Single Nucleotide Polymorphisms, Gene Expression and Economic Evaluation of Parameters Associated with Mastitis Susceptibility in European Cattle Breeds
Abstract
:1. Introduction
2. Material and Methods
2.1. Animals and Experimental Samples
2.2. DNA Extraction and Polymerase Chain Reaction (PCR)
2.3. DNA Sequencing and Polymorphism Detection
2.4. Total RNA Extraction, Reverse Transcription and Quantitative Real-Time PCR
2.5. Economic Parameters
2.5.1. Total Variable Costs (TVC)
2.5.2. Total Fixed Costs (TFC)
2.5.3. Total Costs (TC)
2.5.4. Total Return
2.5.5. Net Income (Net Return)
2.5.6. Reduction Percentage in Net Profit
2.6. Statistical Analysis
3. Results
3.1. PCR-DNA Sequencing of Investigated Genes
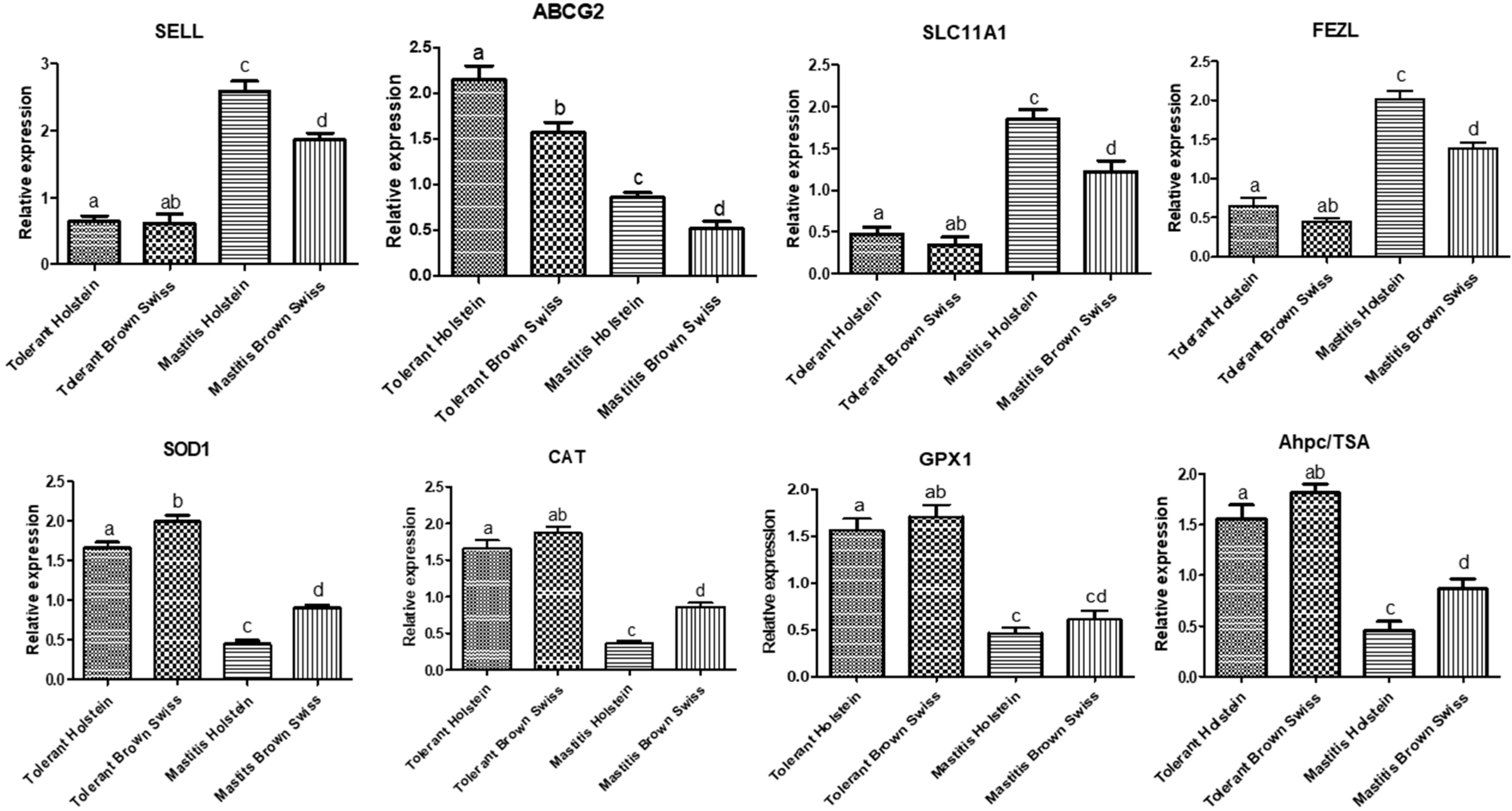
3.2. Gene Expression Pattern of Immune and Antioxidant Markers
3.3. Economic Evaluation of Parameters Associated with Mastitis Susceptibility in Holstein and Brown Swiss Breeds
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schukken, Y.H.; Günther, J.; Fitzpatrick, J.; Fontaine, M.C.; Goetze, L.; Holst, O.; Leigh, J.; Petzl, W.; Schuberth, H.J.; Sipka, A.; et al. Host-response patterns of intramammary infections in dairy cows. Vet. Immunol. Immunopathol. 2011, 144, 270–289. [Google Scholar] [CrossRef] [PubMed]
- Oviedo-Boyso, J.; Valdez-Alarcón, J.J.; Cajero-Juárez, M.; Ochoa-Zarzosa, A.; López-Meza, J.E.; Bravo-Patiño, A.; Baizabal-Aguirre, V.M. Innate immune response of bovine mammary gland to pathogenic bacteria responsible for mastitis. J. Infect. 2007, 54, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Giraudo, J.A.; Calzolari, A.; Rampone, H.; Rampone, A.; Giraudo, A.T.; Cristina Bogni, C.; Larriestra, A.; Nagel, R. Field Trials of a Vaccine Against Bovine Mastitis. 1. Evaluation in Heifers. J. Dairy Sci. 1997, 80, 845–853. [Google Scholar] [CrossRef]
- Pereira, U.P.; Oliveira, D.G.; Mesquita, L.R.; Costa, G.M.; Pereira, L.J. Efficacy of Staphylococcus aureus vaccines for bovine mastitis: A systematic review. Vet. Microbiol. 2011, 148, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Thompson-Crispi, K.; Atalla, H.; Miglior, F.; Mallard, B.A. Bovine mastitis: Frontiers in immunogenetics. Front. Immunol. 2014, 5, 493. [Google Scholar] [CrossRef] [PubMed]
- Albenzio, M.; Taibi, L.; Muscio, A.; Sevi, A. Prevalence and etiology of subclinical mastitis in intensively managed flocks and related changes in the yield and quality of milk. Small Rum. Res. 2002, 43, 219–226. [Google Scholar] [CrossRef]
- Patnaik, S.; Prasad, A.; Ganguly, S. Mastitis, an Infection of Cattle Udder: A Review. J. Chem. Bio. Phys. Sci. Sec. B. 2013, 3, 2676–2678. [Google Scholar]
- Hogeveen, H.; Huijps, K.; Lam, T.J. Economic aspects of mastitis. New developments. New Zealand Vet. J. 2011, 59, 16–23. [Google Scholar] [CrossRef]
- Halasa, T.; Huijps, K.; Sters, O.; Hogeveen, H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q. 2007, 29, 18–31. [Google Scholar] [CrossRef]
- Mekibib, B.; Furgasa, M.; Abunna, F.; Megersa, B.; Regassa, A. Bovine Mastitis: Prevalence, Risk Factors and Major Pathogens in Dairy Farms of Holeta Town, Central Ethiopia. Vet. World 2010, 3, 397–403. [Google Scholar] [CrossRef]
- El-tarabany, M.S.; Ali, M.A. Incidence, Production and Economic Losses of Clinical Mastitis in Egyptian Holstein Cows. Indian J. Appl. Anim. Sci. 2015, 5, 813–818. [Google Scholar]
- Pellegrino, M.; Berardo, N.; Giraudo, J.; Nader-Macías, M.E.F.; Bogni, C. Bovine mastitis prevention: Humoral and cellular response of dairy cows inoculated with lactic acid bacteria at the dry-off period. Benef. Microbes 2017, 8, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Akira, S. Roles of toll-like receptors in innate immune responses. Genes Cells 2001, 6, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Prakash, O.; Kumar, A.; Sonwane, A.; Rathore, R.; Singh, R.V.; Chauhan, A.; Kumar, P.; Renjith, R.; Yadav, R.; Bhaladhare, A.; et al. Polymorphism of cytokine and innate immunity genes associated with bovine brucellosis in cattle. Mol. Biol. Rep. 2014, 41, 2815–2825. [Google Scholar] [CrossRef] [PubMed]
- Heringstad, B.; Rekaya, R.; Gianola, D.; Klemetsdal, G.; Welgel, K.A. Genetic change for clinical mastitis in Norwegian cattle: A threshold model analysis. J. Dairy Sci. 2003, 86, 369–375. [Google Scholar] [CrossRef] [Green Version]
- Heringstad, B.; Klemetsdal, G.; Ruane, J. Selection for mastitis resistance in dairy cattle: A review with focus on the situation in the Nordic countries. Livest. Prod. Sci. 2000, 64, 95–106. [Google Scholar] [CrossRef]
- Negussie, E.; Stranden, I.; Mantysaari, E.A. Genetic analysis of liability to clinical mastitis, with somatic cell score and production traits using bivariate threshold–linear and linear–linear models. Livest. Sci. 2008, 117, 52–59. [Google Scholar] [CrossRef]
- Hayes, B.J.; Pryce, J.; Chamberlain, A.J.; Bowman, P.J.; Goddard, M.E. Genetic architecture of complex traits and accuracy of genomic prediction: Coat colour, milk-fat percentage, and type in Holstein cattle as contrasting model traits. PLoS Genet. 2010, 6, 39. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.-W.; Liao, X.; Stothard, P.; Chung, W.H.; Jeon, H.J.; Miller, S.P.; Choi, S.; Lee, J.K.; Yang, B.; Lee, K.T.; et al. Whole-genome analyses of Korean native and Holstein cattle breeds by massively parallel sequencing. PLoS ONE 2014, 9, 101–127. [Google Scholar] [CrossRef]
- Szyda, J.; Frąszczak, M.; Mielczarek, M.; Giannico, R.; Minozzi, G.; Nicolazzi, E.L.; Kamiński, S.; Wojdak-Maksymiec, K. The assessment of inter-individual variation of whole-genome DNA sequence in 32 cows. Mamm. Genome 2015, 26, 658–665. [Google Scholar] [CrossRef] [Green Version]
- Mielczarek, M.; Fraszczak, M.; Giannico, R.; Minozzi, G.; Williams, J.L.; WojdakMaksymiec, K.; Szyda, J. Analysis of copy number variations in Holstein-Friesian cow genomes based on whole-genome sequence data. J. Dairy Sci. 2017, 100, 5515–5525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VanRaden, P.M.; Tooker, M.E.; O’Connell, J.R.; Cole, J.B.; Bickhart, D.M. Selecting sequence variants to improve genomic predictions for dairy cattle. Genet. Select. Evol. 2017, 49, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez, M.-P.; Govignon-Gion, A.; Croiseau, P.; Fritz, S.; Hozé, C.; Miranda, G.; Martin, P.; Barbat-Leterrier, A.; Letaïef, R.; Rocha, D.; et al. Within-breed and multi-breed GWAS on imputed whole-genome sequence variants reveal candidate mutations affecting milk protein composition in dairy cattle. Genet. Select. Evol. 2017, 49, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez, M.-P.; Govignon-Gion, A.; Ferrand, M.; Gelé, M.; Pourchet, D.; Amigues, Y.; Fritz, S.; Boussaha, M.; Capitan, A.; Rocha, D.; et al. Whole-genome scan to detect quantitative trait loci associated with milk protein composition in 3 French dairy cattle breeds. J. Dairy Sci. 2016, 99, 8203–8215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, W.; Fang, X.; Zhang, C.; Pang, Y.; Xu, H.; Gu, C.; Shao, R.; Lei, C.; Chen, H. Two novel SNPs of the ABCG2 gene and its associations with milk traits in Chinese Holsteins. Mol. Biol. Rep. 2011, 38, 2927–2932. [Google Scholar] [CrossRef]
- Asadollahpour, N.H.; Ansari, M.S.; Edriss, M.A. Effect of LEPR, ABCG2 and SCD1 gene polymorphisms on reproductive traits in the Iranian Holstein cattle. Reprod. Domest. Anim. 2014, 49, 769–774. [Google Scholar] [CrossRef]
- Dusza, M.; Pokorska, J.; Makulska, J.; Kulaj, D.; Cupial, M. L-selectin gene polymorphism and its association with clinical mastitis, somatic cell score, and milk production in Polish Holstein-Friesian cattle. Czech J. Anim. Sci. 2018, 63, 256–262. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Wang, Y.; Chen, H.; Gu, C.; Fang, X. SLC11A1 gene polymorphisms are not associated to somatic cell score and milk yield in Chinese Holstein. Vet. Immunol. Immunopathol. 2009, 127, 389–392. [Google Scholar] [CrossRef]
- Ali, G.E.; Ibrahim, M.A.; Zaki, S.M. Association assessment of single nucleotide polymorphism in Forebrain Embryonic Zinc Finger-Like (FEZL) gene with mastitis susceptibility in Holstein cattle (Bos taurus). Large Anim. Rev. 2019, 25, 163–171. [Google Scholar]
- Boom, R.; Sol, C.J.; Salimans, M.M.; Jansen, C.L.; Wertheim-van Dillen, P.M.; Noordaa, J.V.D. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990, 28, 495–503. [Google Scholar] [CrossRef] [Green Version]
- Boesenberg-Smith, K.A.; Pessarakli, M.M.; Wolk, D.M. Assessment of DNA Yield and Purity: An Overlooked Detail of PCR Troubleshooting. Clin. Microbiol. Newsl. 2012, 34, 1–6. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Atallah, S.T. Economic and Productive Efficiency of Veterinary Management in Dairy Farms. Ph.D. Thesis, Medicine Alexandria University, Alexandria, Egypt, 1997. [Google Scholar]
- Muhammad, D.F. Production Performance and Economic Appraisal of Commercial Layers in District Chakwal. Ph.D. Thesis, NWFP Agriculture University, Peshawar, Pakistan, 2002. [Google Scholar]
- Atallah, S.T. Study the economic and productive efficiency of some broiler farms in relation to ration constituents. Minoufyia Vet. J. 2000, 1, 169–183. [Google Scholar]
- Tom, K. Production and Technical Efficiency on Australian Dairy Farms; ACT: Canberra, Australia, 2000. [Google Scholar]
- Fidan, H. Cattle fattening systems and environmental regulations in Turkey. J. Appl. Biosci. 2010, 25, 1579–1584. [Google Scholar]
- Atallah, S.T. Effect of cattle diseases on reproductive, productive and economic efficiency of dairy farms. Minufiya Vet. J. 2004, 1, 99–114. [Google Scholar]
- El-Tahawy, A.S. Factors Affecting the Profitability of Fish Farms and Their Relation to Veterinary Management. M.V.Sc. Thesis, Alexandria University, Faculty of Veterinary Medicine, Alexandria, Egypt, 2004. [Google Scholar]
- Mallard, B.A.; Emam, M.; Paibomesai, M.; Thompson-Crispi, K.; Wagter-Lesperance, L. Genetic selection of cattle for improved immunity and health. Jpn. J. Vet. Res. 2015, 63, 37–44. [Google Scholar]
- Olsen, H.G.; Knutsen, T.M.; Lewandowska-Sabat, A.M.; Grove, H.; Nome, T.; Svendsen, M.; Arnyasi, M.; Sodeland, M.; Sundsaasen, K.K.; Dahl, S.R.; et al. Fine mapping of a QTL on bovine chromosome 6 using imputed full sequence data suggests a key role for the group specific component (GC) gene in clinical mastitis and milk production. Genet. Select. Evol. 2016, 48, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, X.; Zhao, Z.; Jiang, P.; Yu, H.; Xiao, H.; Yang, R. Identification of the bovine HSL gene expression profiles and its association with fatty acid composition and fat deposition traits. Meat Sci. 2017, 131, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Groeneveld, L.F.; Lenstra, A.J.; Eding, H.; Toro, A.M.; Scherf, B.; Pilling, D.; Negrini, R.; Finlay, K.E.; Jianlin, H.; Groeneveld, E.; et al. Genetic diversity in farm animals e a review. Anim. Genet. 2010, 41, 6–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautier, M.; Faraut, T.; Moazami-Goudarzi, K.; Navratil, V.; Foglio, M.; Grohs, C.; Boland, A.; Garnier, J.G.; Boichard, D.; Lathrop, G.M.; et al. Genetic and haplotypic structure in 14 European and African cattle breeds. Genetics 2007, 177, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- McKay, D.S.; Schnabel, D.R.; Murdoch, M.B.; Matukumalli, K.L.; Aerts, J.; Coppieters, W.; Crews, D.; Neto, D.E.; Gil, A.C.; Gao, C.; et al. An assessment of population structure in eight breeds of cattle using a whole genome SNP panel. BMC Genet. 2008, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Socol, C.T.; Iacob, L.; Mihalca, I.; Criste, F.L. Molecular and population genetics tools for farm animal genetic resources conservation: Brief overview. J. Anim. Sci. Biotechnol. 2015, 48, 95–102. [Google Scholar]
- Svensson, E.; Anderung, C.; Baubliene, J. Tracing genetic change over time using nuclear SNPs in ancient and modern cattle. Anim. Genet. 2007, 38, 378–383. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, S.; Cheng, Z.; Cooke, J.S.; Werling, D.; Wathes, D.C.; Pollott, G.E. Polymorphisms in the selectin gene cluster are associated with fertility and survival time in a population of Holstein Friesian cows. PLoS ONE 2017, 1812, 0175555. [Google Scholar] [CrossRef] [Green Version]
- Komisarek, J.; Dorynek, Z. Effect of ABCG2, PPARGC1A, OLR1 and SCD1 gene polymorphism on estimated breeding values for functional and production traits in Polish Holstein-Friesian bulls. J. Appl. Genet. 2009, 50, 125–132. [Google Scholar] [CrossRef]
- Soltani-Ghombavani, M.; Ansari-Mahyari, S.; Rostami, M.; Ghanbari-Baghenoei, S.; Edriss, M.A. Effect of polymorphisms in the ABCG2, LEPR and SCD1 genes on milk production traits in Holstein cows. S. Afr. J. Anim. Sci. 2016, 46, 196–203. [Google Scholar] [CrossRef] [Green Version]
- Alima, M.A.; Xiea, Y.; Fana, Y.; Wua, X.; Suna, D.; Zhanga, Y.; Zhanga, S.; Zhanga, Y.; Zhanga, Q.; Liub, L. Genetic effects of ABCG2 polymorphism on milk production traits in the Chinese Holstein cattle. Appl. Anim. Res. 2013, 41, 333–338. [Google Scholar] [CrossRef]
- Holder, A.; Garty, R.; Elder, C.; Mesnard, P.; Laquerbe, C.; Bartens, M.-C.; Salavati, M.; Shabbir, M.Z.; Tzelos, T.; Connelly, T.; et al. Analysis of Genetic Variation in the Bovine SLC11A1 Gene, Its Influence on the Expression of NRAMP1 and Potential Association with Resistance to Bovine Tuberculosis. Front. Microbiol. 2020, 11, 1420. [Google Scholar] [CrossRef] [PubMed]
- Kadarmideen, H.N.; Ali, A.A.; Thomson, P.C.; Müller, B.; Zinsstag, J. Polymorphisms of the SLC11A1 gene and resistance to bovine tuberculosis in African Zebu cattle. Anim. Genet. 2011, 42, 656–658. [Google Scholar] [CrossRef] [PubMed]
- Somasundaram, R.K.; Gupta, I.K.; Raja, N.; Periasamy, K.; Ramasamy, S. Polymorphism of Bovine Forebrain Embryonic Zinc Finger Like (FEZL) Gene Associated with Resistance to Mastitis in Indian Cattle. Inter. J. Livest. Res. 2020, 10, 144–149. [Google Scholar] [CrossRef]
- Darwish, A.; Ebissy, E.; Ateya, A.; El-Sayed, A. Single nucleotide polymorphisms, gene expression and serum profile of immune and antioxidant markers associated with postpartum disorders susceptibility in Barki sheep. Anim. Biotech. 2021, 1–13. Available online: https://pubmed.ncbi.nlm.nih.gov/34406916/ (accessed on 20 September 2021). [CrossRef]
- Asadpour, R.; Zangiband, P.; Nofouzi, K.; Saberivand, A. Differential expression of antioxidant genes during clinical mastitis of cow caused by Staphylococcus aureus and Escherichia coli. Vet. Arh. 2021, 91, 451–458. [Google Scholar] [CrossRef]
- Ateya, A.; El-Sayed, A.; Mohamed, R. Gene expression and serum profile of antioxidant markers discriminate periparturient period time in dromedary camels. Mamm. Res. 2021, 66, 603–613. [Google Scholar] [CrossRef]
- Kishimoto, T.K.; Rothlein, R. Integrins, ICAMs, and selectins: Role and regulation of adhesion molecules in neutrophil recruitment to inflammatory sites. Adv. Pharmacol. 1994, 25, 117–169. [Google Scholar]
- Diez-Fraille, A.; Mehrzad, J.; Meyer, E.; Duchateau, L.; Burvenich, C. Comparison of L-selectin and Mac-1 expression on blood and milk neutrophils during experimental Escherichia coli-induced mastitis in cows. Am. J. Vet Res. 2004, 65, 1164–1171. [Google Scholar] [CrossRef]
- Litman, T.; Brangi, M.; Hudson, E.; Fetch, P.; Abati, A.; Ross, D.D.; Miyake, K.; Resau, J.H.; Bates, S.E. The multidrug-resistant phenotype associated with overexpresion of the new ABC half-transporter, MXR (ABCG2). J. Cell. Sci. 2000, 113, 2011–2021. [Google Scholar] [CrossRef]
- Farke, C.; Mayer, H.H.; Bruckmaier, R.M.; Albrecht, C. Differential expression of ABC transporters and their regulatory genes during lactation and dry period in bovine mammary tissue. J. Dairy Sci. 2008, 75, 406–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, N.; Ganguly, I.; Singh, R.; Deb, S.M.; Kumar, S.; Sharma, A.; Mitra, A. DNA polymorphism in SLC11A1 Gene and its association with Brucellosis resistance in Indian Zebu (Bos Indicus) and crossbred (Bos Indicus × Bos Taurus) Cattle. Asian-Australas. J. Anim. Sci. 2011, 24, 898–904. [Google Scholar] [CrossRef]
- Sugimoto, M.; Itoh, T.; Gotoh, Y.; Kawahara, T.; Moriya, H.; Uchimura, Y.; Sugimoto, Y. Short communication: Enhanced clinical mastitis resistance in Holsteins with a FEZL p.Gly105 (12_13) polymorphism. J. Dairy Sci. 2010, 94, 2103–2107. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Uchiza, M.; Kuniyuki, M. Effects of a Forebrain Embryonic Zinc Finger-Like p.Gly105 (12_13) polymorphism on mastitis resistance: An embryo-transfer study. J. Dairy Sci. 2011, 94, 2103–2107. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Fujikawa, A.; Womack, J.E.; Sugimoto, Y. Evidence that bovine forebrain embryonic zinc finger-like gene influences immune response associated with mastitis resistance. Proc. Natl. Acad. Sci. USA 2006, 103, 6454–6459. [Google Scholar] [CrossRef] [Green Version]
- Tam, K.J.; Hui, D.H.F.; Lee, W.W.; Dong, M.; Tombe, T.; Jiao, I.Z.F.; Khosravi, S.; Takeuchi, A.; Peacock, J.W.; Ivanova, L.; et al. Semaphorin 3C drives epithelial-to-mesenchymal transition, invasiveness, and stem-like characteristics in prostate cells. Sci. Rep. 2017, 7, 11501. [Google Scholar] [CrossRef] [Green Version]
- Masella, R.; Di Benedetto, R.; Varì, R.; Filesi, C.; Giovannini, C. Novel mechanisms of natural antioxidant compounds in biological systems: Involvement of glutathione and glutathione-related enzymes. J. Nutr. Biochem. 2005, 16, 577–586. [Google Scholar] [CrossRef]
- Glasauer, A.; Chandel, N.S. Targeting antioxidants for cancer therapy. Biochem. Pharmacol. 2014, 92, 90–101. [Google Scholar] [CrossRef]
- Caverly, J.M.; Diamond, G.; Gallup, J.M.; Brogden, K.A.; Dixon, R.A.; Ackermann, M.R. Coordinated Expression of Tracheal Antimicrobial Peptide and Inflammatory-Response Elements in the Lungs of Neonatal Calves with Acute Bacterial Pneumonia. Infect. Immun. 2003, 71, 2950–2955. [Google Scholar] [CrossRef] [Green Version]
- Wadleya, A.J.; Aldred, S.; Coles, S.J. An unexplored role for Peroxiredoxin in exercise-induced redox signalling? Redox Biol. 2016, 8, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Celi, P. The role of oxidative stress in small ruminants’ health and production. Rev. Bras. Zootec. 2010, 39, 348–363. [Google Scholar] [CrossRef] [Green Version]
- Cuschieri, J.; Maier, R.V. Oxidative stress, lipid rafts, and macrophage reprogramming. Antioxid. Redox Signal 2007, 9, 1485–1497. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, M.; Moroni, P.; Paape, M.J.; Bannerman, D.D. Evaluation of assays for the measurement of bovine neutrophil reactive oxygen species. Vet. Immunol. Immunopathol. 2007, 115, 107–125. [Google Scholar] [CrossRef] [PubMed]
- Sordillo, L.M.; Aitken, S.L. Impact of oxidative stress on the health and immune function of dairy cattle. Vet. Immunol. Immunopathol. 2009, 128, 104–109. [Google Scholar] [CrossRef]
- Blöttner, S.; Heins, B.J.; Wensch-Dorendorf, M.; Hansen, L.B.; Swalve, H.H. Short communication: A comparison between purebred Holstein and Brown Swiss × Holstein cows for milk production, somatic cell score, milking speed, and udder measurements in the first 3 lactations. J. Dairy Sci. 2011, 94, 5212–5216. [Google Scholar] [CrossRef]
- Wolfova, M.; Stıpkova, M.; Wolf, J. Incidence and economics of clinical mastitis in five Holstein herds in the Czech Republic. Prevent. Vet. Med. 2006, 77, 48–64. [Google Scholar] [CrossRef]
| Primer | Forward | Reverse | Annealing Temperature (°C) | Length of PCR Product (bp) | Reference |
|---|---|---|---|---|---|
| SELL | 5’-GAAAGAAAGTAAGCCTTTCTGG -3′ | 5′-CCAGAAAGGCTTACTTTCTTTC-3′ | 60 | 809 | Current study |
| ABCG2 | 5’-AAAGCTTGCGAAGTGAGGCTGA-3′ | 5′-GTAATAAGCTCCATTGCAATAC-3′ | 62 | 756 | Current study |
| SLC11A1 | 5′-GCTTGCCATGCCCGTGAGGGGCT-3′ | 5′-TAGTAGAGATGGCAGACCTCGC-3′ | 64 | 450 | Current study |
| FEZL | 5’-GATTGGACCGTCTCAATTATACA-3′ | 5′-CTGTGTGTTGAGGAGACCGGAC-3′ | 62 | 813 | Current study |
| SOD1 | 5′-GCTTGCCATGCCCGTGAGGGGCT-3′ | 5′-GAATCCAGCCACAGCCCCAGC-3′ | 60 | 334 | Current study |
| CAT | 5′-CTATCCTGACACTCACCGCCAC-3′ | 5′-GAAAGTCCGCACCTGAGTGACAT-3′ | 64 | 268 | Current study |
| GPX1 | 5′-GGTCGCCCGCCTTTTAAAAGCAG-3′ | 5′-TCGGTCATGAGAGCAGTGGCG-3′ | 64 | 534 | Current study |
| AhpC/TSA | 5′-TAAGAATTGTTTAAACTGAAA-3′ | 5′-TATGATTCAGCAGTTTTAAGTC-3′ | 62 | 480 | Current study |
| Gene | Primer | Product Length (bp) | Annealing Temperature (°C) | Accession Number | Source |
|---|---|---|---|---|---|
| SELL | F5′-CAACAGGAAGAGTAAGGAGGAC-3 R5′-TTGTCCATGGCCGCTGCATGAC-3′ | 151 | 60 | NM_174182.1 | Current study |
| ABCG2 | F5′-CTGAAGGAGCTGTGTTAAGT-3′ R5′-CCAGAATGGCATTGAGGCCAG-3′ | 144 | 62 | EU570105.1 | Current study |
| SLC11A1 | F5′-TGTGGCTGGATTCAAACTGCTC-3′ R5′-AGATGGCAGACCTCGCCCAAGT-3′ | 123 | 62 | NM_174652.2 | Current study |
| FEZL | F5′-CGTGTGCTGCAAGGCCGAGCTG-3′ R5′-GCGGAGTCCAGGTAGTTGAAGTA-3′ | 138 | 62 | NM_001038198.2 | Current study |
| SOD1 | F5′-GGAAGCTGTGGGCCTTCACGG-3′ R5′-CCAGCCTGAAGATCCGACTCA-3′ | 88 | 64 | NM_174615.2 | Current study |
| CAT | F5′-TATCCTGACACTCACCGCCA-3′ R5′-CGCTGGTAGTTGGCCACTCGA-3′ | 92 | 62 | MK423993.1 | Current study |
| GPX1 | F5′-CTGGATTCGGAAACGGATACC-3′ R5′-ACGTTCTCAATGAGCAGCACCT-3′ | 164 | 60 | NM_174076.3 | Current study |
| AhpC/TSA | F5′-TCTGAATCTATTTTCATGTGTA-3′ R5′-CCACCAATGTTTCCTTACTTA-3′ | 124 | 62 | XM_005210409.4 | Current study |
| ß. actin | F5′-CTAGGCACCAGGGCGTAATG-3′ R5′-CCGTGCTCAATGGGGTACTT-3′ | 109 | 60 | AF191490.1 | Current study |
| Gene | SNPs | Tolerant n = 120 | Mastitic n = 120 | Total | Type of Mutation | Amino Acid Number and Type | Chi Value | p-Value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Holstein n = 60 | Brown Swiss n = 60 | Holstein n = 60 | Brown Swiss n = 60 | |||||||
| SELL | A226G | 34 | - | - | 34/240 | Non-synonymous | 77 M to V | 21.26 | <0.0001 | |
| C260T | 41 | - | - | 41/240 | Non-synonymous | 87 P to L | 25.64 | <0.0001 | ||
| G338A | - | 38 | - | 38/240 | Non-synonymous | 113 R to K | 23.76 | <0.0001 | ||
| T695C | - | 28 | - | 28/240 | Non-synonymous | 232 L to P | 17.51 | <0.0001 | ||
| ABCG2 | A91G | - | - | 18 | 18/240 | Non-synonymous | 31 T o A | 11.26 | <0.0001 | |
| T108G | - | 32 | 32/240 | Non-synonymous | 36 H to Q | 20.01 | <0.0001 | |||
| G630A | - | 38 | - | 38/240 | Synonymous | 210 T | 23.76 | <0.0001 | ||
| SLC11A1 | A160 G | - | 33 | - | 33/240 | Non-synonymous | 54 T to A | 20.63 | <0.0001 | |
| A218G | - | 29 | - | 29/240 | Non-synonymous | 73 Y to C | 18.13 | <0.0001 | ||
| A230C | - | 23 | - | 23/240 | Non-synonymous | 77 E to A | 14.38 | <0.0001 | ||
| FEZL | T262A | 22 | 31 | - | 53/240 | Non-synonymous | 88 C to N | 33.14 | <0.0001 | |
| G263A | - | 29 | - | - | 29/240 | 18.13 | <0.0001 | |||
| T760C | 27 | - | - | 27/240 | Non-synonymous | 254 S to P | 16.88 | <0.0001 | ||
| SOD1 | G88A | - | 42 | - | 42/240 | Non-synonymous | 30 G to R | 26.26 | <0.0001 | |
| A160G | 26 | 38 | - | 64/240 | Non-synonymous | 54 T to A | 40.02 | <0.0001 | ||
| A218G | - | 33 | - | 33/240 | Non-synonymous | 73 Y to C | 20.63 | <0.0001 | ||
| A230C | - | 28 | - | 28/240 | Non-synonymous | 77 E to A | 17.51 | <0.0001 | ||
| CAT | C202T | - | 36 | 23 | 59/240 | Non-synonymous | 68 L to F | 36.89 | <0.0001 | |
| GPX1 | G33A | - | 19 | - | 19/240 | Synonymous | 11 P | 11.88 | <0.0001 | |
| T375C | - | 27 | 27/240 | Synonymous | 125 P | 16.88 | <0.0001 | |||
| AhpC/TSA | G256T | 29 | 41 | - | 70/240 | Non-synonymous | 86 V to F | 43.77 | <0.0001 | |
| A298G | - | 27 | - | 27/240 | Non-synonymous | 100 K to E | 16.88 | <0.0001 | ||
| Economic Parameters | Holstein n = 120 | Brown Swiss n = 120 | ||
|---|---|---|---|---|
| Tolerant n = 60 | Mastitic n = 60 | Tolerant n = 60 | Mastitic n = 60 | |
| Service cost (EGP) | 197.30 ± 9.15 b | 445.60 ± 7.58 a | 203.40 ± 8.47 b | 415.20 ± 11.75 a |
| Treatment cost (EGP) | - | 480.65± 8.46 | - | 395.84 ± 13.90 |
| Veterinary management cost (EGP) | 690.50 ± 9.45 b | 850.38 ± 14.59 a | 670.73 ± 13.98 b | 795.35 ± 17.65 a |
| Total cost (EGP) | 39835.24 ± 199.79 b | 40978.56 ± 263.82 a | 36750.53 ± 220.29 b | 38367.78 ± 264.18 a |
| Total return (EGP) | 68579.20 ± 219.26 a | 58534.90 ± 128.55 c | 59341.10 ± 123.59 b | 54757.18 ± 139.51 c |
| Net Return (EGP) | 28743.96 ± 157.19 a | 17556.34 ± 113.75 d | 22590.57 ± 189.42 b | 16389.40 ± 120.36 c |
| Reduction % in Net profit | - | 39 | - | 27 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ateya, A.I.; Ibrahim, S.S.; Al-Sharif, M.M. Single Nucleotide Polymorphisms, Gene Expression and Economic Evaluation of Parameters Associated with Mastitis Susceptibility in European Cattle Breeds. Vet. Sci. 2022, 9, 294. https://doi.org/10.3390/vetsci9060294
Ateya AI, Ibrahim SS, Al-Sharif MM. Single Nucleotide Polymorphisms, Gene Expression and Economic Evaluation of Parameters Associated with Mastitis Susceptibility in European Cattle Breeds. Veterinary Sciences. 2022; 9(6):294. https://doi.org/10.3390/vetsci9060294
Chicago/Turabian StyleAteya, Ahmed I., Samer S. Ibrahim, and Mona M. Al-Sharif. 2022. "Single Nucleotide Polymorphisms, Gene Expression and Economic Evaluation of Parameters Associated with Mastitis Susceptibility in European Cattle Breeds" Veterinary Sciences 9, no. 6: 294. https://doi.org/10.3390/vetsci9060294
APA StyleAteya, A. I., Ibrahim, S. S., & Al-Sharif, M. M. (2022). Single Nucleotide Polymorphisms, Gene Expression and Economic Evaluation of Parameters Associated with Mastitis Susceptibility in European Cattle Breeds. Veterinary Sciences, 9(6), 294. https://doi.org/10.3390/vetsci9060294




