Differences in Behavior between Normal and Atopic Keratinocytes in Culture: Pilot Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Biopsy Collection
2.3. Cell Culture
2.4. Transepithelial Electrical Resistance (TEER) Measurement
2.5. Real Time Polymerase Chain Reaction (RT-PCR)
2.6. Protein Extraction
2.7. Western Blot
2.8. Immunofluorescence (IF) Staining
2.9. Imaging and Data Analysis
2.10. Confocal Microscopy
2.11. Data Analysis and Statistics
2.11.1. TEER
2.11.2. RT-PCR
2.11.3. Immunofluorescent Staining Picture Analysis, Videos and Western Blot
3. Results
3.1. TEER
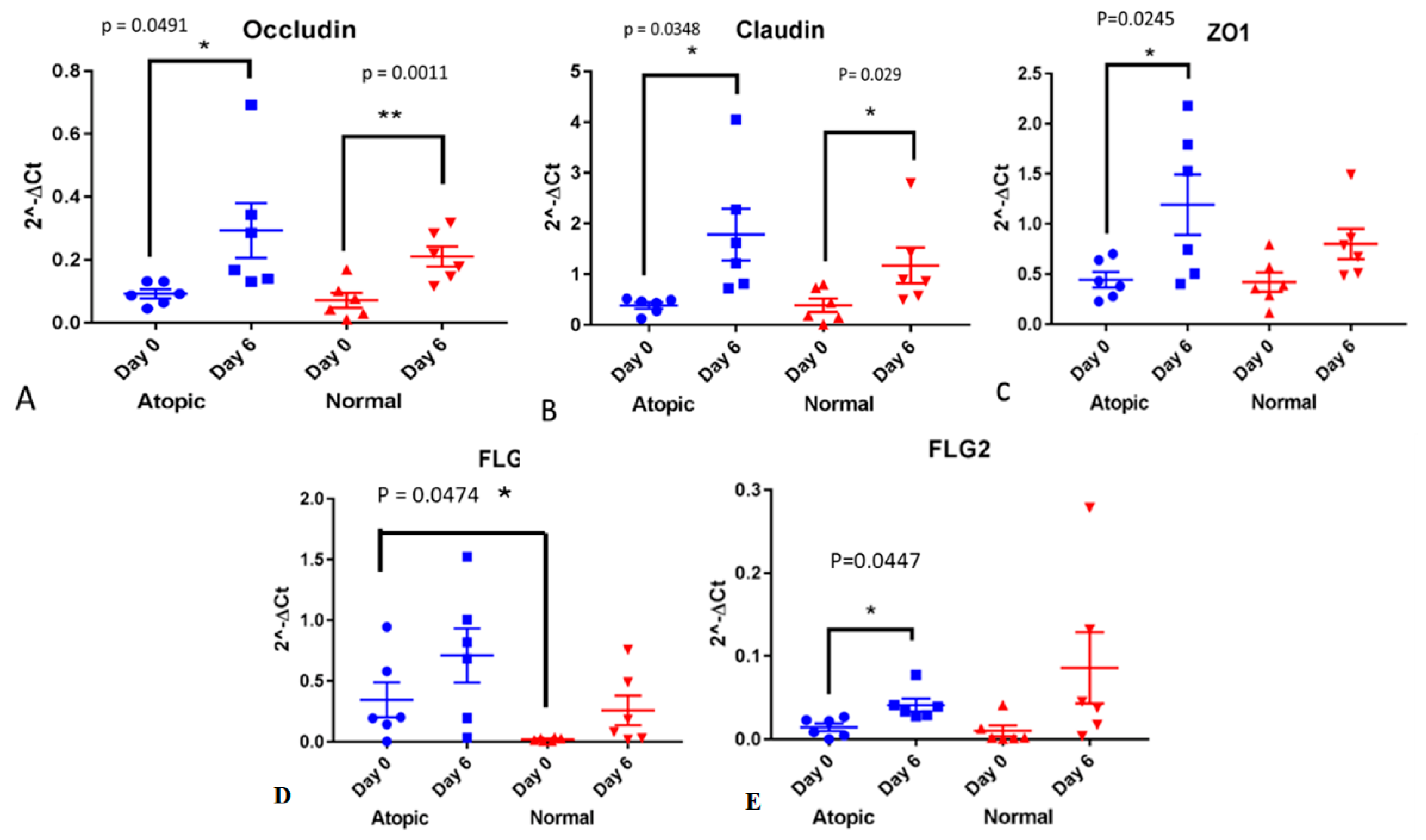
3.2. RT-PCR
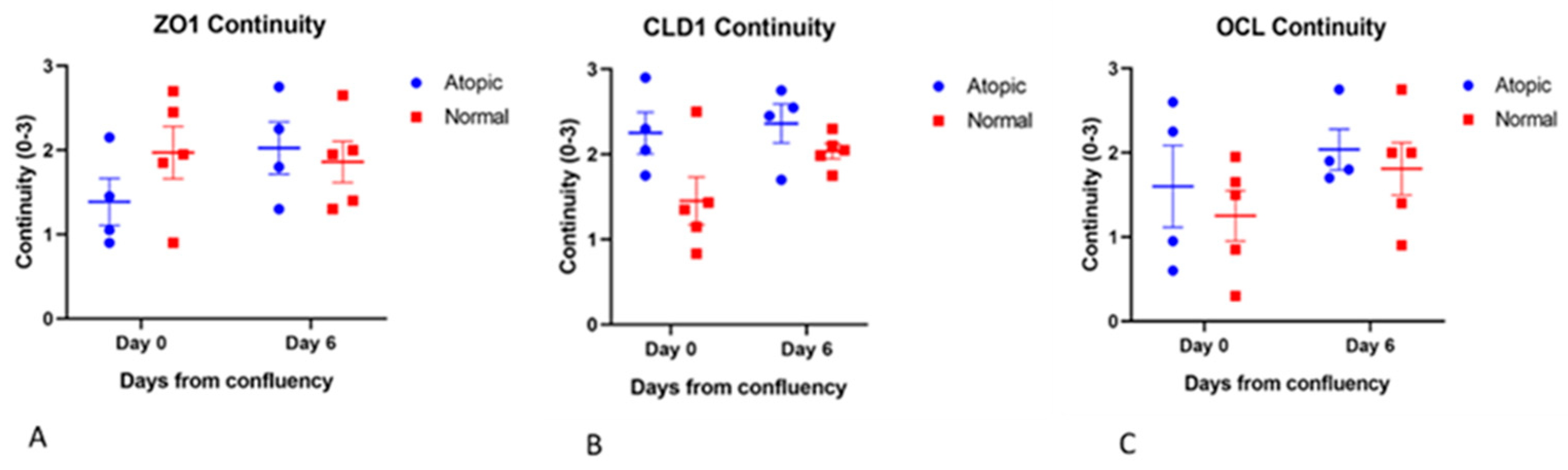
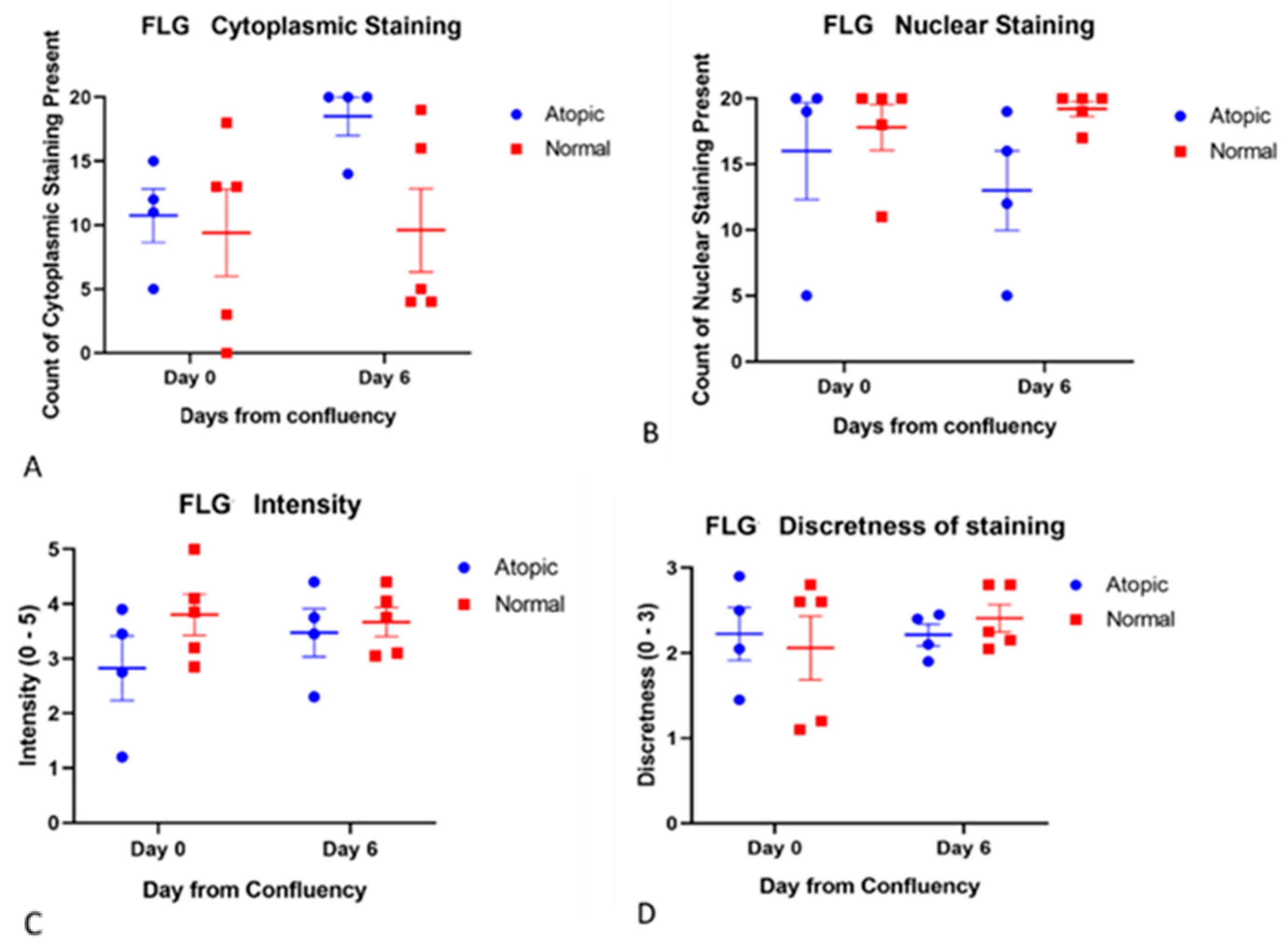
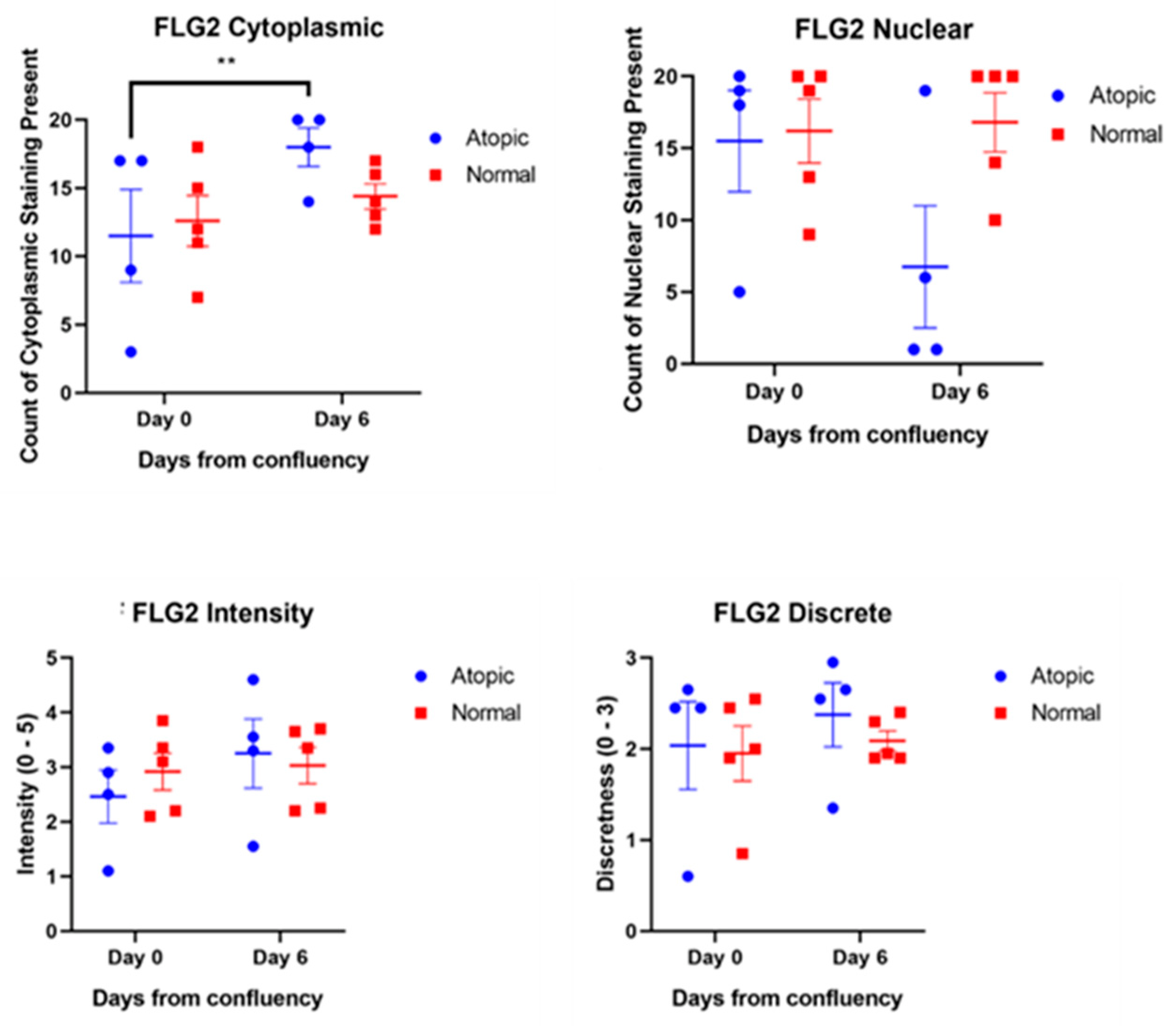
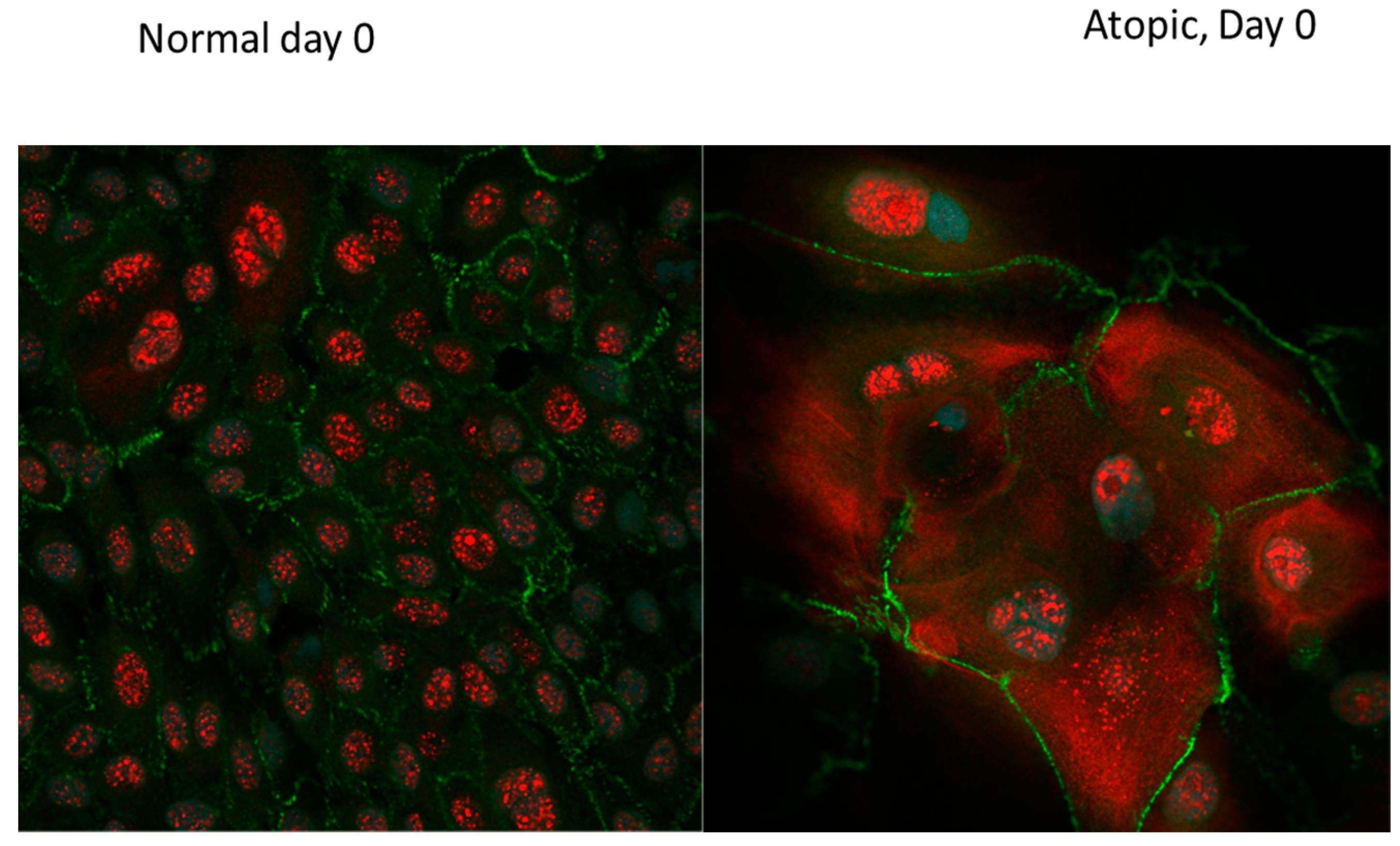
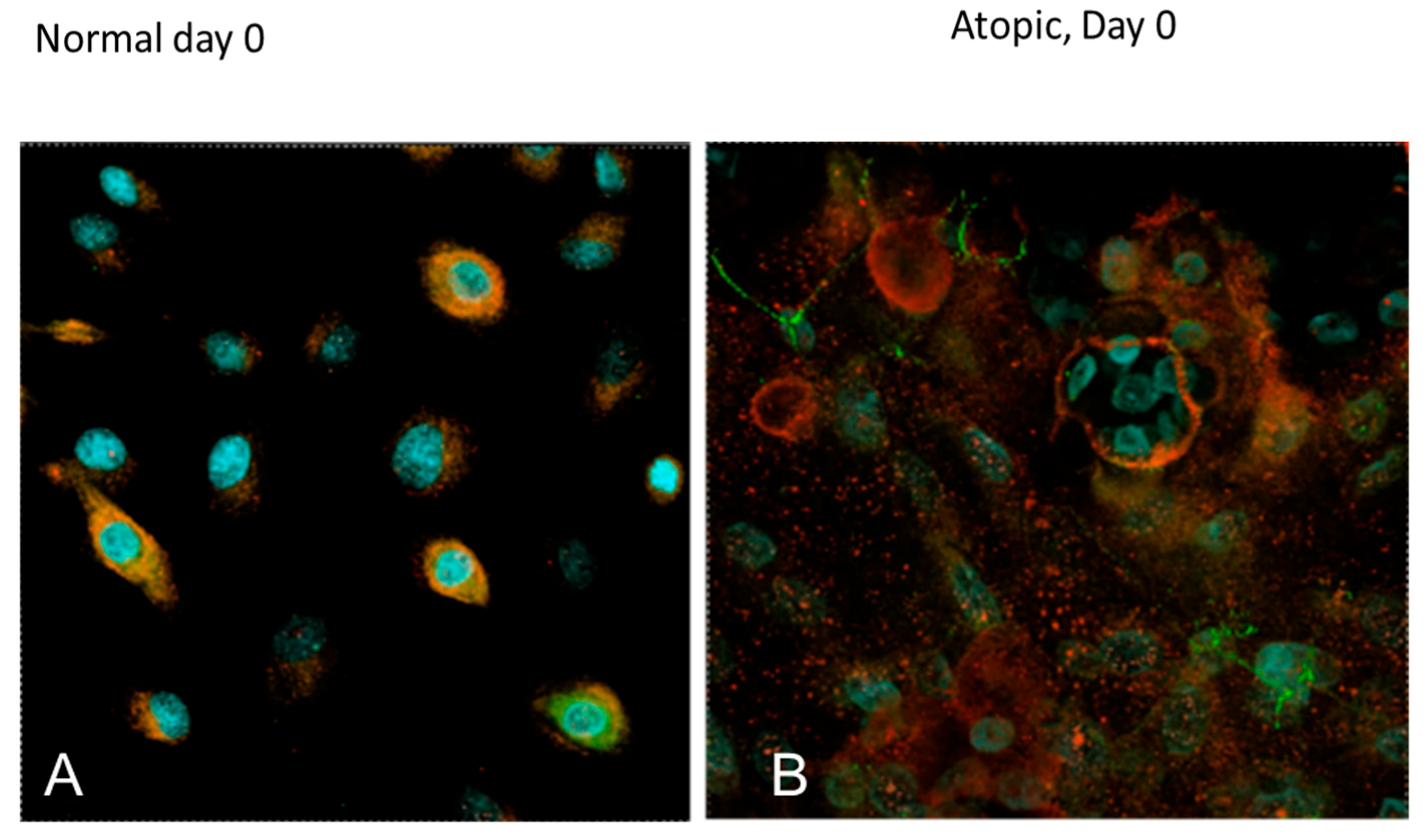
3.3. Subjective Evaluation of IF Images
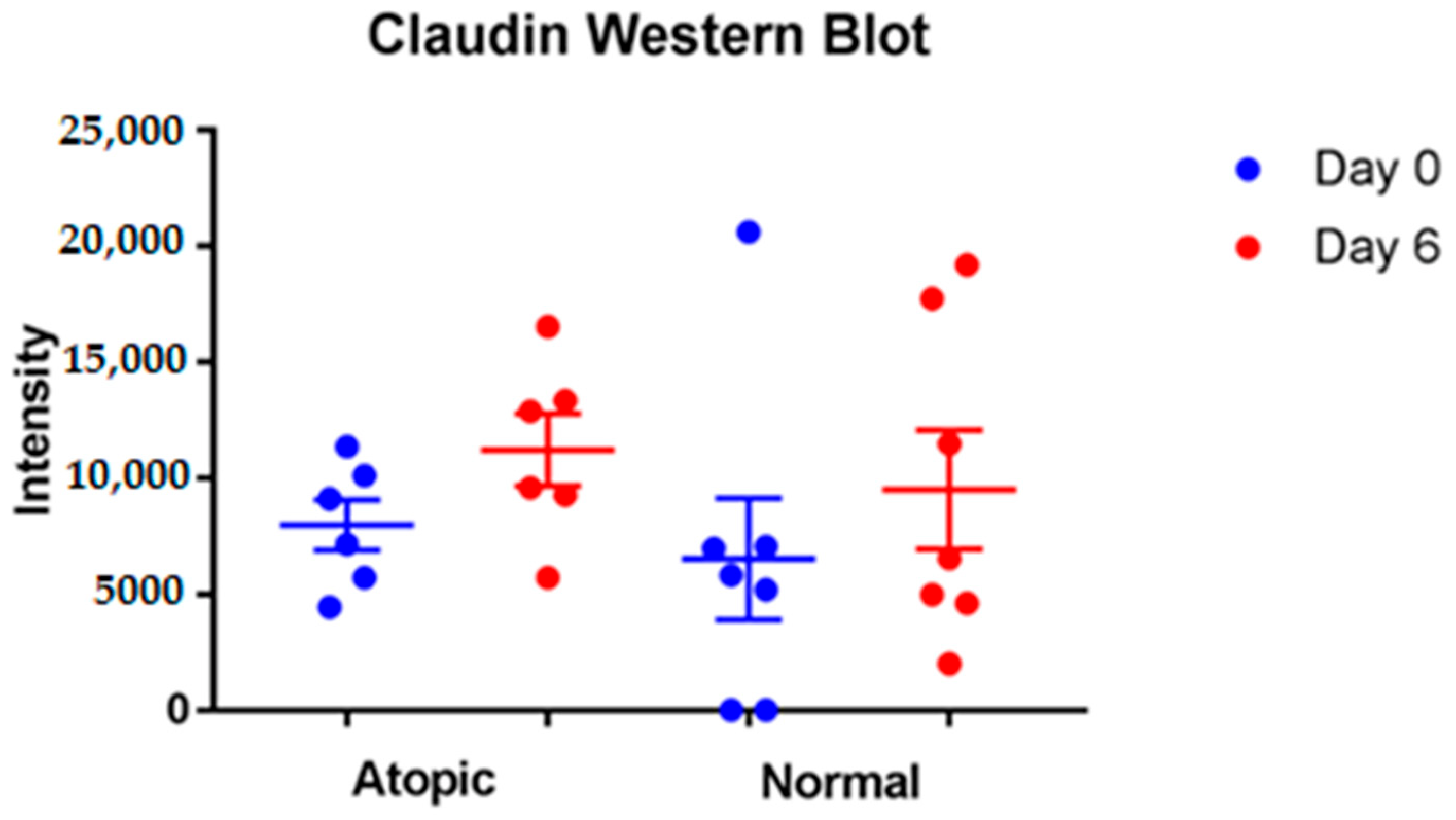
3.3.1. Tight Junction Proteins
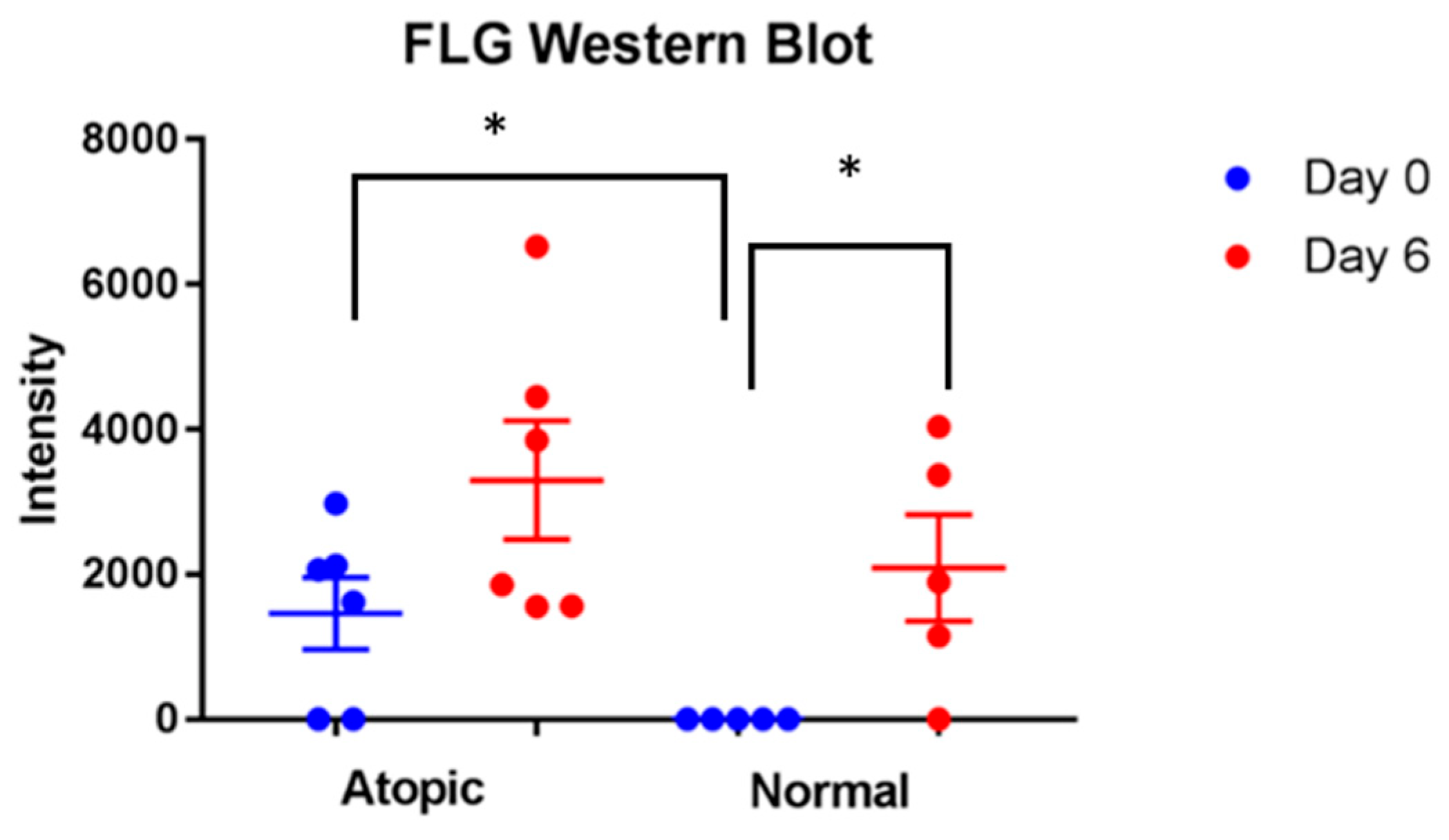
3.3.2. Filaggrins
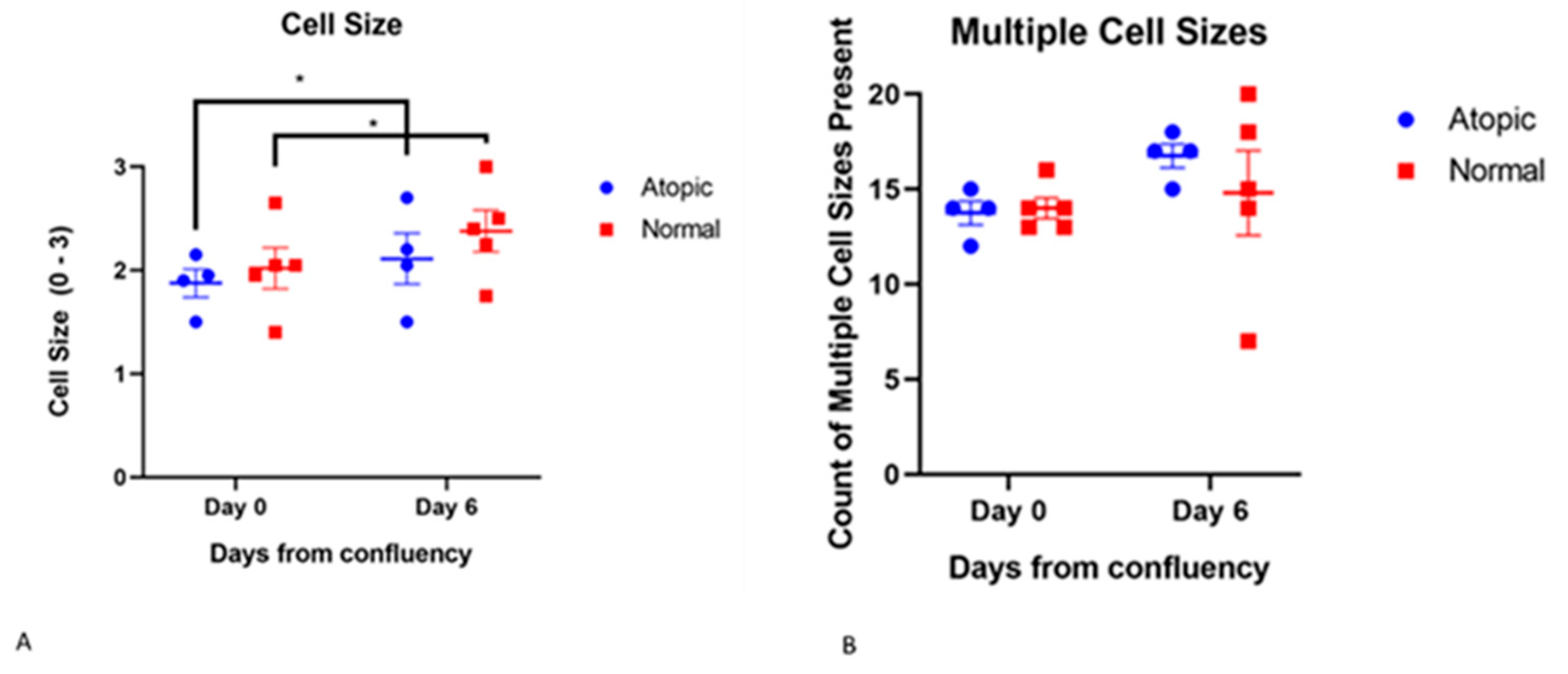
3.3.3. Cell Size
3.4. Western Blot
3.5. Images and Threedismensional Videos Made Using Confocal Miscroscope
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Santoro, D.; Marsella, R.; Pucheu-Haston, C.M.; Eisenschenk, M.N.C.; Nuttall, T.; Bizikova, P. Review: Pathogenesis of canine atopic dermatitis: Skin barrier and host-micro-organism interaction. Vet. Dermatol. 2015, 26, 84-e25. [Google Scholar] [CrossRef] [PubMed]
- Marsella, R. Advances in our understanding of canine atopic dermatitis. Vet. Dermatol. 2021, 32, 547-e151. [Google Scholar] [CrossRef] [PubMed]
- Combarros, D.; Cadiergues, M.-C.; Simon, M. Update on canine filaggrin: A review. Vet. Q. 2020, 40, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.H.; Ollier, W.E.; Nuttall, T.; McEwan, N.A.; Carter, S. Despite identifying some shared gene associations with human atopic dermatitis the use of multiple dog breeds from various locations limits detection of gene associations in canine atopic dermatitis. Vet. Immunol. Immunopathol. 2010, 138, 193–197. [Google Scholar] [CrossRef]
- Suriyaphol, G.; Suriyaphol, P.; Sarikaputi, M.; Theerawatanasirikul, S.; Sailasuta, A. Association of filaggrin (FLG) gene polymorphism with canine atopic dermatitis in small breed dogs. Thai J. Vet. Med. 2011, 41, 509–517. [Google Scholar]
- Olivry, T. Is the skin barrier abnormal in dogs with atopic dermatitis? Vet. Immunol. Immunopathol. 2011, 144, 11–16. [Google Scholar] [CrossRef]
- Olivry, T.; Paps, J.S.; Amalric, N. Transient and reversible reduction of stratum corneum filaggrin degradation products after allergen challenge in experimentally mite-sensitised atopic dogs. Vet. Dermatol. 2021, 33, 62-e20. [Google Scholar] [CrossRef]
- Bäsler, K.; Bergmann, S.; Heisig, M.; Naegel, A.; Zorn-Kruppa, M.; Brandner, J.M. The role of tight junctions in skin barrier function and dermal absorption. J. Control. Release 2016, 242, 105–118. [Google Scholar] [CrossRef]
- Brandner, J.M. Importance of Tight Junctions in Relation to Skin Barrier Function. Curr. Probl. Dermatol. 2016, 49, 27–37. [Google Scholar] [CrossRef]
- Brandner, J.; Zorn-Kruppa, M.; Yoshida, T.; Moll, I.; Beck, L.; De Benedetto, A. Epidermal tight junctions in health and disease. Tissue Barriers 2015, 3, e974451. [Google Scholar] [CrossRef]
- Yokouchi, M.; Kubo, A.; Kawasaki, H.; Yoshida, K.; Ishii, K.; Furuse, M.; Amagai, M. Epidermal tight junction barrier function is altered by skin inflammation, but not by filaggrin-deficient stratum corneum. J. Dermatol. Sci. 2014, 77, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Hönzke, S.; Wallmeyer, L.; Ostrowski, A.; Radbruch, M.; Mundhenk, L.; Schäfer-Korting, M.; Hedtrich, S. Influence of Th2 Cytokines on the Cornified Envelope, Tight Junction Proteins, and ß-Defensins in Filaggrin-Deficient Skin Equivalents. J. Investig. Dermatol. 2016, 136, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Steelant, B.; Seys, S.F.; Van Gerven, L.; Van Woensel, M.; Farre, R.; Wawrzyniak, P.; Krohn, I.K.; Bullens, D.M.; Talavera, K.; Raap, U.; et al. Histamine and T helper cytokine–driven epithelial barrier dysfunction in allergic rhinitis. J. Allergy Clin. Immunol. 2018, 141, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Bäsler, K.; Brandner, J.M. Tight junctions in skin inflammation. Pflugers Arch. 2017, 469, 3–14. [Google Scholar] [CrossRef]
- Kim, H.-J.; Cronin, M.; Ahrens, K.; Papastavros, V.; Santoro, D.; Marsella, R. A comparative study of epidermal tight junction proteins in a dog model of atopic dermatitis. Vet. Dermatol. 2015, 27, 40-e11. [Google Scholar] [CrossRef]
- Olivry, T.; Dunston, S.M. Expression patterns of superficial epidermal adhesion molecules in an experimental dog model of acute atopic dermatitis skin lesions. Veter-Dermatol. 2015, 26, 53–56. [Google Scholar] [CrossRef]
- Wu, L.; Oshima, T.; Li, M.; Tomita, T.; Fukui, H.; Watari, J.; Miwa, H. Filaggrin and tight junction proteins are crucial for IL-13-mediated esophageal barrier dysfunction. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 315, G341–G350. [Google Scholar] [CrossRef]
- Presland, R.B.; Kuechle, M.K.; Lewis, S.; Fleckman, P.; Dale, B.A. Regulated Expression of Human Filaggrin in Keratinocytes Results in Cytoskeletal Disruption, Loss of Cell–Cell Adhesion, and Cell Cycle Arrest. Exp. Cell Res. 2001, 270, 199–213. [Google Scholar] [CrossRef]
- Sandilands, A.; Sutherland, C.; Irvine, A.D.; McLean, W.H.I. Filaggrin in the frontline: Role in skin barrier function and disease. J. Cell Sci. 2009, 122, 1285–1294. [Google Scholar] [CrossRef]
- Wu, Z.; Hansmann, B.; Meyer-Hoffert, U.; Gläser, R.; Schröder, J.-M. Molecular Identification and Expression Analysis of Filaggrin-2, a Member of the S100 Fused-Type Protein Family. PLoS ONE 2009, 4, e5227. [Google Scholar] [CrossRef]
- Hansmann, B.; Ahrens, K.; Wu, Z.; Proksch, E.; Meyer-Hoffert, U.; Schröder, J.-M. Murine filaggrin-2 is involved in epithelial barrier function and down-regulated in metabolically induced skin barrier dysfunction. Exp. Dermatol. 2012, 21, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Pellerin, L.; Henry, J.; Hsu, C.-Y.; Balica, S.; Jean-Decoster, C.; Méchin, M.-C.; Hansmann, B.; Rodriguez, E.; Weindinger, S.; Schmitt, A.-M.; et al. Defects of filaggrin-like proteins in both lesional and nonlesional atopic skin. J. Allergy Clin. Immunol. 2013, 131, 1094–1102. [Google Scholar] [CrossRef] [PubMed]
- Furue, M. Regulation of Filaggrin, Loricrin, and Involucrin by IL-4, IL-13, IL-17A, IL-22, AHR, and NRF2: Pathogenic Implications in Atopic Dermatitis. Int. J. Mol. Sci. 2020, 21, 5382. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER Measurement Techniques for In Vitro Barrier Model Systems. J. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef]
- Chen, S.; Einspanier, R.; Schoen, J. Transepithelial electrical resistance (TEER): A functional parameter to monitor the quality of oviduct epithelial cells cultured on filter supports. Histochem. Cell Biol. 2015, 144, 509–515. [Google Scholar] [CrossRef]
- Fanton, N.; Santoro, D.; Cornegliani, L.; Marsella, R. Increased filaggrin-metabolizing enzyme activity in atopic skin: A pilot study using a canine model of atopic dermatitis. Vet. Dermatol. 2017, 28, 479-e111. [Google Scholar] [CrossRef]
- Johnson, L.G. Applications of imaging techniques to studies of epithelial tight junctions. Adv. Drug Deliv. Rev. 2005, 57, 111–121. [Google Scholar] [CrossRef]
- Zhang, D.; Karunaratne, S.; Kessler, M.; Mahony, D.; Rothnagel, J.A. Characterization of Mouse Profilaggrin: Evidence for Nuclear Engulfment and Translocation of the Profilaggrin B-Domain during Epidermal Differentiation. J. Investig. Dermatol. 2002, 119, 905–912. [Google Scholar] [CrossRef]
- Gutowska-Owsiak, D.; De La Serna, J.B.; Fritzsche, M.; Naeem, A.; Podobas, E.I.; Leeming, M.; Colin-York, H.; O’Shaughnessy, R.; Eggeling, C.; Ogg, G.S. Orchestrated control of filaggrin–actin scaffolds underpins cornification. Cell Death Dis. 2018, 9, 412. [Google Scholar] [CrossRef]
- Santoro, D.; Ahrens, K.; Marsella, R.; Segre, M. Evaluation of antimicrobial peptides and cytokine production in primary keratinocyte cell culture from healthy and atopic beagles. Exp. Dermatol. 2015, 24, 317–319. [Google Scholar] [CrossRef]
- Yoon, J.; Nishifuji, K.; Iwasaki, T. Development of an in vitro submerged culture system to synthesize epidermal ceramides in canine keratinocytes. Res. Vet. Sci. 2020, 130, 48–51. [Google Scholar] [CrossRef] [PubMed]
- Ide, K.; Waka, Y.; Masafumi, S.; Atsushi, Y.; Toshiroh, I. Koji NishifujiExpression Analysis of Desmosomal Components of the Novel Canine Epidermal Keratinocyte Cell Line (MSCEK). J. Vet. Med. Sci. 2010, 72, 1479–1482. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cerrato, S.; Ramió-Lluch, L.; Brazís, P.; Fondevila, D.; Segarra, S.; Puigdemont, A. Effects of sphingolipid extracts on the morphological structure and lipid profile in an in vitro model of canine skin. Vet. J. 2016, 212, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Marsella, R.; Wilkes, R.; Ahrens, K. Canine Epidermal Keratinocytes (CPEK) Grown in Monolayer Are Not Representative of Normal Canine Keratinocytes for Permeability Studies: Pilot Studies. Vet. Sci. 2022, 9, 25. [Google Scholar] [CrossRef]
- Kiatsurayanon, C.; Niyonsaba, F.; Smithrithee, R.; Akiyama, T.; Ushio, H.; Hara, M.; Okumura, K.; Ikeda, S.; Ogawa, H. Host Defense (Antimicrobial) Peptide, Human β-Defensin-3, Improves the Function of the Epithelial Tight-Junction Barrier in Human Keratinocytes. J. Investig. Dermatol. 2014, 134, 2163–2173. [Google Scholar] [CrossRef]
- Akiyama, T.; Niyonsaba, F.; Kiatsurayanon, C.; Nguyen, T.T.; Ushio, H.; Fujimura, T.; Ueno, T.; Okumura, K.; Ogawa, H.; Ikeda, S. The human cathelicidin LL-37 host defense peptide upregulates tight junction-related proteins and increases human epidermal keratinocyte barrier function. J. Innate Immun. 2014, 6, 739–753. [Google Scholar] [CrossRef]
- Ryu, W.-I.; Lee, H.; Bae, H.C.; Jeon, J.; Ryu, H.J.; Kim, J.; Kim, J.H.; Son, J.W.; Imai, Y.; Yamanishi, K.; et al. IL-33 down-regulates CLDN1 expression through the ERK/STAT3 pathway in keratinocytes. J. Dermatol. Sci. 2018, 90, 313–322. [Google Scholar] [CrossRef]
- Niehues, H.; Rikken, G.; van Vlijmen-Willems, I.M.J.J.; Rodijk-Olthuis, D.; van Erp, P.E.J.; Zeeuwen, P.L.J.M.; Schalkwijk, J.; van den Bogaard, E.H. Identification of Keratinocyte Mitogens: Implications for Hyperproliferation in Psoriasis and Atopic Dermatitis. JID Innov. 2021, 22, 100066. [Google Scholar] [CrossRef]
- Schirmacher, P.; Mann, A.; Breuhahn, K.; Blessing, M. Keratinocyte-Derived Granulocyte-Macrophage Colony Stimulating Factor Accelerates Wound Healing: Stimulation of Keratinocyte Proliferation, Granulation Tissue Formation, and Vascularization. J. Investig. Dermatol. 2001, 117, 1382–1390. [Google Scholar] [CrossRef]
- Tanaka, M.; Dykes, P.J.; Marks, R. Keratinocyte growth stimulation by granulocyte-macrophage colony-stimulating factor (GM-CSF). Keio J. Med. 1997, 46, 184–187. [Google Scholar] [CrossRef][Green Version]
- Pastore, S.; Giustizieri, M.L.; Mascia, F.; Giannetti, A.; Kaushansky, K.; Girolomoni, G. Dysregulated activation of activator protein 1 in keratinocytes of atopic dermatitis patients with enhanced expression of granulocyte/macrophage-colony stimulating factor. J. Investig. Dermatol. 2000, 115, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- Pastore, S.; Fanales-Belasio, E.; Albanesi, C.; Chinni, L.M.; Giannetti, A.; Girolomoni, G. Granulocyte macrophage colony-stimulating factor is overproduced by keratinocytes in atopic dermatitis. Implications for sustained dendritic cell activation in the skin. J. Clin. Investig. 1997, 99, 3009–3017. [Google Scholar] [CrossRef] [PubMed]
- Bratton, D.L.; Hamid, Q.; Boguniewicz, M.; E Doherty, D.; Kailey, J.M.; Leung, D.Y. Granulocyte macrophage colony-stimulating factor contributes to enhanced monocyte survival in chronic atopic dermatitis. J. Clin. Investig. 1995, 95, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Sekido, M.; Chimura, N.; Shibata, S.; Kondo, N.; Kamishina, H.; Kamishina, H.; Maeda, S. Production of GM-CSF Mediated by Cysteine Protease of Der f in Canine Keratinocytes. J. Vet. Med Sci. 2012, 74, 1033–1036. [Google Scholar] [CrossRef]
- Shibata, S.; Maeda, S.; Kondo, N.; Inoue, A.; Maeda, S.; Chimura, N.; Fukata, T. Effect of recombinant canine interferon-γ on granulocyte-macrophage colony-stimulating factor, transforming growth factor-β and CC chemokine ligand 17 mRNA transcription in a canine keratinocyte cell line (CPEK). Vet. Dermatol. 2011, 22, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.D.; Kim, J.P.; Zhang, K.; Sarret, Y.; Wynn, K.C.; Kramer, R.H.; Woodley, D.T. Epidermal Growth Factor (EGF) Promotes Human Keratinocyte Locomotion on Collagen by Increasing the α2 Integrin Subunit. Exp. Cell Res. 1993, 209, 216–223. [Google Scholar] [CrossRef]
- Blumenberg, M. Profiling and metaanalysis of epidermal keratinocytes responses to epidermal growth factor. BMC Genom. 2013, 14, 85. [Google Scholar] [CrossRef]
- Gibbs, S.; Pinto, A.N.S.; Murli, S.; Huber, M.; Hohl, D.; Ponec, M. Epidermal growth factor and keratinocyte growth factor differentially regulate epidermal migration, growth, and differentiation. Wound Repair Regen. 2000, 8, 192–203. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiao, C.; Gibson, A.M.; Bass, S.A.; Hershey, G.K.K. EGFR Signaling Blunts Allergen-Induced IL-6 Production and Th17 Responses in the Skin and Attenuates Development and Relapse of Atopic Dermatitis. J. Immunol. 2014, 192, 859–866. [Google Scholar] [CrossRef]
- Haase, I.; Evans, R.; Pofahl, R.; Watt, F. Regulation of keratinocyte shape, migration and wound epithelialization by IGF-1- and EGF-dependent signalling pathways. J. Cell Sci. 2003, 116, 3227–3238. [Google Scholar] [CrossRef]
- Choi, S.Y.; Lee, Y.J.; Kim, J.M.; Kang, H.J.; Cho, S.H.; Chang, S.E. Epidermal Growth Factor Relieves Inflammatory Signals in Staphylococcus aureus-Treated Human Epidermal Keratinocytes and Atopic Dermatitis-Like Skin Lesions in Nc/Nga Mice. BioMed Res. Int. 2018, 2018, 9439182. [Google Scholar] [CrossRef] [PubMed]
- Sääf, A.; Pivarcsi, A.; Winge, M.C.G.; Wahlgren, C.-F.; Homey, B.; Nordenskjöld, M.; Tengvall-Linder, M.; Bradley, M. Characterization of EGFR and ErbB2 expression in atopic dermatitis patients. Arch Dermatol. Res. 2012, 304, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Mascia, F.; Cataisson, C.; Lee, T.-C.; Threadgill, D.; Mariani, V.; Amerio, P.; Chandrasekhara, C.; Adeva, G.S.; Girolomoni, G.; Yuspa, S.H.; et al. EGFR Regulates the Expression of Keratinocyte-Derived Granulocyte/Macrophage Colony-Stimulating Factor In Vitro and In Vivo. J. Investig. Dermatol. 2010, 130, 682–693. [Google Scholar] [CrossRef] [PubMed]
- Roussel, A.J.J.; Bruet, V.; Marsella, R.; Knol, A.C.; Bourdeau, P.J. Tight junction proteins in the canine epidermis: A pilot study on their distribution in normal and in high IgE-producing canines. Can. J. Vet. Res. Rev. Can. Rech. Vet. 2015, 79, 46–51. [Google Scholar]
- Wise, S.K.; Laury, A.M.; Katz, E.H.; Bs, K.A.D.B.; Parkos, C.A.; Nusrat, A. Interleukin-4 and interleukin-13 compromise the sinonasal epithelial barrier and perturb intercellular junction protein expression. Int. Forum Allergy Rhinol. 2014, 4, 361–370. [Google Scholar] [CrossRef]
- Strid, J.; McLean, W.H.I.; Irvine, A.D. Too Much, Too Little or Just Enough: A Goldilocks Effect for IL-13 and Skin Barrier Regulation? J. Investig. Dermatol. 2016, 136, 561–564. [Google Scholar] [CrossRef]
- Santoro, D.; Marsella, R.; Ahrens, K.; Graves, T.K.; Bunick, D. Altered mRNA and protein expression of filaggrin in the skin of a canine animal model for atopic dermatitis. Vet. Dermatol. 2013, 24, 329-e73. [Google Scholar] [CrossRef]
- Chervet, L.; Galichet, A.; McLean, W.H.I.; Chen, H.; Suter, M.M.; Roosje, P.J.; Müller, E.J. Missing C-terminal filaggrin expression, NFkappaB activation and hyperproliferation identify the dog as a putative model to study epidermal dysfunction in atopic dermatitis. Exp. Dermatol. 2010, 19, e343–e346. [Google Scholar] [CrossRef] [PubMed]
- Donovan, M.; Salamito, M.; Thomas-Collignon, A.; Simonetti, L.; Desbouis, S.; Rain, J.-C.; Formstecher, E.; Bernard, D. Filaggrin and filaggrin 2 processing are linked together through skin aspartic acid protease activation. PLoS ONE 2020, 15, e0232679. [Google Scholar] [CrossRef] [PubMed]
- Pendaries, V.; Le Lamer, M.; Cau, L.; Hansmann, B.; Malaisse, J.; Kezic, S.; Serre, G.; Simon, M. In a three-dimensional reconstructed human epidermis filaggrin-2 is essential for proper cornification. Cell Death Dis. 2015, 6, e1656. [Google Scholar] [CrossRef]
- Cobiella, D.; Archer, L.; Bohannon, M.; Santoro, D. Pilot study using five methods to evaluate skin barrier function in healthy dogs and in dogs with atopic dermatitis. Vet. Dermatol. 2019, 30, 121-e34. [Google Scholar] [CrossRef] [PubMed]
- Marsella, R.; Samuelson, D.; Harrington, L. Immunohistochemical evaluation of filaggrin polyclonal antibody in atopic and normal beagles. Veter- Dermatol. 2009, 20, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Marsella, R.; Papastavros, V.; Ahrens, K.; Santoro, D. Decreased expression of caspase-14 in an experimental model of canine atopic dermatitis. Vet. J. 2016, 209, 201–203. [Google Scholar] [CrossRef] [PubMed]
- Nuttall, T. The genomics revolution: Will canine atopic dermatitis be predictable and preventable? Vet. Dermatol. 2013, 24, 10-8.e3-4. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marsella, R.; Ahrens, K.; Wilkes, R. Differences in Behavior between Normal and Atopic Keratinocytes in Culture: Pilot Studies. Vet. Sci. 2022, 9, 329. https://doi.org/10.3390/vetsci9070329
Marsella R, Ahrens K, Wilkes R. Differences in Behavior between Normal and Atopic Keratinocytes in Culture: Pilot Studies. Veterinary Sciences. 2022; 9(7):329. https://doi.org/10.3390/vetsci9070329
Chicago/Turabian StyleMarsella, Rosanna, Kim Ahrens, and Rachel Wilkes. 2022. "Differences in Behavior between Normal and Atopic Keratinocytes in Culture: Pilot Studies" Veterinary Sciences 9, no. 7: 329. https://doi.org/10.3390/vetsci9070329
APA StyleMarsella, R., Ahrens, K., & Wilkes, R. (2022). Differences in Behavior between Normal and Atopic Keratinocytes in Culture: Pilot Studies. Veterinary Sciences, 9(7), 329. https://doi.org/10.3390/vetsci9070329






