The Genomic and Genetic Evolution Analysis of Rabbit Astrovirus
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. RT-PCR
2.3. Virus Isolation
2.4. Viral Genome Sequencing and Analysis
2.5. Bioinformatics Analysis of Cap Protein
3. Results
3.1. RT-PCR Detection of RAstV
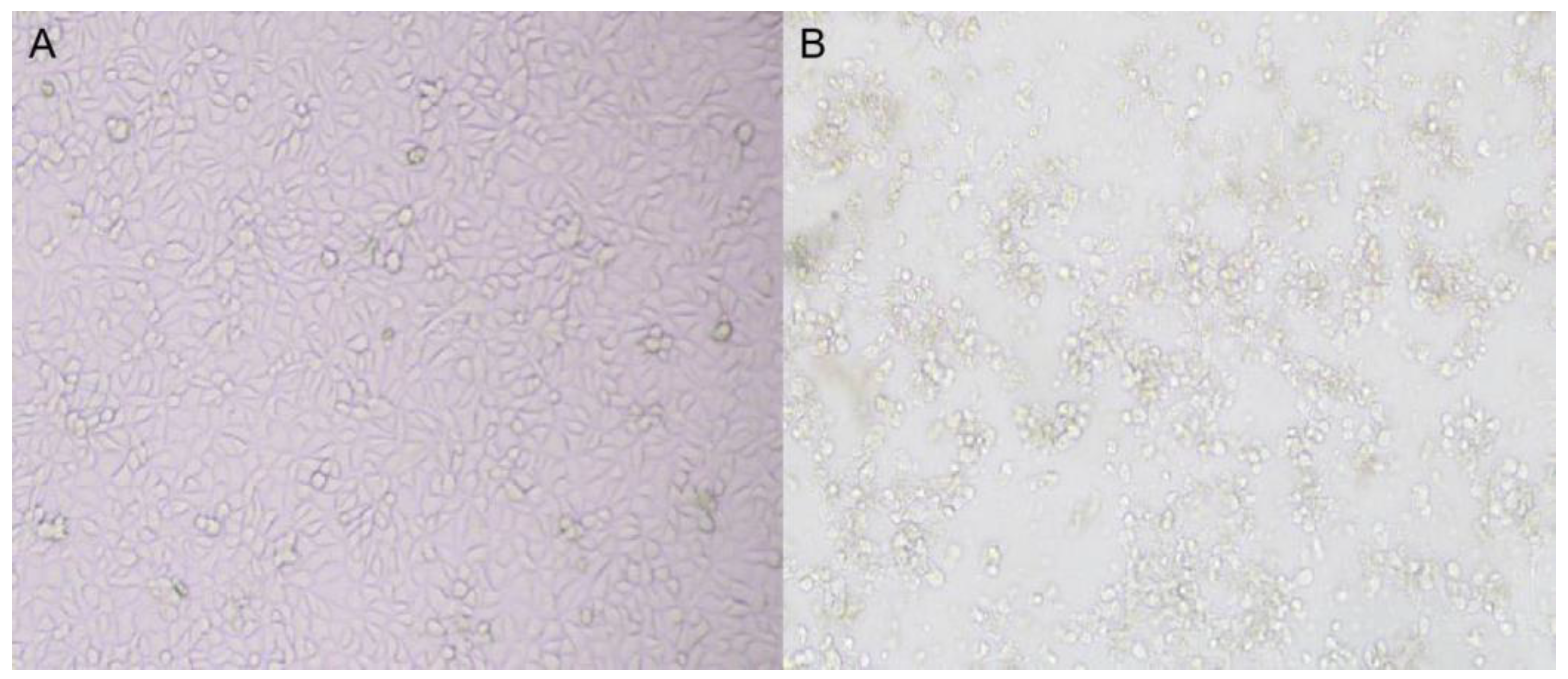
3.2. Virus Isolation
3.3. Genome Composition
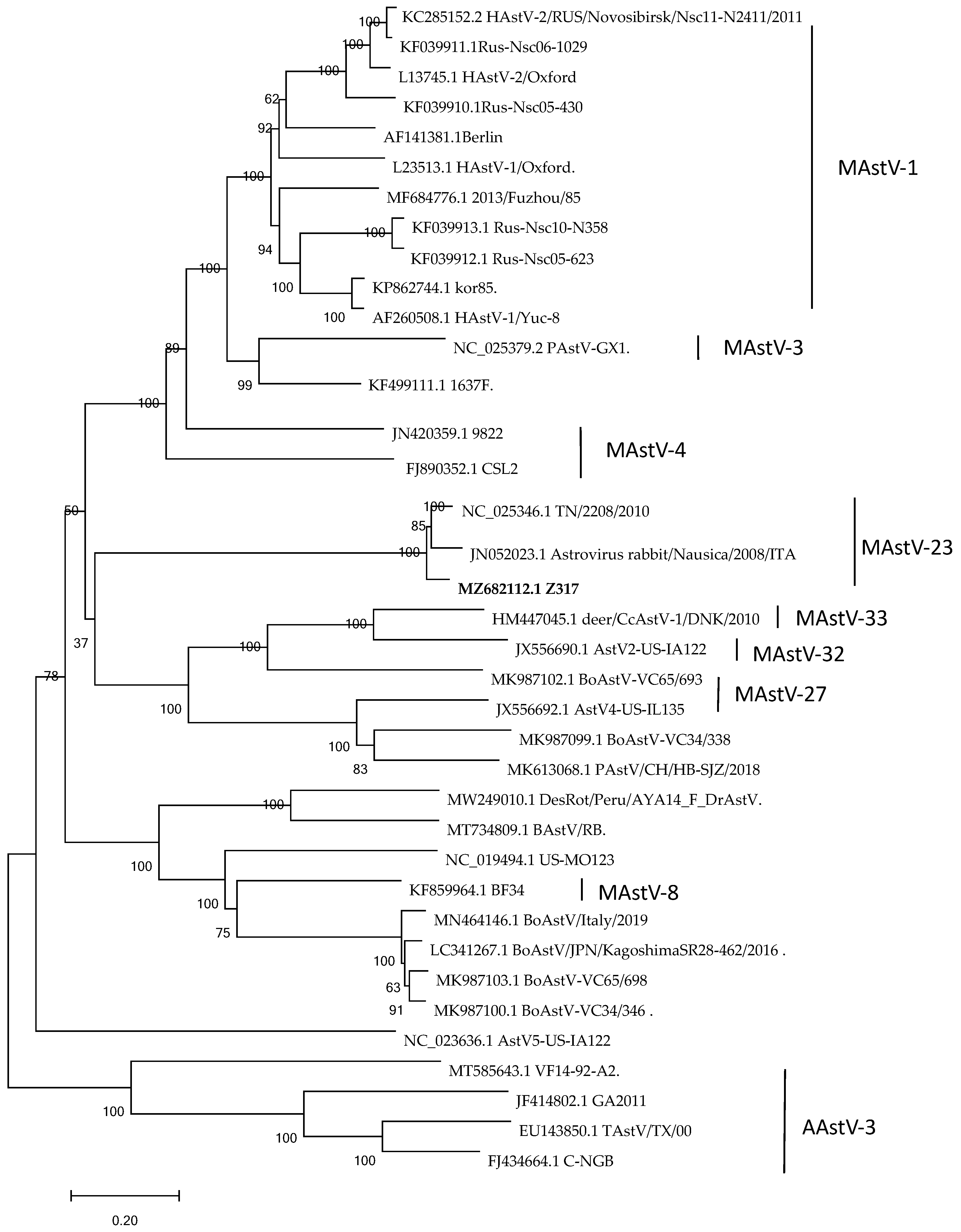
3.4. Nucleotide Sequence Characteristics and Genetic Evolution Analysis
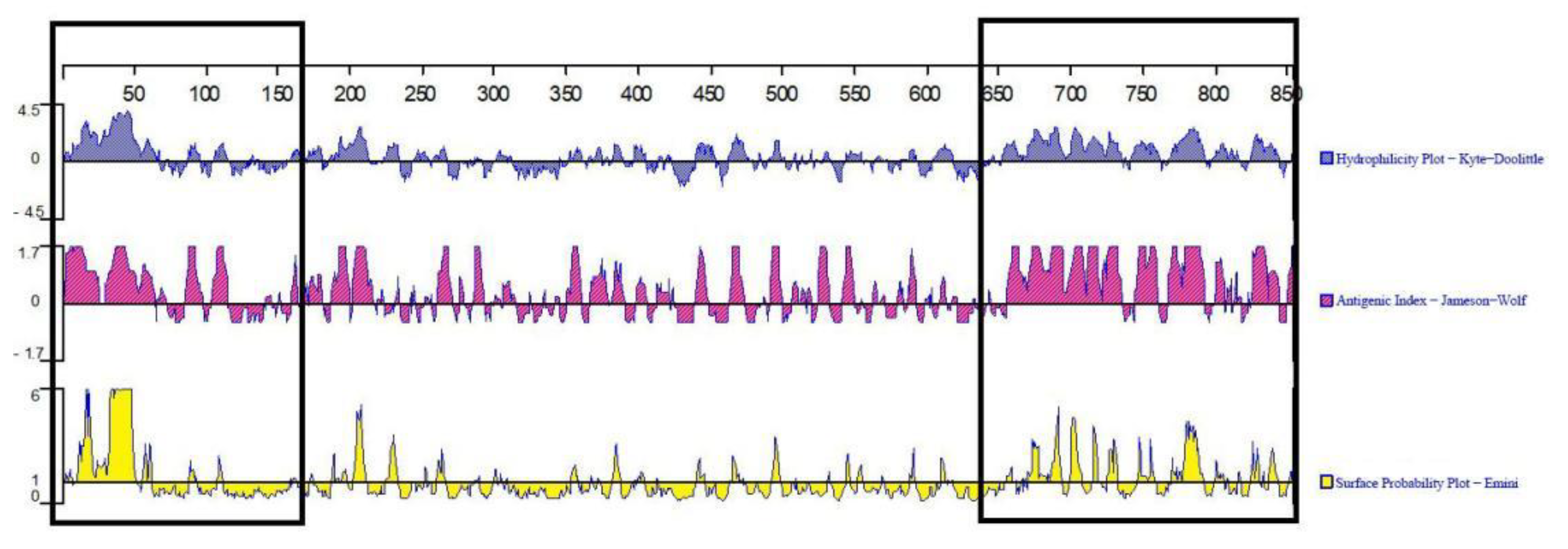
3.5. Bioinformatics Analysis of Cap Protein
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Madeley, C.R.; Cosgrove, B.P. Letter: 28 nm particles in faeces in infantile gastroenteritis. Lancet 1975, 2, 451–452. [Google Scholar] [CrossRef]
- Snodgrass, D.R.; Gray, E.W. Detection and transmission of 30 nm virus particles (astroviruses) in faeces of lambs with diarrhoea. Arch. Virol. 1977, 55, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Imada, T.; Yamaguchi, S.; Mase, M.; Tsukamoto, K.; Kubo, M.; Morooka, A. Avian nephritis virus (ANV) as a new member of the family Astroviridae and construction of infectious ANV cDNA. J. Virol. 2000, 74, 8487–8493. [Google Scholar] [CrossRef]
- Chu, D.K.W.; Poon, L.L.M.; Guan, Y.; Peiris, J.S.M. Novel astroviruses in insectivorous bats. J. Virol. 2008, 82, 9107–9114. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.; Seifert, M.; Bartenschlager, R.; Seitz, S. Discovery of highly divergent lineages of plant-associated astro-like viruses sheds light on the emergence of potyviruses. Virus Res. 2019, 260, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cao, Y.; Wang, J.; Fu, G.; Sun, M.; Zhang, L.; Meng, L.; Cui, G.; Huang, Y.; Hu, X.; et al. Isolation and characterization of an astrovirus causing fatal visceral gout in domestic goslings. Emerg. Microbes Infect. 2018, 7, 1–11. [Google Scholar] [CrossRef]
- Niu, X.; Tian, J.; Yang, J.; Jiang, X.; Wang, H.; Chen, H.; Yi, T.; Diao, Y. Novel Goose Astrovirus Associated Gout in Gosling, China. Vet. Microbiol. 2018, 220, 53–56. [Google Scholar] [CrossRef]
- Martella, V.; Moschidou, P.; Pinto, P.; Catella, C.; Desario, C.; Larocca, V.; Circella, E.; Bànyai, K.; Lavazza, A.; Al, V.M.E.; et al. Astroviruses in rabbits. Emerg. Infect. Dis. 2011, 17, 2287–2293. [Google Scholar] [CrossRef]
- Xie, X.; Bil, J.; Shantz, E.; Hammermueller, J.; Nagy, E.; Turner, P.V. Prevalence of lapine rotavirus, astrovirus, and hepatitis E virus in Canadian domestic rabbit populations. Vet. Microbiol. 2017, 208, 146–149. [Google Scholar] [CrossRef]
- Rosell, J.M.; de la Fuente, L.F.; Parra, F.; Dalton, K.P.; Sáiz, J.I.B.; de Rozas, A.P.; Díez, J.J.B.; de Luco, D.F.; Casal, J.; Majó, N.; et al. Myxomatosis and Rabbit Haemorrhagic Disease: A 30-Year Study of the Occurrence on Commercial Farms in Spain. Animals 2019, 9, 780. [Google Scholar] [CrossRef]
- Lau Susanna, K.P.; Woo Patrick, C.Y.; Yip Cyril, C.Y.; Fan Rachel, Y.Y.; Huang, Y. Isolation and characterization of a novel Betacoronavirus subgroup A coronavirus, rabbit coronavirus HKU14, from domestic rabbits. J. Virol. 2012, 86, 5481–5496. [Google Scholar] [CrossRef]
- Huang, P.; Song, Y.H.; Hu, B.; Wei, H.J.; Fan, Z.Y.; Qiu, R.L.; Chen, M.M.; Xue, J.B.; Wang, F.; Yong-Sam, J. Generation of a RK13 cell line stably expressing Lam R and its effect on absorption of RHDV. Anim. Husb. Vet. Med. 2018, 50, 44–48. (In Chinese) [Google Scholar]
- Almeida, M.A.; Almeida-Paes, R.; Guimarães, A.J.; Valente, R.H.; Soares, C.M.D.A.; Zancopé-Oliveira, R.M. Immunoproteomics Reveals Pathogen’s Antigens Involved in Homo sapiens-Histoplasma capsulatum Interaction and Specific Linear B-Cell Epitopes in Histoplasmosis. Front. Cell. Infect. Microbiol. 2020, 10, 591121. [Google Scholar] [CrossRef] [PubMed]
- Lewis, T.L.; Greenberg, H.B.; Herrmann, J.E.; Smith, L.S.; Matsui, S.M. Analysis of astrovirus serotype 1 RNA, identification of the viral RNA-dependent RNA polymerase motif, and expression of a viral structural protein. J. Virol. 1994, 68, 77–83. [Google Scholar] [CrossRef]
- Arruda, B.; Arruda, P.; Hensch, M.; Chen, Q.; Zheng, Y.; Yang, C.; Gatto, I.R.H.; Ferreyra, F.M.; Gauger, P.; Schwartz, K.; et al. Porcine Astrovirus Type 3 in Central Nervous System of Swine with Polioencephalomyelitis. Emerg. Infect. Dis. 2017, 23, 2097–2100. [Google Scholar] [CrossRef]
- Jonassen, C.M.; Jonassen, T.O.; Grinde, B. A common RNA motif in the 3’ end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. J. Gen. Virol. 1998, 79, 715–718. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Michael, L.; Christina, K.; Koichiro, T. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tse, H.; Chan, W.-M.; Tsoi, H.-W.; Fan, R.Y.Y.; Lau, C.C.Y.; Lau, S.K.P.; Woo, P.C.Y.; Yuen, K.-Y. Rediscovery and genomic characterization of bovine astroviruses. J. Gen. Virol. 2011, 92, 1888–1898. [Google Scholar] [CrossRef]
- Pantin-Jackwood, M.J.; Spackman, E.; Woolcock, P.R. Phylogenetic Analysis of Turkey Astroviruses Reveals Evidence of Recombination. Virus Genes 2006, 32, 187–192. [Google Scholar] [CrossRef]
- Lee, T.W.; Kurtz, J.B. Serial Propagation of Astrovirus in Tissue Culture with the Aid of Trypsin. J. Gen. Virol. 1981, 57, 421–424. [Google Scholar] [CrossRef]
- Mi, S.J.; Guo, S.B.; Xing, C.N.; Xiao, C.T.; He, B.; Wu, B.; Xia, X.Z.; Tu, C.C.; Gong, W.J. Isolation and Characterization of Porcine Astrovirus 5 from a Classical Swine Fever Virus-Infected Specimen. J. Virol. 2020, 95, e01513-20. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Zhang, S.; Xie, J.; Gu, L.; Wu, S.; Wu, Z.; Liu, L.; Feng, Q.; Dong, H.; Zhu, S. Isolation and characterization of a goose astrovirus 1 strain causing fatal gout in goslings, China. Poult. Sci. 2021, 100, 101432. [Google Scholar] [CrossRef] [PubMed]
- Toh, Y.; Harper, J.; Dryden, K.A.; Yeager, M.; Arias, C.F.; Méndez, E.; Tao, Y.J. Crystal Structure of the Human Astrovirus Capsid Protein. J. Virol. 2016, 90, 9008–9017. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, S.R.; Kirkwood, C.D.; Wang, D. Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. BioMed Cent. 2008, 5, 117. [Google Scholar] [CrossRef]
- Chu, D.K.W.; Chin, A.W.H.; Smith, G.J.; Chan, K.-H.; Guan, Y.; Peiris, J.S.M.; Poon, L.L.M. Detection of novel astroviruses in urban brown rats and previously known astroviruses in humans. J. Gen. Virol. 2010, 91, 2457–2462. [Google Scholar] [CrossRef]
- Monceyron, C.; Grinde, B.; Jonassen, T.Ø. Molecular characterisation of the 3′-end of the astrovirus genome. Arch. Virol. 1997, 142, 699–706. [Google Scholar] [CrossRef]
- Zhou, N.; Zhou, L.; Wang, B. Molecular Evolution of Classic Human Astrovirus, as Revealed by the Analysis of the Capsid Protein Gene. Viruses 2019, 11, 707. [Google Scholar] [CrossRef]
- Lan, J.; Zhang, R.; Li, P.; Chen, J.; Xie, Z.; Jiang, S. Identification of a Type-Specific Epitope in the ORF2 Protein of Duck Astrovirus Type 1. Animals 2019, 9, 1069. [Google Scholar] [CrossRef]
- Fanfan, Z.; Haiqin, L.; Qipeng, W.; Quan, X.; Yanbing, Z.; Chengcheng, W.; Qun, Y.; Jia, T.; Meifang, T.; Zhaofeng, K. Isolation and phylogenetic analysis of goose astrovirus type 1 from goslings with gout in Jiangxi province, China. Poult. Sci. 2022, 101, 101800. [Google Scholar] [CrossRef]
- Jie, T.; Benqiang, L.; Jinghua, C.; Ying, S.; Changtao, Q.; Zhi, L.; Huili, L. Genomic Divergence Characterization and Quantitative Proteomics Exploration of Type 4 Porcine Astrovirus. Viruses 2022, 14, 1383. [Google Scholar] [CrossRef]
- Hasan, M.; Azim, K.F.; Imran, M.; Chowdhury, I.M.; Ahmed, S. Comprehensive genome based analysis of Vibrio parahaemolyticus for identifying novel drug and vaccine molecules: Subtractive proteomics and vaccinomics approach. PLoS ONE 2020, 15, e0237181. [Google Scholar] [CrossRef] [PubMed]
| Genome Component | Size | Deduced Amino Acid | Position |
|---|---|---|---|
| ORF1a | 3201 | 1067aa | 3–3203 |
| ORF1b | 1464 | 488aa | 3224–4687 |
| ORF2 | 2700 | 900aa | 4539–7238 |
| 3′UTR | 90 | 30aa | 7239–7328 |
| Name | Conserved Sequence Characteristics | Function | Position |
|---|---|---|---|
| Frameshift signal [14] | AAAAAAC | Induced ribosome translocation | 2324–2330 |
| Junction motif [15] | UUUGGAGNGGNGGACCNAAN4-8AUGNC | Subgenomic RNA transcription promoter | 4656–4687 |
| stem-loop Ⅱ motif [16] | stem-loop Ⅱ motif | Plays an important role in virus replication | - |
| Rank | Location | Epitope | Score |
|---|---|---|---|
| 1 | 128–147 | RCVQAHIRFTPLVGSSAVSG | 1.000 |
| 2 | 821–840 | VSRFDYNRVERGMSNLEAKK | 0.878 |
| 3 | 308–327 | TGTSVSSTIFQVVDASVSTA | 0.738 |
| 4 | 445–464 | LNGWIQNHVNAVVGLWIQDS | 0.614 |
| 5 | 186–205 | LARKQLAGPRESWWLTNTND | 0.594 |
| 6 | 655–674 | EWQRASGARVDLRTVRFRDD | 0.578 |
| 7 | 88–107 | SSDRVELEMASMLNPALVKE | 0.513 |
| 8 | 792–811 | QSALGAQFDPETAAHRAMRA | 0.471 |
| 9 | 45–64 | QQRRTRAVARSEVKREVHRL | 0.416 |
| 10 | 257–276 | ATLSKQEAPASSVVIDAATA | 0.402 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Q.; Tian, Y.; Liu, L.; Jiang, Y.; Sun, H.; Tan, S.; Huang, B. The Genomic and Genetic Evolution Analysis of Rabbit Astrovirus. Vet. Sci. 2022, 9, 603. https://doi.org/10.3390/vetsci9110603
Zhao Q, Tian Y, Liu L, Jiang Y, Sun H, Tan S, Huang B. The Genomic and Genetic Evolution Analysis of Rabbit Astrovirus. Veterinary Sciences. 2022; 9(11):603. https://doi.org/10.3390/vetsci9110603
Chicago/Turabian StyleZhao, Qiaoya, Ye Tian, Liping Liu, Yifei Jiang, Haitao Sun, Shanjie Tan, and Bing Huang. 2022. "The Genomic and Genetic Evolution Analysis of Rabbit Astrovirus" Veterinary Sciences 9, no. 11: 603. https://doi.org/10.3390/vetsci9110603
APA StyleZhao, Q., Tian, Y., Liu, L., Jiang, Y., Sun, H., Tan, S., & Huang, B. (2022). The Genomic and Genetic Evolution Analysis of Rabbit Astrovirus. Veterinary Sciences, 9(11), 603. https://doi.org/10.3390/vetsci9110603





