Updating Sacbrood Virus Quantification PCR Method Using a TaqMan-MGB Probe
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collections
2.2. RNA Extraction and Reverse Transcription
2.3. Quantification PCR with the TaqMan-MGB (Minor Groove Binder) Probe
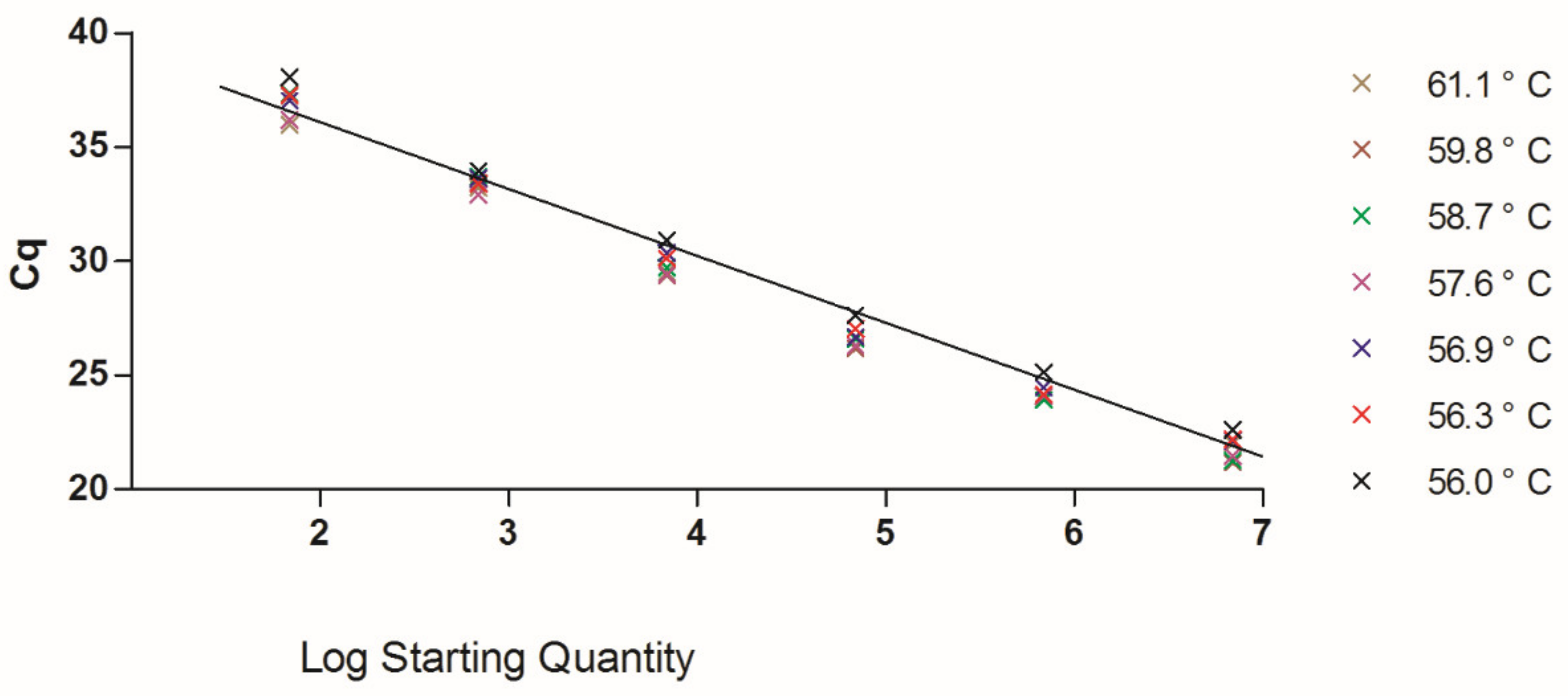
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Bailey, L. The multiplication and spread of sacbrood virus of bees. Ann. Appl. Biol. 1969, 63, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Levitt, A.L.; Rajotte, E.G.; Holmes, E.C.; Ostiguy, N.; van Engelsdorp, D.; Lipkin, W.I.; de Pamphilis, C.W.; Toth, A.L.; Cox-Foster, D.L. RNA viruses in hymenopteran pollinators: Evidence of inter-taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLoS ONE 2010, 5, e14357. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-F.; Mehmood, S.; Huang, S.; Chen, Y.-W.; Ko, C.-Y.; Su, S. Phylogenetic analysis and survey of Apis cerana strain of Sacbrood virus (AcSBV) in Taiwan suggests a recent introduction. J. Invertebr. Pathol. 2017, 146, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, Z.; Du, Z.L. The pathogen identification of the Chinese sacbrood bee larvae disease. APIC China 1979, 5, 15–17. [Google Scholar]
- Mingxiao, M.; Yanna, Y.; Xiaoli, X.; Lin, Z.; Yongfei, L.; Zhidong, L. Genetic characterization of VP1 gene of seven Sacbrood virus isolated from three provinces in northern China during the years 2008–2012. Virus Res. 2013, 176, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Mingxiao, M.; Ming, L.; Jian, C.; Song, Y.; Shude, W.; Pengfei, L. Molecular and biological characterization of Chinese sacbrood virus LN isolate. Comp. Funct. Genom. 2011, 2011, 409386. [Google Scholar] [CrossRef] [PubMed]
- Bailey, L.; Carpenter, J.M.; Woods, R.D. A strain of sacbrood virus from Apis cerana. J. Invertebr. Pathol. 1982, 39, 264–265. [Google Scholar] [CrossRef]
- Verma, L.R.; Rana, B.S.; Verma, S. Observations on Apis cerana colonies surviving from Thai sacbrood virus infestation. Apidologie 1990, 21, 169–174. [Google Scholar] [CrossRef]
- Aruna, R.; Srinivasan, M.R.; Balasubramanian, V.; Selvarajan, R. Complete genome sequence of sacbrood virus isolated from Asiatic honey bee Apis cerana indica in India. VirusDisease 2018, 29, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Ahn, A.-J.; Ahn, K.-S.; Suh, G.-H.; Noh, J.-H.; Kim, Y.-H.; Yoo, M.-S.; Kang, S.-W.; Shin, S.-S. Efficacy of silver ions against sacbrood virus infection in the eastern honey bee Apis cerana. J. Vet. Sci. 2015, 16, 289–295. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sun, L.; Li, M.; Fei, D.; Hu, Y.; Ma, M. Chinese sacbrood virus infection in apis mellifera, Shandong, China, 2016. Virus Res. 2017, 242, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Gayral, P.; Zhao, H.; Wu, Y.; Jiang, X.; Wu, Y.; Bigot, D.; Wang, X.; Yang, D.; Herniou, E.A.; et al. Occurrence and molecular phylogeny of honey bee viruses in vespids. Viruses 2020, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, P.; Guillot, S.; Antùnez, K.; Köglberger, H.; Kryger, P.; de Miranda, J.R.; Franco, S.; Chauzat, M.-P.; Thiéry, R.; Ribière, M. Development and validation of a real-time two-step RT-qPCR TaqMan® assay for quantitation of sacbrood virus (SBV) and its application to a field survey of symptomatic honey bee colonies. J. Virol. Methods 2014, 197, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Mingxiao, M.; Jinhua, L.; Yingjin, S.; Li, L.; Yongfei, L. TaqMan MGB probe fluorescence real-time quantitative PCR for rapid detection of Chinese sacbrood virus. PLoS ONE 2013, 8, e52670. [Google Scholar] [CrossRef] [PubMed]
- Kukielka, D.; Sánchez-Vizcaíno, J.M. One-step real-time quantitative PCR assays for the detection and field study of Sacbrood honeybee and Acute bee paralysis viruses. J. Virol. Methods 2009, 161, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Jin-Long, Y.; Rui, Y.; Ke-Fei, S.; Xiang-Wei, P.; Tao, X.; Zuo-Hua, L. Rapid detection of sacbrood virus (SBV) by one-step reverse transcription loop-mediated isothermal amplification assay. Virol. J. 2012, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Ma, C.; Li, M.; Wang, S.; Yang, S.; Wang, S. Loop-mediated isothermal amplification for rapid detection of Chinese sacbrood virus. J. Virol. Methods 2011, 176, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Mehmood, S.; Zhang, G.; Song, Y.; Su, S.; Huang, S.; Huang, H.; Zhang, Y.; Geng, H.; Huang, W.-F. Visualizing sacbrood virus of honey bees via transformation and coupling with enhanced green fluorescent protein. Viruses 2020, 12, 224. [Google Scholar] [CrossRef] [PubMed]
- Tajadini, M.; Panjehpour, M.; Javanmard, S.H. Comparison of SYBR green and TaqMan methods in quantitative real-time polymerase chain reaction analysis of four adenosine receptor subtypes. Adv. Biomed. Res. 2014, 3, 85. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, W.-F.; Zhang, Y.; Mehmood, S.; Wang, Z.; Hou, C.; Li, Z. Updating Sacbrood Virus Quantification PCR Method Using a TaqMan-MGB Probe. Vet. Sci. 2021, 8, 63. https://doi.org/10.3390/vetsci8040063
Huang W-F, Zhang Y, Mehmood S, Wang Z, Hou C, Li Z. Updating Sacbrood Virus Quantification PCR Method Using a TaqMan-MGB Probe. Veterinary Sciences. 2021; 8(4):63. https://doi.org/10.3390/vetsci8040063
Chicago/Turabian StyleHuang, Wei-Fone, Yakun Zhang, Shahid Mehmood, Zhengwei Wang, Chunsheng Hou, and Zhiguo Li. 2021. "Updating Sacbrood Virus Quantification PCR Method Using a TaqMan-MGB Probe" Veterinary Sciences 8, no. 4: 63. https://doi.org/10.3390/vetsci8040063
APA StyleHuang, W.-F., Zhang, Y., Mehmood, S., Wang, Z., Hou, C., & Li, Z. (2021). Updating Sacbrood Virus Quantification PCR Method Using a TaqMan-MGB Probe. Veterinary Sciences, 8(4), 63. https://doi.org/10.3390/vetsci8040063






