Molecular Diversity of Hard Tick Species from Selected Areas of a Wildlife-Livestock Interface Ecosystem at Mikumi National Park, Morogoro Region, Tanzania
Abstract
1. Introduction
2. Methods
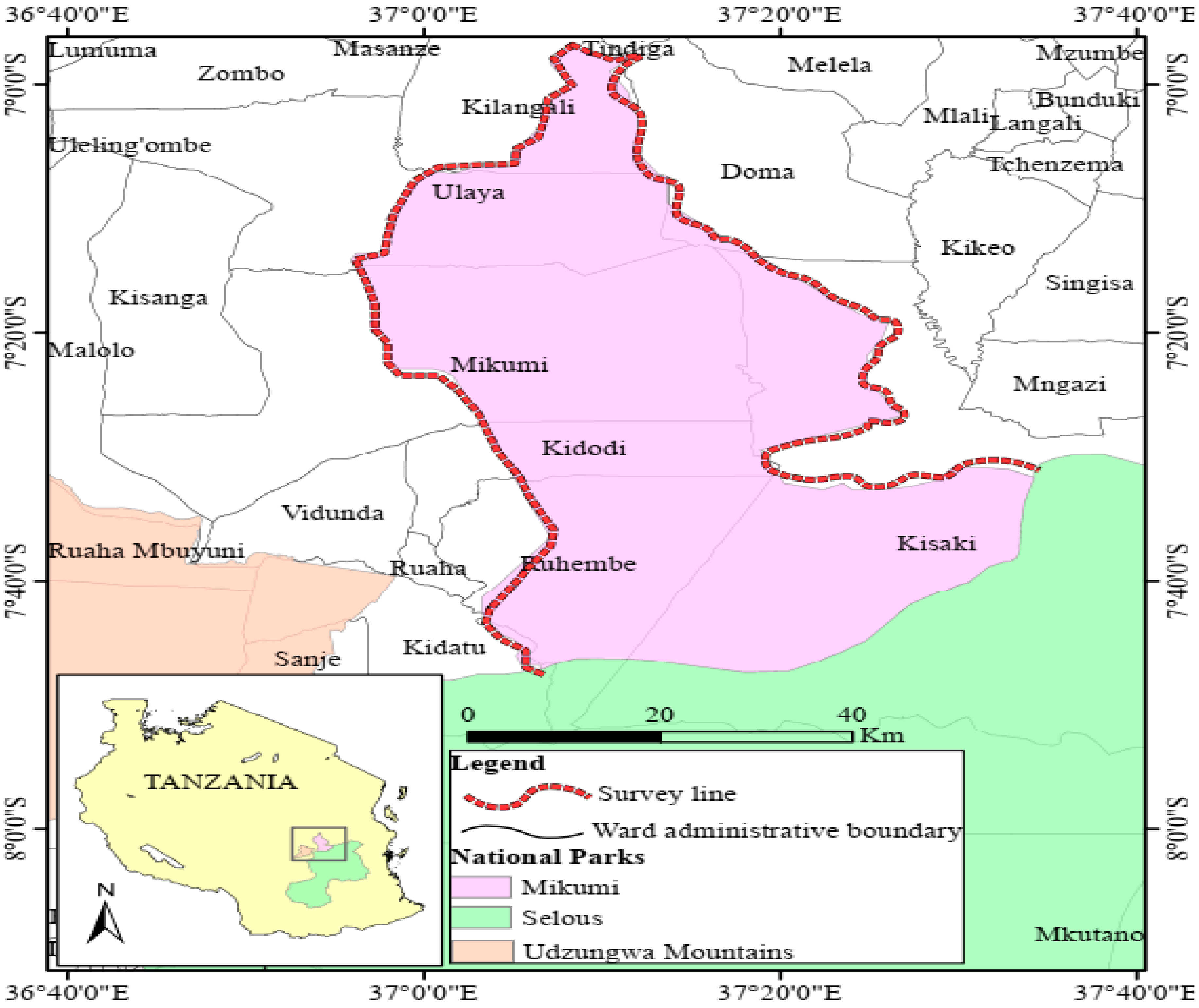
2.1. Study Site
2.2. Collection of Ticks
2.3. Tick Identification
2.3.1. Morphological Identification of Ticks
2.3.2. Molecular Identification of Ticks
DNA Extraction
DNA Amplification
Agarose Gel Analysis and Sequencing of the 16S rRNA Amplicons
Sequences Editing, Blast Analysis, and Alignment
Evolutionary Relationships of Taxa
3. Results
3.1. Data Acquisition and Morphological Identification of Ticks
3.2. Molecular Identification and Classification of Ticks
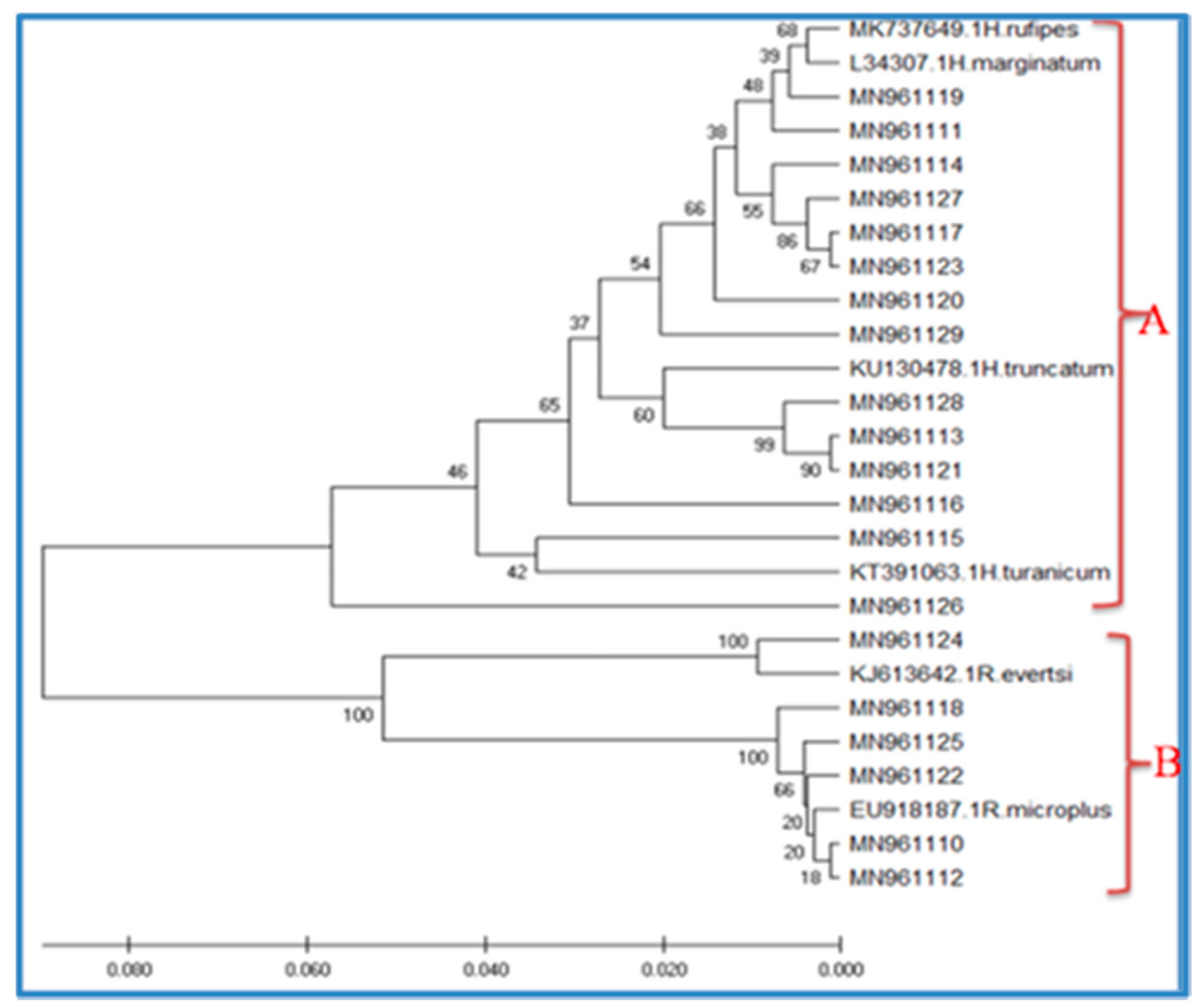
3.3. EvolutionaryRelationships of Taxa(Phylogenetic Analysis)
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Yu, Z.; Wang, H.; Wang, T.; Sun, W.; Yang, X.; Liu, J.-Z. Tick-borne pathogens and the vector potential of ticks in China. Parasites Vectors 2015, 8, 24–28. [Google Scholar] [CrossRef]
- Mansfield, K.L.; Jizhou, L.; Phipps, L.P.; Johnson, N. Emerging Tick-Borne Viruses in the Twenty-First Century. Front. Cell. Infect. Microbiol. 2017, 7, 298. [Google Scholar] [CrossRef]
- Jongejan, F.; Uilenberg, G. The global importance of ticks. Parasitology 2004, 129, S3–S14. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Peña, A.; Gray, J.S.; Kahl, O.; Lane, R.S.; Nijhof, A.M. Research on the ecology of ticks and tick-borne pathogens-methodological principles and caveats. Front. Cell. Infect. Microbiol. 2013, 3, 29. [Google Scholar] [CrossRef]
- Walker, A.R.; Bouattour, A.; Camicas, J.J.; Estrada-Pena, A.; Horak, I.G.; Latif, A.A.; Pegram, R.G.; Preston, P.M. Ticks of Domestic Animals in Africa: A Guide to Identification of Species; Bioscience Reports: Edinburgh, UK, 2003; pp. 1–1221. [Google Scholar]
- Kerario, I.; Muleya, W.; Chenyambuga, S.; Koski, M.; Seong-Gu Hwang, S.; Simuunza, M. Abundance and distribution of Ixodid tick speciesinfesting cattle reared under traditional farmingsystems in Tanzania. Afr. J. Agric. Sci. 2017, 12, 286–299. [Google Scholar]
- Kwak, Y.S.; Kim, T.Y.; Man, S.H.; Lee, I.Y.; Kim, H.P.; Mduma, S.; Keyyu, J.; Fyumagwa, R.; Yong, T.S. Ixodid tick infestation in cattle and w ild animals in Maswa and Iringa, Tanzania. Korean J. Parasitol. 2014, 5, 565–568. [Google Scholar] [CrossRef]
- Laisser, E.L.K.; Kipanyula, M.J.; Msalya, G.; Mdegela, R.H.; Karimuribo, E.D.; Mwilawa, A.J.; Mwega, E.D.; Kusiluka, L.J.M.; Chenyambuga, S.W. Tick burden and prevalence of Theileriaparvainfection in Tarime zebu cattle in the Lake zone of Tanzania. Trop. Anim. Health Prod. 2014, 46, 1391–1396. [Google Scholar] [CrossRef]
- Swai, E.S.; Mbise, A.N.; Kessy, V.; Kaaya, E.; Sanka, P.; Loomu, P.M. Farm Constraints, Cattle Disease Perception and Tick Managementpractices in Pastoral Maasai Community, Ngorongoro, Tanzania. 2005. Available online: http://www.lrrd.org/17/2/cont1702 (accessed on 18 October 2019).
- Labuda, M.; Nuttall, P.A. Tick-borneviruses. Parasitology 2004, 129, S221–S245. [Google Scholar] [CrossRef]
- Muruthi, C.W.; Lwande, O.W.; Makumi, J.N.; Runo, S.; Otiende, M.; Makori, W.A. Phenotypic and genotypicidentification of ticks sampled from wildlife species in selected conservation sites of Kenya. Vet. Sci. Technol. 2016, 7, 1–8. [Google Scholar]
- Man, S.Q.; Qiao, K.; Cui, J.; Feng, M.; Fu, Y.F.; Cheng, X.J. A case of human infection with a novel Babesia species in China. Infect. Dis. Poverty 2016, 5, 28. [Google Scholar] [CrossRef]
- Apanaskevich, M.A.; Apanaskevich, D.A. Description of new Dermacentor(Acari: Ixodidae) speciesfrom Malaysia and Vietnam. J. Med. Entomol. 2015, 52, 156–162. [Google Scholar] [CrossRef][Green Version]
- Wallace, D.M. Large and Small Scale Phenol Extractions Guide to Molecular Cloning Techniques; Academic Press: Orlando, FL, USA, 1987; pp. 31–41. [Google Scholar]
- Hill, C.A.; Gutierrez, J.A. Method for extraction and analysis of high quality genomicDNA from ixodid ticks. Med. Vet. Entomol. 2003, 12, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Mauel, M.J.; Carlton, S.J.; Mather, T.N. Polymerase chain reaction detection efficiency of thehuman granulocytic ehrlichiosis agent (Rickettsiaceae: Ehrlichieae) in ticks (Acari: Ixodidae) is dependent on the DNA extraction method. J. Med. Entomol. 1999, 36, 649–652. [Google Scholar] [CrossRef]
- Zemtsova, G.E.; Apanaskevich, D.A.; Reeves, W.K.; Hahn, M.; Snellgrove, A.; Levin, M.L. Phylogeography of Rhipicephalus sanguineussensulato and its relationships with climatic factors. Exp. Appl. Acarol. 2016, 69, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Takano, A.; Fujita, H.; Kadosaka, T.; Takahashi, M.; Yamauchi, T.; Ishiguro, F. Construction of a DNA database for ticks collected in Japan: Application of molecular identification based on the mitochondrial 16S rDNA gene. Med. Entomol. Zool. 2014, 65, 13–21. [Google Scholar] [CrossRef]
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput. Appl. Biosci. 1994, 10, 189–191. [Google Scholar] [CrossRef]
- Zeryehun, T.; Atomsa, M. Ectoparasiotes infestations of sheep and goats. Eurasian J. Vet. Sci. 2012, 28, 185–189. [Google Scholar]
- Kaiser, M.N.; Surtherst, R.W.; Bourne, A.S. Relationshipbetweenticks and Zebu in southern Uganda. Trop. Anim. Health Prod. 1982, 14, 63–74. [Google Scholar] [CrossRef]
- Moges, N.; Bogale, B.; Fentahun, T. Hard Ticks (Ixodidae) Species Composition, Seasonal Dynamics and Body Site Distribution on cattle in Chilga District, Northwest Ethiopia. Asian J. Agric. Sci. 2012, 4, 341–345. [Google Scholar]
- Awaa, D.N.; Adakald, H.; Luogboua, N.D.D.; Wachonga, K.H.; Leinyuya, I.; Achkwiaa, M.D. Is Ripicephalus (Boophilus) microplus absentin Cameroon and the central African region? Ticks Tick-Borne Dis. 2015, 6, 117–122. [Google Scholar] [CrossRef]
- Tønnesen, M.; Penzhorn, B.; Bryson, N.; Stoltsz, W.; Masibigiri, T. Displacement of Boophilusdecoloratus by Boophilus microplus in theSoutpansberg region, Limpopo Province, South Africa. Exp. Appl. Acarol. 2004, 32, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Lynen, G.; Zeman, P.; Bakuname, C.; Di Giulio, G.; Mtui, P.; Sanka, P. Shifts in the distributional ranges of Boophilusticks in Tanzania: Evidence that a parapatric boundary between Boophilus microplus and B. decoloratus follows climate gradients. Exp. Appl. Acarol. 2008, 44, 147–164. [Google Scholar] [CrossRef]
- Nyangiwe, N.; Harrison, A.; Horak, I.G. Displacement of Rhipicephalus decoloratus by Rhipicephalusmicroplus(Acari: Ixodidae) in the Eastern Cape Province, South Africa. Exp. Appl. Acarol. 2013, 61, 371–382. [Google Scholar] [CrossRef]
- Madder, M.; Thys, E.; Achi, L.; Touré, A.; De Deken, R. Rhipicephalus (Boophilus) microplus: A most successful invasive tick species in West-Africa. Exp. Appl. Acarol. 2011, 53, 139–145. [Google Scholar] [CrossRef]
- Busch, J.D.; Stone, N.E.; Nottingham, R.; Araya-Anchetta, A.; Lewis, J.; Hochhalter, C. Widespread movement of invasive cattle fever ticks (Rhipicephalus microplus) in southern Texas leads to shared local infestations on cattle and deer. Parasites Vectors 2014, 7, 188. [Google Scholar] [CrossRef]
- Bock, R.; Jackson, L.; de Vos, A.; Jorgensen, W. Babesiosis ofcattle. Parasitology 2004, 129, S247–S269. [Google Scholar] [CrossRef]
- Baffi, M.A.; de Sauza, G.R.; de Sauza, G.S.; Ceron, C.R.; Bonetti, A.M. Esterase enzymes involved in pyrethroid and organophosphateresistance in a Brazilian population of Rhipicephalus (Boophilus) microplus(Acari, Ixodidae). Mol. Biochem. Parasitol. 2008, 160, 70–73. [Google Scholar] [CrossRef]
- Kocan, K.M.; de La Fuente, J.; Blouin, E.F.; Garcia, J.C. Anaplasmamarginale (Rickettsiales: Anaplasmataceae): Resent advances in defining host-pathogen adaptation of a tick-borne rickettisia. Parasitology 2004, 129, S285–S300. [Google Scholar] [CrossRef] [PubMed]
- Abebe, R.; Fantahun, T.; Abera, M.; Bekele, J. Survey ofticks (Acari:Ixodidae) infesting cattle in two districts of SomaliRegional State. Ethiopia. Vet. World 2010, 3, 539–543. [Google Scholar]
- Potgieter, F.T. Epidemiology and control of Anaplasmosis in South Africa. J. S. Afr. Vet. Assoc. 1979, 504, 367–372. [Google Scholar]
- Tadesse, F.; Abadfaji, G.; Girma, S.; Kumsa, B.; Jibat, T. Identification of tick species and their preferred site on cattle’s body in and around MizanTeferi, Southwestern Ethiopia. J. Vet. Med. Anim. Health 2012, 4, 1–5. [Google Scholar]
- Perveen, N.; Muzaffar, S.; Al-Deeb, M.A. Ticks and Tick-borne Diseases of Livestock in the Middle East and North Africa. Insects 2021, 12, 83. [Google Scholar] [CrossRef]
| Animals | Type | Total | |
|---|---|---|---|
| Cattle N (%) | Goats N (%) | ||
| Examined | 260 | 176 | 436 |
| Infested with ticks | 134 (51.5) | 78 (44.3) | 212 (48.6) |
| Category | Cattle | Goats | Total |
|---|---|---|---|
| Animals infested | 134 | 78 | 212 |
| Ticks collected | 632 | 187 | 819 |
| Mean tick intensity | 4.7 | 2.4 | 3.9 |
| Tick Genus | Source | Total | ||
|---|---|---|---|---|
| Cattle N (%) | Goats N (%) | Free N (%) | ||
| Rhipicephalus | 378 (59.8) | 36 (19.3) | 16 (57.1) | 430 (51) |
| Hyalomma | 254 (40.2) | 151 (80.7) | 12 (42.9) | 417 (49) |
| Total | 632 (100) | 187 (100) | 28 (100) | 847 (100) |
| Category | Cattle | Goats | Total |
|---|---|---|---|
| Animal analyzed | 260 | 176 | 436 |
| Ticks collected | 632 | 187 | 819 |
| Mean abundance | 2.4 | 1.1 | 1.8 |
| Sample Accession | Source | Site/Ward | GeneBank Reference | Identity % | Species |
|---|---|---|---|---|---|
| MN961110 | Cattle | Doma | EU918187.1 | 100 | Rhipicephalusmicroplus |
| MN961111 | Environment | Mikumi | KU170517.1 | 98.76 | Hyalomma refipes |
| MN961112 | Cattle | Mangae | KT428016.1 | 100 | Rhipicephalusmicroplus |
| MN961113 | Cattle | Kidodi | KU13478.1 | 97.15 | Hyalomma truncatun |
| MN961114 | Cattle | Mkata | KP776645.1 | 98.99 | Hyalomma marginatum |
| MN961115 | Cattle | Mikumi | KT391063.1 | 93.98 | Hyalomma turanicum |
| MN961116 | Cattle | Doma | MK737649.1 | 94.70 | Hyalomma rufipes |
| MN961117 | Cattle | Ulaya | KP776645.1 | 98.74 | Hyalomma marginatum |
| MN961118 | Cattle | Melela | KT428016.1 | 99.75 | Rhipicephalusmicroplus |
| MN961119 | Environment | Kilangali | KU170517.1 | 98.99 | Hyalomma rufipes |
| MN961120 | Goat | Doma | KU170517 | 99.97 | Hyalomma rufipes |
| MN961121 | Cattle | Ruhembe | KU130478.1 | 96.63 | Hyalomma truncatum |
| MN961122 | Environment | Mangae | KT428016.1 | 99.26 | Rhipicephalusmicroplus |
| MN961123 | Goat | Mikumi | KT391060.1 | 98.50 | Hyalomma marginatum |
| MN961124 | Cattle | Mkata | KJ613642.1 | 98.42 | Rhipicephalus evertsi |
| MN961125 | Goat | Tindiga | KC170742.1 | 99.50 | Rhipicephalusmicroplus |
| MN961126 | Cattle | Melela | KP776645 | 91.54 | Hyalomma turanicum |
| MN961127 | Cattle | Kisaki | KP776645.1 | 98.50 | Hyalomma marginatum |
| MN961128 | Goat | Mangae | KU130478.1 | 96.37 | Hyalomma truncatum |
| MN961129 | Cattle | Kidatu | MK737650.1 | 97.47 | Hyalomma rufipes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Damian, D.; Damas, M.; Wensman, J.J.; Berg, M. Molecular Diversity of Hard Tick Species from Selected Areas of a Wildlife-Livestock Interface Ecosystem at Mikumi National Park, Morogoro Region, Tanzania. Vet. Sci. 2021, 8, 36. https://doi.org/10.3390/vetsci8030036
Damian D, Damas M, Wensman JJ, Berg M. Molecular Diversity of Hard Tick Species from Selected Areas of a Wildlife-Livestock Interface Ecosystem at Mikumi National Park, Morogoro Region, Tanzania. Veterinary Sciences. 2021; 8(3):36. https://doi.org/10.3390/vetsci8030036
Chicago/Turabian StyleDamian, Donath, Modester Damas, Jonas Johansson Wensman, and Mikael Berg. 2021. "Molecular Diversity of Hard Tick Species from Selected Areas of a Wildlife-Livestock Interface Ecosystem at Mikumi National Park, Morogoro Region, Tanzania" Veterinary Sciences 8, no. 3: 36. https://doi.org/10.3390/vetsci8030036
APA StyleDamian, D., Damas, M., Wensman, J. J., & Berg, M. (2021). Molecular Diversity of Hard Tick Species from Selected Areas of a Wildlife-Livestock Interface Ecosystem at Mikumi National Park, Morogoro Region, Tanzania. Veterinary Sciences, 8(3), 36. https://doi.org/10.3390/vetsci8030036









