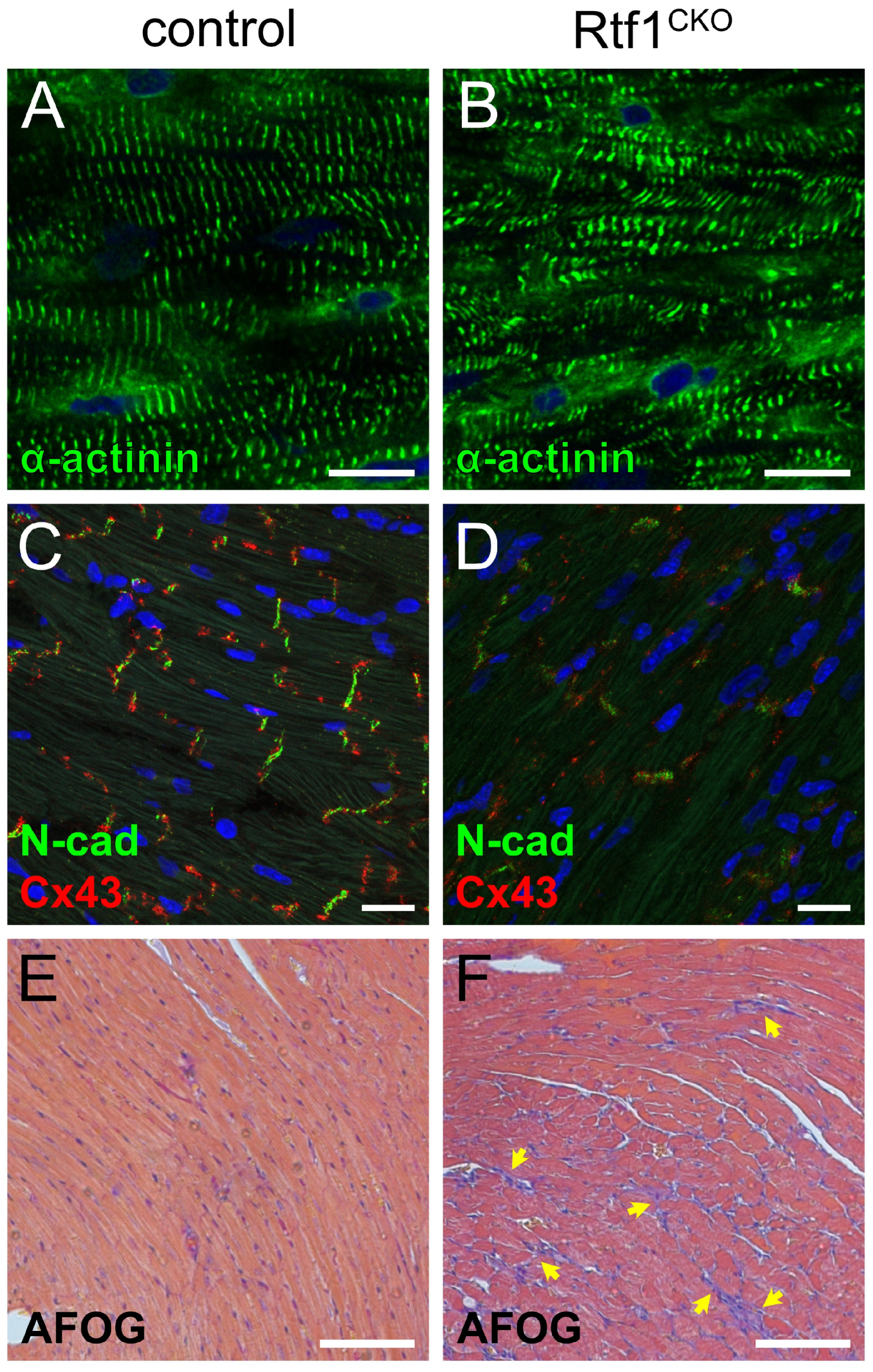
3.3. Rtf1-Deficient LV Exhibits Structural Deterioration and Disruption of Intercalated Discs
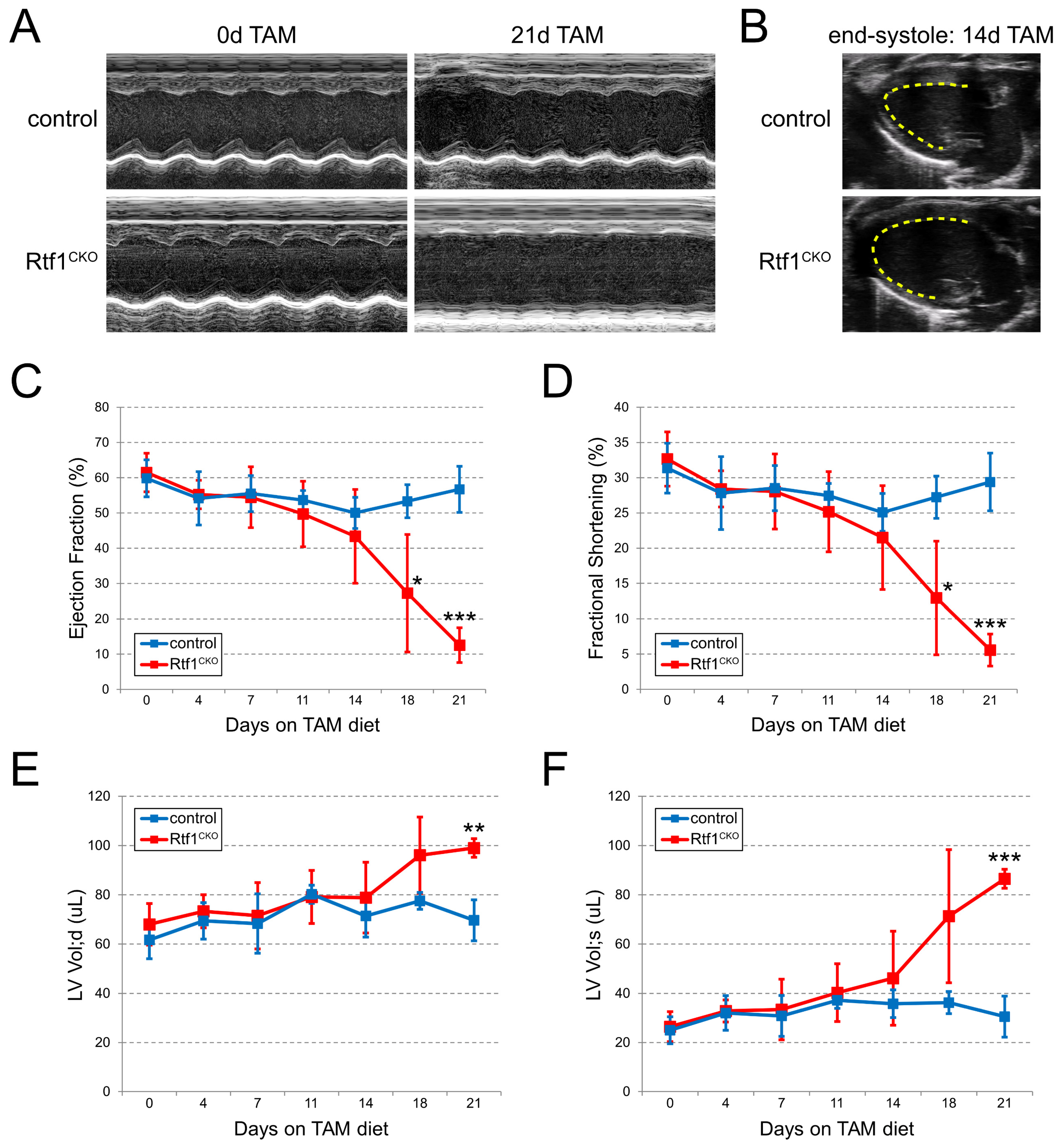
3.4. Myocardial Knockout of Rtf1 Causes Progressive Dilated Cardiomyopathy and Severe LV Systolic Dysfunction
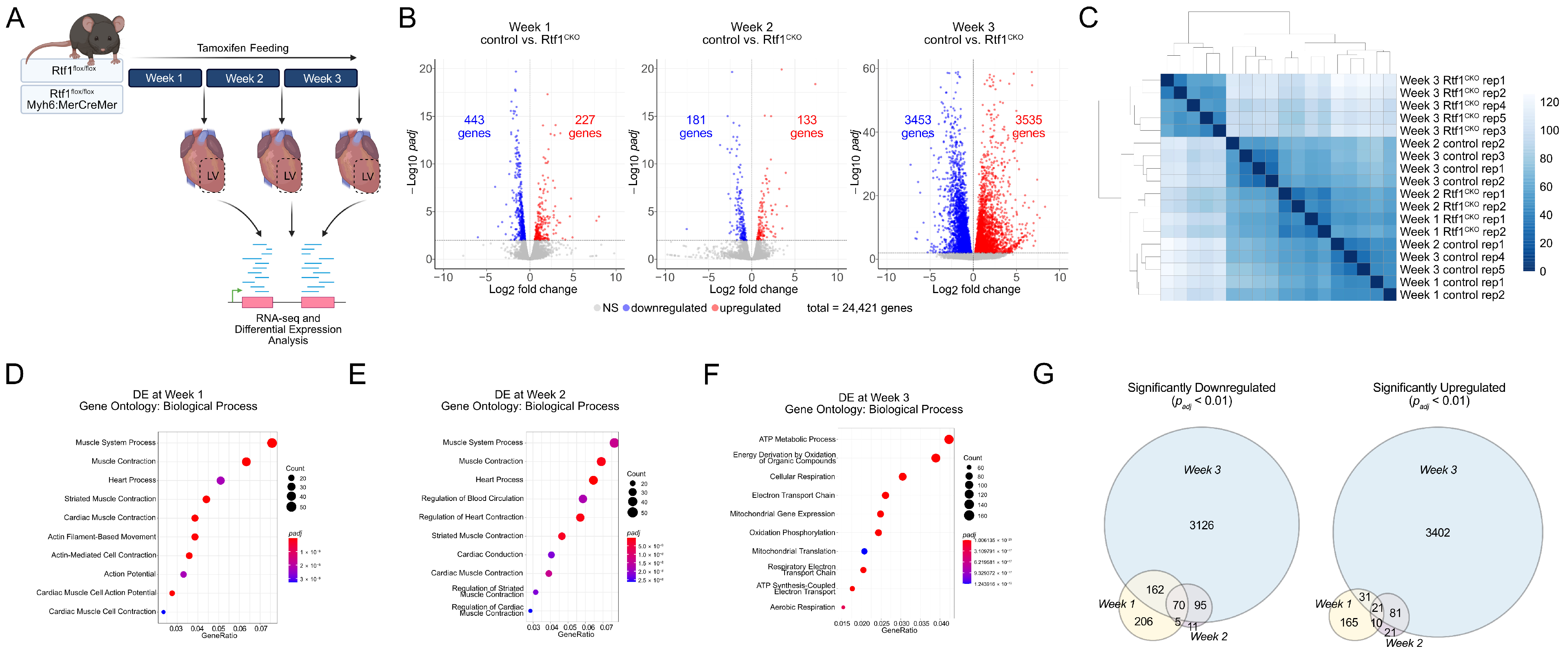
3.5. Rtf1 Knockout LV Exhibits Progressive Transcriptional Dysregulation
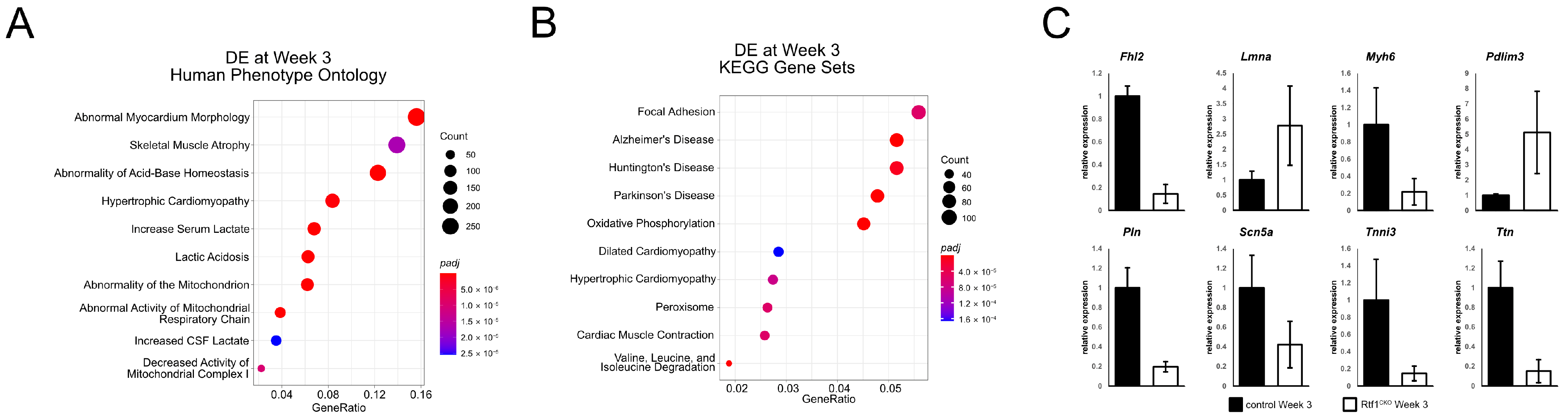
3.6. Failing Rtf1 Knockout Hearts Exhibit Gene Expression Changes Similar to Those Observed in Dilated Cardiomyopathy
3.7. Rtf1 Transcriptionally Regulates a Core Set of Critical Cardiac Genes
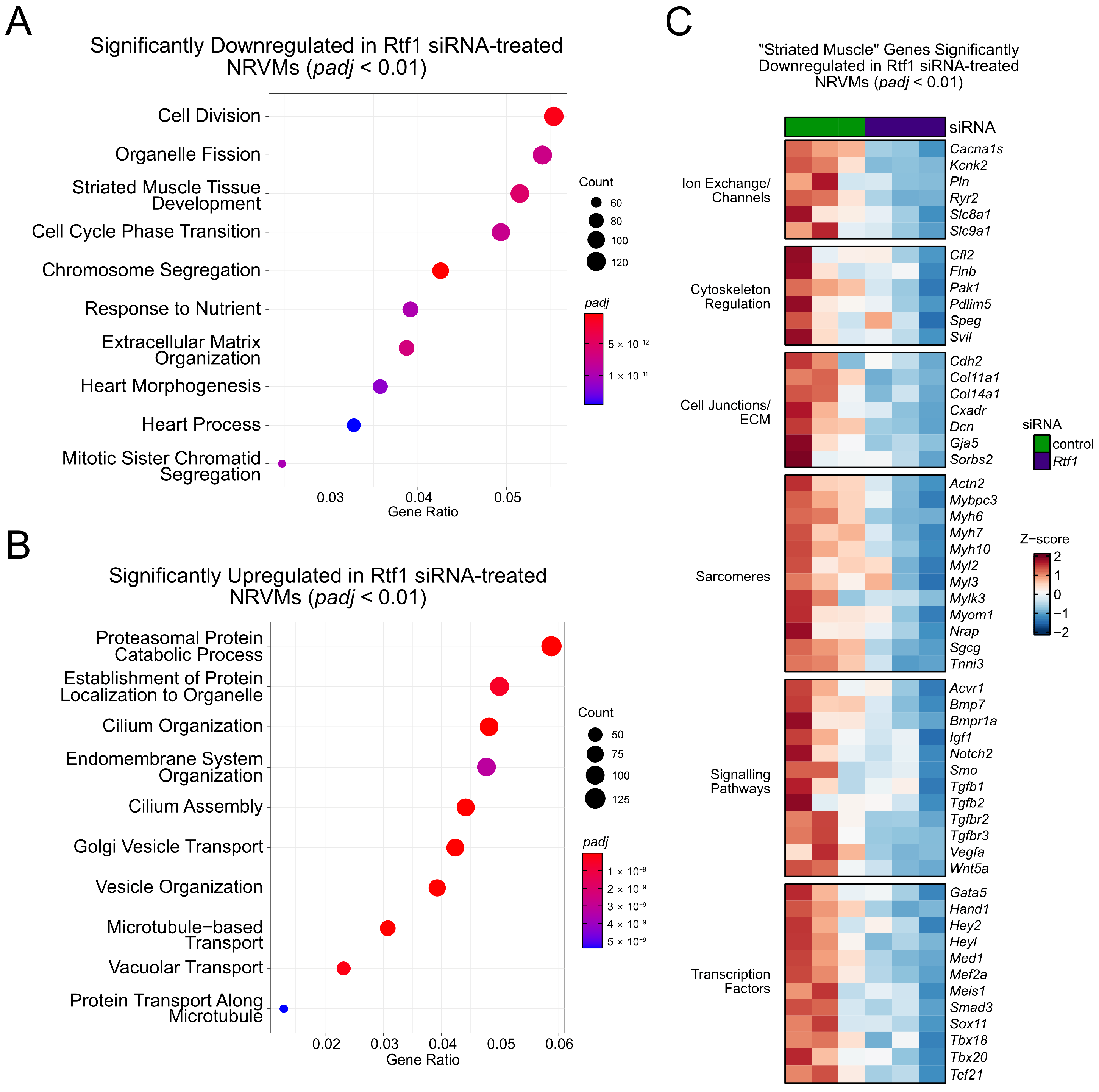
3.8. Rtf1 Regulates Genes Essential for Neonatal Cardiomyocyte Biology
4. Discussion
4.1. Genetic Regulation of Dilated Cardiomyopathy
4.2. Maturation of Differentiated Cardiomyocytes
4.3. Transcriptional Regulation of Cardiac Gene Expression
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coulon, A.; Chow, C.C.; Singer, R.H.; Larson, D.R. Eukaryotic transcriptional dynamics: From single molecules to cell populations. Nat. Rev. Genet. 2013, 14, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Dent, S.Y. Chromatin modifiers and remodellers: Regulators of cellular differentiation. Nat. Rev. Genet. 2014, 15, 93–106. [Google Scholar] [CrossRef]
- Core, L.; Adelman, K. Promoter-Proximal pausing of RNA polymerase II: A nexus of gene regulation. Genes Dev. 2019, 33, 960–982. [Google Scholar] [CrossRef] [PubMed]
- Francette, A.M.; Tripplehorn, S.A.; Arndt, K.M. The paf1 complex: A keystone of nuclear regulation operating at the interface of transcription and chromatin. J. Mol. Biol. 2021, 433, 166979. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Park, S.; Lee, J.S. Role of the paf1 complex in the maintenance of stem cell pluripotency and development. FEBS J. 2023, 290, 951–961. [Google Scholar] [CrossRef]
- Van Oss, S.B.; Cucinotta, C.E.; Arndt, K.M. Emerging insights into the roles of the paf1 complex in gene regulation. Trends Biochem. Sci. 2017, 42, 788–798. [Google Scholar] [CrossRef]
- Jaehning, J.A. The paf1 complex: Platform or player in RNA polymerase II transcription? Biochim. Biophys. Acta 2010, 1799, 379–388. [Google Scholar] [CrossRef]
- Zhu, B.; Mandal, S.S.; Pham, A.D.; Zheng, Y.; Erdjument-Bromage, H.; Batra, S.K.; Tempst, P.; Reinberg, D. The human paf complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005, 19, 1668–1673. [Google Scholar] [CrossRef]
- Mueller, C.L.; Jaehning, J.A. Ctr9, rtf1, and leo1 are components of the paf1/rna polymerase II complex. Mol. Cell Biol. 2002, 22, 1971–1980. [Google Scholar] [CrossRef]
- Rozenblatt-Rosen, O.; Hughes, C.M.; Nannepaga, S.J.; Shanmugam, K.S.; Copeland, T.D.; Guszczynski, T.; Resau, J.H.; Meyerson, M. The parafibromin tumor suppressor protein is part of a human paf1 complex. Mol. Cell Biol. 2005, 25, 612–620. [Google Scholar] [CrossRef]
- Adelman, K.; Wei, W.; Ardehali, M.B.; Werner, J.; Zhu, B.; Reinberg, D.; Lis, J.T. Drosophila paf1 modulates chromatin structure at actively transcribed genes. Mol. Cell Biol. 2006, 26, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Kubota, Y.; Ota, N.; Takatsuka, H.; Unno, T.; Onami, S.; Sugimoto, A.; Ito, M. The paf1 complex cell autonomously promotes oogenesis in caenorhabditis elegans. Genes Cells 2022, 27, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Kubota, Y.; Tsuyama, K.; Takabayashi, Y.; Haruta, N.; Maruyama, R.; Iida, N.; Sugimoto, A. The paf1 complex is involved in embryonic epidermal morphogenesis in caenorhabditis elegans. Dev. Biol. 2014, 391, 43–53. [Google Scholar] [CrossRef]
- Akanuma, T.; Koshida, S.; Kawamura, A.; Kishimoto, Y.; Takada, S. Paf1 complex homologues are required for notch-regulated transcription during somite segmentation. EMBO Rep. 2007, 8, 858–863. [Google Scholar] [CrossRef] [PubMed]
- Tenney, K.; Gerber, M.; Ilvarsonn, A.; Schneider, J.; Gause, M.; Dorsett, D.; Eissenberg, J.C.; Shilatifard, A. Drosophila rtf1 functions in histone methylation, gene expression, and notch signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 11970–11974. [Google Scholar] [CrossRef]
- Mosimann, C.; Hausmann, G.; Basler, K. Parafibromin/hyrax activates wnt/wg target gene transcription by direct association with beta-catenin/armadillo. Cell 2006, 125, 327–341. [Google Scholar] [CrossRef]
- Bahrampour, S.; Thor, S. Ctr9, a key component of the paf1 complex, affects proliferation and terminal differentiation in the developing Drosophila nervous system. G3 Genes Genomes Genet. 2016, 6, 3229–3239. [Google Scholar]
- Jurynec, M.J.; Bai, X.; Bisgrove, B.W.; Jackson, H.; Nechiporuk, A.; Palu, R.A.S.; Grunwald, H.A.; Su, Y.C.; Hoshijima, K.; Yost, H.J.; et al. The paf1 complex and p-tefb have reciprocal and antagonist roles in maintaining multipotent neural crest progenitors. Development 2019, 146, dev180133. [Google Scholar] [CrossRef]
- Langenbacher, A.D.; Nguyen, C.T.; Cavanaugh, A.M.; Huang, J.; Lu, F.; Chen, J.N. The paf1 complex differentially regulates cardiomyocyte specification. Dev. Biol. 2011, 353, 19–28. [Google Scholar] [CrossRef]
- Nguyen, C.T.; Langenbacher, A.; Hsieh, M.; Chen, J.N. The paf1 complex component leo1 is essential for cardiac and neural crest development in zebrafish. Dev. Biol. 2010, 341, 167–175. [Google Scholar] [CrossRef]
- Bai, X.; Kim, J.; Yang, Z.; Jurynec, M.J.; Akie, T.E.; Lee, J.; LeBlanc, J.; Sessa, A.; Jiang, H.; DiBiase, A.; et al. Tif1gamma controls erythroid cell fate by regulating transcription elongation. Cell 2010, 142, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, J.D.; Chung, A.Y.; Kim, H.S.; Kim, Y.S.; Kim, M.J.; Koun, S.; Lee, Y.M.; Rhee, M.; Park, H.C.; et al. Antagonistic regulation of paf1c and p-tefb is required for oligodendrocyte differentiation. J. Neurosci. 2012, 32, 8201–8207. [Google Scholar] [CrossRef]
- Ding, L.; Paszkowski-Rogacz, M.; Nitzsche, A.; Slabicki, M.M.; Heninger, A.K.; de Vries, I.; Kittler, R.; Junqueira, M.; Shevchenko, A.; Schulz, H.; et al. A genome-scale RNAi screen for oct4 modulators defines a role of the paf1 complex for embryonic stem cell identity. Cell Stem. Cell 2009, 4, 403–415. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, Z.Y.; Wu, R.; Zhang, C.M.; Cao, K.; Shan, W.G.; Liu, Z.; Ji, M.; Tian, Z.L.; Sethi, G.; et al. Transcriptional regulator ctr9 promotes hepatocellular carcinoma progression and metastasis via increasing peg10 transcriptional activity. Acta Pharm. Sin. 2022, 43, 2109–2118. [Google Scholar] [CrossRef]
- Nimmakayala, R.K.; Ogunleye, A.O.; Parte, S.; Krishna Kumar, N.; Raut, P.; Varadharaj, V.; Perumal, N.K.; Nallasamy, P.; Rauth, S.; Cox, J.L.; et al. Paf1 cooperates with yap1 in metaplastic ducts to promote pancreatic cancer. Cell Death Dis. 2022, 13, 839. [Google Scholar] [CrossRef] [PubMed]
- Carpten, J.D.; Robbins, C.M.; Villablanca, A.; Forsberg, L.; Presciuttini, S.; Bailey-Wilson, J.; Simonds, W.F.; Gillanders, E.M.; Kennedy, A.M.; Chen, J.D.; et al. Hrpt2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat. Genet. 2002, 32, 676–680. [Google Scholar] [CrossRef]
- Vos, S.M.; Farnung, L.; Linden, A.; Urlaub, H.; Cramer, P. Structure of complete pol II-dsif-paf-spt6 transcription complex reveals rtf1 allosteric activation. Nat. Struct. Mol. Biol. 2020, 27, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J.; Mbogning, J.; Hancock, M.A.; Majdpour, D.; Madhok, M.; Nassour, H.; Dallagnol, J.C.; Page, V.; Chatenet, D.; Tanny, J.C. Spt5 phosphorylation and the rtf1 plus3 domain promote rtf1 function through distinct mechanisms. Mol. Cell Biol. 2020, 40, e00150-20. [Google Scholar] [CrossRef] [PubMed]
- Zumer, K.; Maier, K.C.; Farnung, L.; Jaeger, M.G.; Rus, P.; Winter, G.; Cramer, P. Two distinct mechanisms of RNA polymerase II elongation stimulation in vivo. Mol. Cell 2021, 81, 3096–3109.e8. [Google Scholar] [CrossRef]
- Cao, Q.F.; Yamamoto, J.; Isobe, T.; Tateno, S.; Murase, Y.; Chen, Y.; Handa, H.; Yamaguchi, Y. Characterization of the human transcription elongation factor rtf1: Evidence for nonoverlapping functions of rtf1 and the paf1 complex. Mol. Cell Biol. 2015, 35, 3459–3470. [Google Scholar] [CrossRef]
- Cucinotta, C.E.; Hildreth, A.E.; McShane, B.M.; Shirra, M.K.; Arndt, K.M. The nucleosome acidic patch directly interacts with subunits of the paf1 and fact complexes and controls chromatin architecture in vivo. Nucleic. Acids Res. 2019, 47, 8410–8423. [Google Scholar] [CrossRef] [PubMed]
- Van Oss, S.B.; Shirra, M.K.; Bataille, A.R.; Wier, A.D.; Yen, K.; Vinayachandran, V.; Byeon, I.L.; Cucinotta, C.E.; Heroux, A.; Jeon, J.; et al. The histone modification domain of paf1 complex subunit rtf1 directly stimulates h2b ubiquitylation through an interaction with rad6. Mol. Cell 2016, 64, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Warner, M.H.; Roinick, K.L.; Arndt, K.M. Rtf1 is a multifunctional component of the paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol. Cell Biol. 2007, 27, 6103–6115. [Google Scholar] [CrossRef] [PubMed]
- Tomson, B.N.; Davis, C.P.; Warner, M.H.; Arndt, K.M. Identification of a role for histone h2b ubiquitylation in noncoding RNA 3’-end formation through mutational analysis of rtf1 in saccharomyces cerevisiae. Genetics 2011, 188, 273–289. [Google Scholar] [CrossRef]
- Piro, A.S.; Mayekar, M.K.; Warner, M.H.; Davis, C.P.; Arndt, K.M. Small region of rtf1 protein can substitute for complete paf1 complex in facilitating global histone h2b ubiquitylation in yeast. Proc. Natl. Acad. Sci. USA 2012, 109, 10837–10842. [Google Scholar] [CrossRef]
- Crisucci, E.M.; Arndt, K.M. The paf1 complex represses arg1 transcription in saccharomyces cerevisiae by promoting histone modifications. Eukaryot. Cell 2011, 10, 712–723. [Google Scholar] [CrossRef]
- Tomson, B.N.; Crisucci, E.M.; Heisler, L.E.; Gebbia, M.; Nislow, C.; Arndt, K.M. Effects of the paf1 complex and histone modifications on snorna 3’-end formation reveal broad and locus-specific regulation. Mol. Cell Biol. 2013, 33, 170–182. [Google Scholar] [CrossRef]
- Ng, H.H.; Dole, S.; Struhl, K. The rtf1 component of the paf1 transcriptional elongation complex is required for ubiquitination of histone h2b. J. Biol. Chem. 2003, 278, 33625–33628. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. Tophat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. Featurecounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with deseq2. Genome Biol 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. Clusterprofiler: An r package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Blighe, K.; Rana, S.; Lewis, M. Enhancedvolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 20 April 2023).
- Gu, Z. Complex heatmap visualization. iMeta 2022, 1, e43. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Sohal, D.S.; Nghiem, M.; Crackower, M.A.; Witt, S.A.; Kimball, T.R.; Tymitz, K.M.; Penninger, J.M.; Molkentin, J.D. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible cre protein. Circ. Res. 2001, 89, 20–25. [Google Scholar] [CrossRef]
- Jordan, E.; Peterson, L.; Ai, T.; Asatryan, B.; Bronicki, L.; Brown, E.; Celeghin, R.; Edwards, M.; Fan, J.; Ingles, J.; et al. Evidence-based assessment of genes in dilated cardiomyopathy. Circulation 2021, 144, 7–19. [Google Scholar] [CrossRef]
- McNally, E.M.; Golbus, J.R.; Puckelwartz, M.J. Genetic mutations and mechanisms in dilated cardiomyopathy. J. Clin. Investig. 2013, 123, 19–26. [Google Scholar] [CrossRef]
- Tayal, U.; Prasad, S.; Cook, S.A. Genetics and genomics of dilated cardiomyopathy and systolic heart failure. Genome Med. 2017, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Yoshida, M.; Masuda, H.; Maeda, D.; Kudo-Asabe, Y.; Umakoshi, M.; Nanjo, H.; Goto, A. Disorganization of intercalated discs in dilated cardiomyopathy. Sci. Rep. 2021, 11, 11852. [Google Scholar] [CrossRef] [PubMed]
- Ortega, A.; Tarazon, E.; Gil-Cayuela, C.; Garcia-Manzanares, M.; Martinez-Dolz, L.; Lago, F.; Gonzalez-Juanatey, J.R.; Cinca, J.; Jorge, E.; Portoles, M.; et al. Intercalated disc in failing hearts from patients with dilated cardiomyopathy: Its role in the depressed left ventricular function. PLoS ONE 2017, 12, e0185062. [Google Scholar] [CrossRef]
- Guo, Y.; Pu, W.T. Cardiomyocyte maturation: New phase in development. Circ. Res. 2020, 126, 1086–1106. [Google Scholar] [CrossRef] [PubMed]
- Karbassi, E.; Fenix, A.; Marchiano, S.; Muraoka, N.; Nakamura, K.; Yang, X.; Murry, C.E. Cardiomyocyte maturation: Advances in knowledge and implications for regenerative medicine. Nat. Rev. Cardiol. 2020, 17, 341–359. [Google Scholar] [CrossRef]
- Uosaki, H.; Cahan, P.; Lee, D.I.; Wang, S.; Miyamoto, M.; Fernandez, L.; Kass, D.A.; Kwon, C. Transcriptional landscape of cardiomyocyte maturation. Cell Rep. 2015, 13, 1705–1716. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, R.E.; Tokuyama, T.; Anzai, T.; Chanthra, N.; Uosaki, H. Sarcomere maturation: Function acquisition, molecular mechanism, and interplay with other organelles. Philos Trans R Soc. Lond B Biol. Sci. 2022, 377, 20210325. [Google Scholar] [CrossRef]
- Fan, X.; Hughes, B.G.; Ali, M.A.; Cho, W.J.; Lopez, W.; Schulz, R. Dynamic alterations to alpha-actinin accompanying sarcomere disassembly and reassembly during cardiomyocyte mitosis. PLoS ONE 2015, 10, e0129176. [Google Scholar]
- Wang, Z.; Song, A.; Xu, H.; Hu, S.; Tao, B.; Peng, L.; Wang, J.; Li, J.; Yu, J.; Wang, L.; et al. Coordinated regulation of RNA polymerase II pausing and elongation progression by paf1. Sci. Adv. 2022, 8, eabm5504. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Wang, Y.; Liu, Y.; Zhang, N.; Shamovsky, I.; Nudler, E.; Tian, B.; Dynlacht, B.D. Paf1c regulates RNA polymerase II progression by modulating elongation rate. Proc. Natl. Acad. Sci. USA 2019, 116, 14583–14592. [Google Scholar] [CrossRef]
- Czeschik, J.C.; Bauer, P.; Buiting, K.; Dufke, C.; Guillen-Navarro, E.; Johnson, D.S.; Koehler, U.; Lopez-Gonzalez, V.; Ludecke, H.J.; Male, A.; et al. X-linked intellectual disability type nascimento is a clinically distinct, probably underdiagnosed entity. Orphanet. J. Rare Dis. 2013, 8, 146. [Google Scholar] [CrossRef] [PubMed]
- Core, L.J.; Waterfall, J.J.; Lis, J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 2008, 322, 1845–1848. [Google Scholar] [CrossRef]
- Li, J.; Gilmour, D.S. Promoter proximal pausing and the control of gene expression. Curr. Opin. Genet. Dev. 2011, 21, 231–235. [Google Scholar] [CrossRef]
- Adelman, K.; Kennedy, M.A.; Nechaev, S.; Gilchrist, D.A.; Muse, G.W.; Chinenov, Y.; Rogatsky, I. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc. Natl. Acad. Sci. USA 2009, 106, 18207–18212. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, D.A.; Fromm, G.; dos Santos, G.; Pham, L.N.; McDaniel, I.E.; Burkholder, A.; Fargo, D.C.; Adelman, K. Regulating the regulators: The pervasive effects of pol II pausing on stimulus-responsive gene networks. Genes Dev. 2012, 26, 933–944. [Google Scholar] [CrossRef]
- Gilchrist, D.A.; Dos Santos, G.; Fargo, D.C.; Xie, B.; Gao, Y.; Li, L.; Adelman, K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 2010, 143, 540–551. [Google Scholar] [CrossRef] [PubMed]
- Henriques, T.; Gilchrist, D.A.; Nechaev, S.; Bern, M.; Muse, G.W.; Burkholder, A.; Fargo, D.C.; Adelman, K. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol. Cell 2013, 52, 517–528. [Google Scholar] [CrossRef]
- Henriques, T.; Scruggs, B.S.; Inouye, M.O.; Muse, G.W.; Williams, L.H.; Burkholder, A.B.; Lavender, C.A.; Fargo, D.C.; Adelman, K. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 2018, 32, 26–41. [Google Scholar] [CrossRef]
- Gaertner, B.; Zeitlinger, J. Rna polymerase II pausing during development. Development 2014, 141, 1179–1183. [Google Scholar] [CrossRef]
- Bothma, J.P.; Magliocco, J.; Levine, M. The snail repressor inhibits release, not elongation, of paused pol II in the Drosophila embryo. Curr. Biol. 2011, 21, 1571–1577. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Langenbacher, A.D.; Lu, F.; Crisman, L.; Huang, Z.Y.S.; Chapski, D.J.; Vondriska, T.M.; Wang, Y.; Gao, C.; Chen, J.-N. Rtf1 Transcriptionally Regulates Neonatal and Adult Cardiomyocyte Biology. J. Cardiovasc. Dev. Dis. 2023, 10, 221. https://doi.org/10.3390/jcdd10050221
Langenbacher AD, Lu F, Crisman L, Huang ZYS, Chapski DJ, Vondriska TM, Wang Y, Gao C, Chen J-N. Rtf1 Transcriptionally Regulates Neonatal and Adult Cardiomyocyte Biology. Journal of Cardiovascular Development and Disease. 2023; 10(5):221. https://doi.org/10.3390/jcdd10050221
Chicago/Turabian StyleLangenbacher, Adam D., Fei Lu, Lauren Crisman, Zi Yi Stephanie Huang, Douglas J. Chapski, Thomas M. Vondriska, Yibin Wang, Chen Gao, and Jau-Nian Chen. 2023. "Rtf1 Transcriptionally Regulates Neonatal and Adult Cardiomyocyte Biology" Journal of Cardiovascular Development and Disease 10, no. 5: 221. https://doi.org/10.3390/jcdd10050221
APA StyleLangenbacher, A. D., Lu, F., Crisman, L., Huang, Z. Y. S., Chapski, D. J., Vondriska, T. M., Wang, Y., Gao, C., & Chen, J.-N. (2023). Rtf1 Transcriptionally Regulates Neonatal and Adult Cardiomyocyte Biology. Journal of Cardiovascular Development and Disease, 10(5), 221. https://doi.org/10.3390/jcdd10050221







