The Genetic Relatedness and Antimicrobial Resistance Patterns of Mastitis-Causing Staphylococcus aureus Strains Isolated from New Zealand Dairy Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection, Microbiology, and Whole-Genome Sequencing
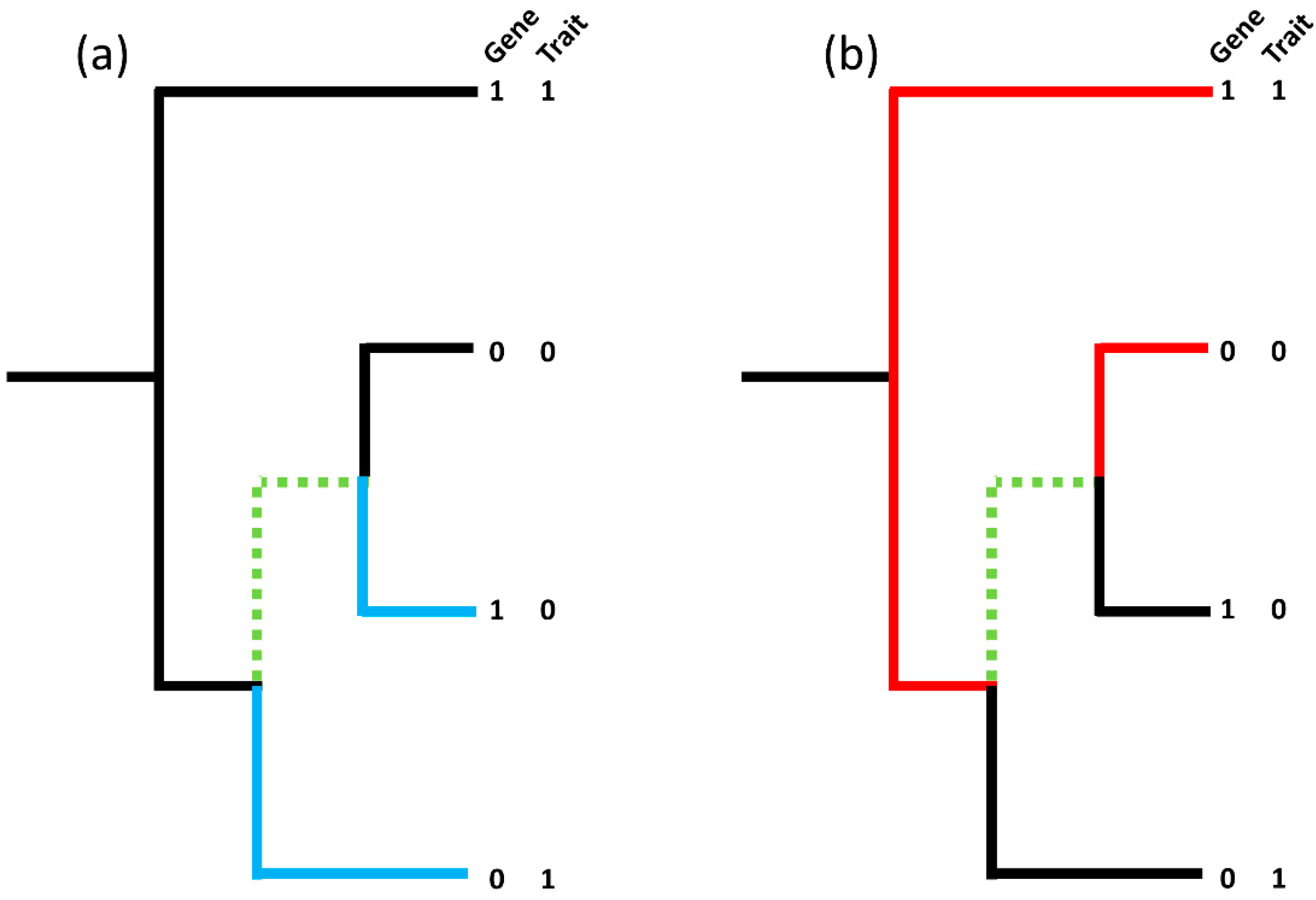
2.2. Genomic Analyses
2.3. Antimicrobial Susceptibility
2.4. Dry Cow Therapy and Antimicrobial Usage
2.5. Statistical Analyses
3. Results
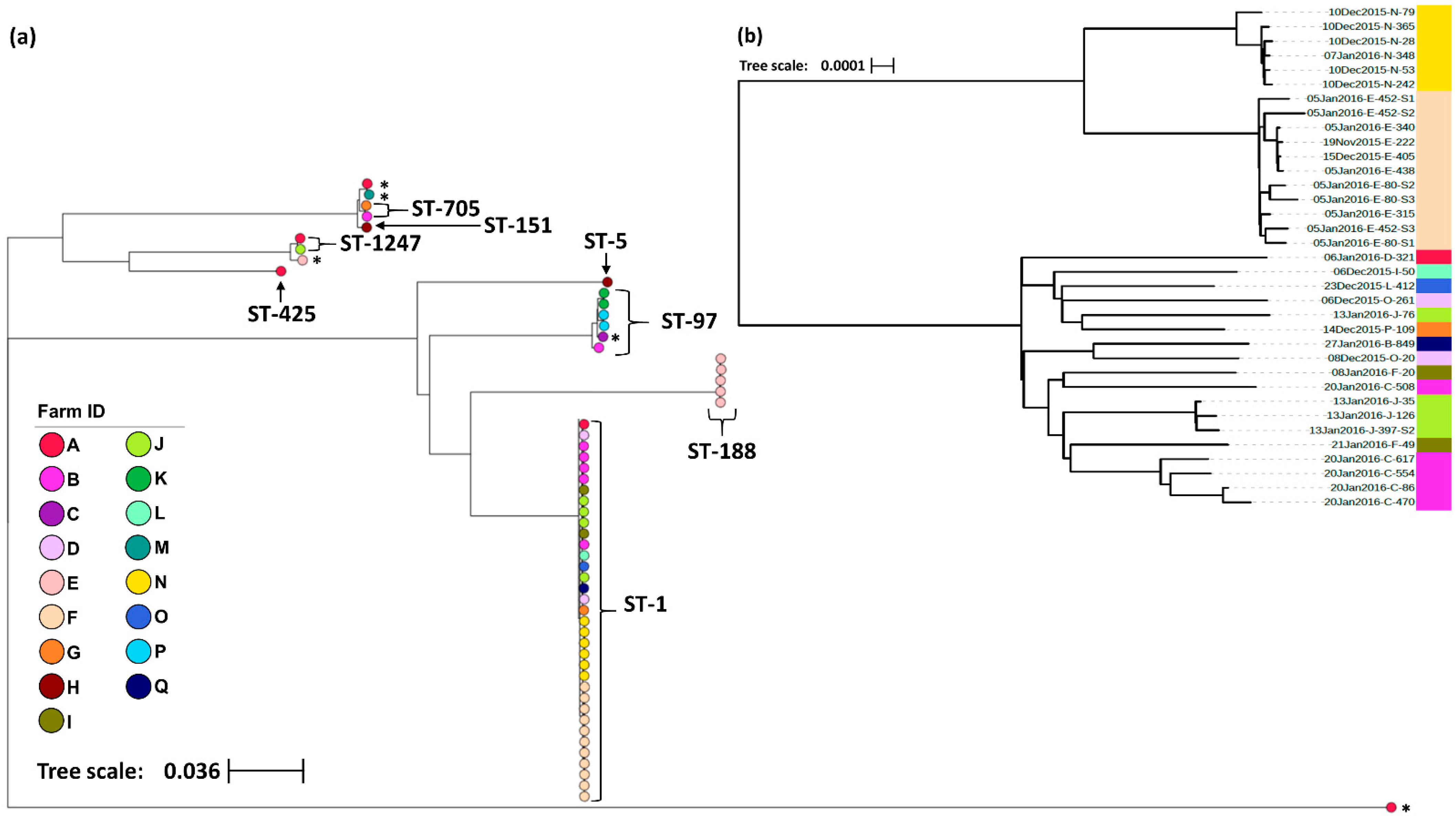
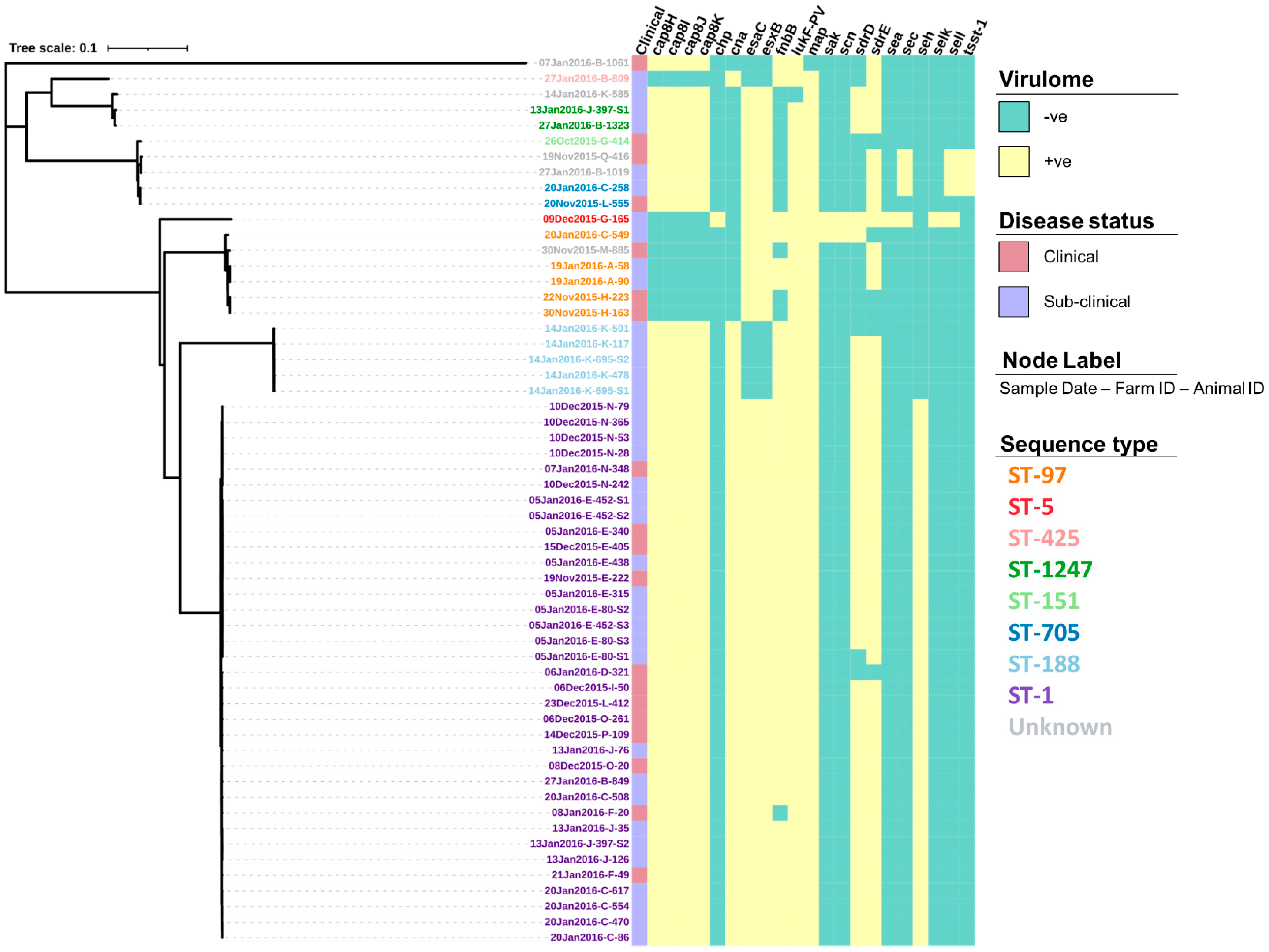
3.1. Genomic Analysis
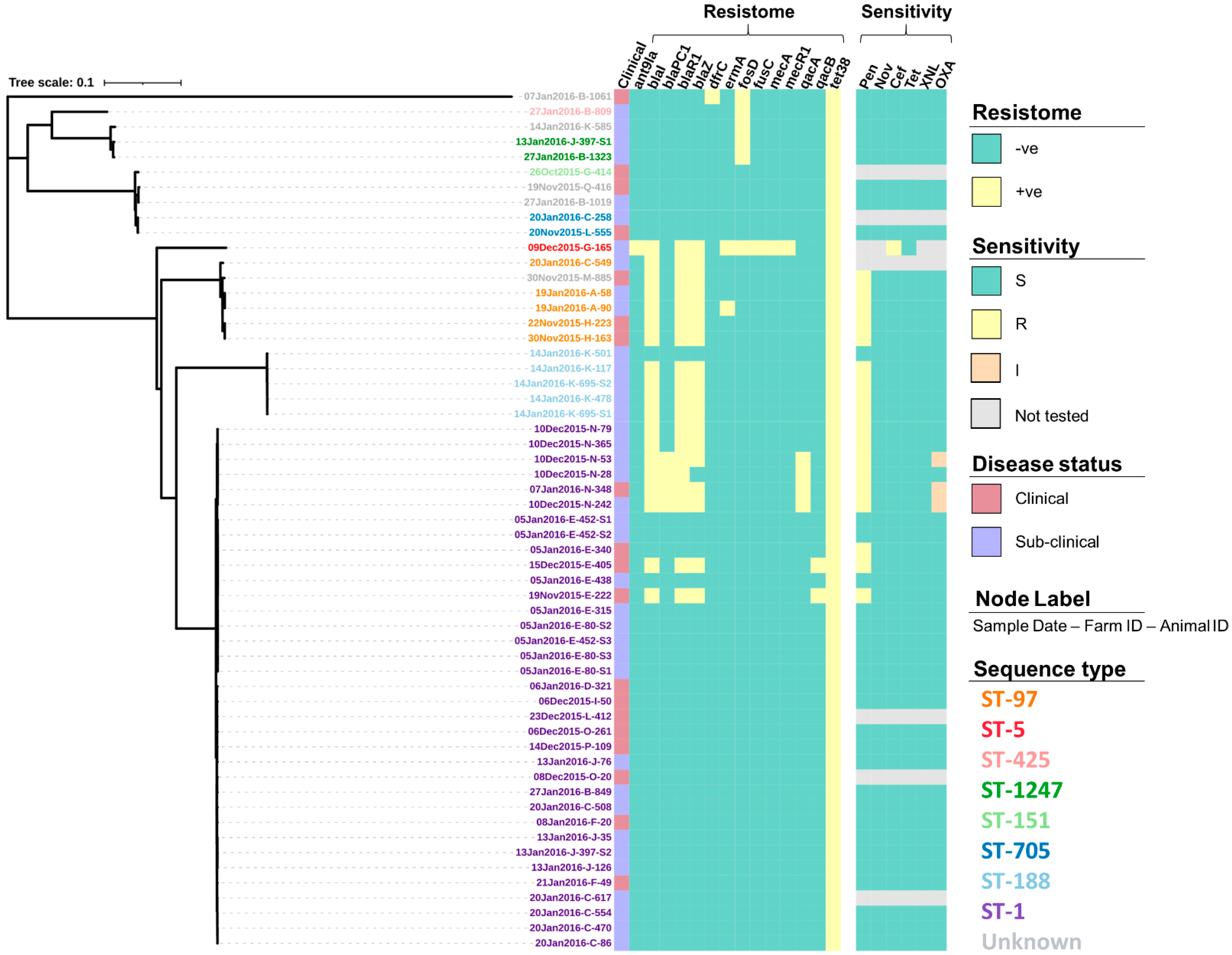
3.2. Antimicrobial Susceptibility
3.3. Dry Cow Therapy and Antimicrobial Usage
3.4. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seegers, H.; Fourichon, C.; Beaudeau, F. Production Effects Related to Mastitis and Mastitis Economics in Dairy Cattle Herds. Vet. Res. 2003, 34, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Petrovski, K.R.; Trajcev, M.; Buneski, G. A Review of the Factors Affecting the Costs of Bovine Mastitis. J. S. Afr. Vet. Assoc. 2006, 77, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.L. Etiological Agents of Bovine Mastitis. Vet. Microbiol. 1988, 16, 41–66. [Google Scholar] [CrossRef]
- Bradley, A.J. Bovine Mastitis: An Evolving Disease. Vet. J. 2002, 164, 116–128. [Google Scholar] [CrossRef] [PubMed]
- McDougall, S. Bovine Mastitis: Epidemiology, Treatment and Control. N. Z. Vet. J. 2002, 50, 81–84. [Google Scholar] [CrossRef]
- Petrovski, K.R.; Heuer, C.; Parkinson, T.J.; Williamson, N.B. The Incidence and Aetiology of Clinical Bovine Mastitis on 14 Farms in Northland, New Zealand. N. Z. Vet. J. 2009, 57, 109–115. [Google Scholar] [CrossRef]
- Heffernan, H.; Bakker, S.; Woodhouse, R.; Dyet, K.; Williamson, D.A. Demographics, Antimicrobial Susceptibility and Molecular Epidemiology of Staphylococcus aureus in New Zealand, 2014; Institute of Environmental Science and Research Limited: Wellington, New Zealand, 2015. [Google Scholar]
- McDougall, S.; Arthur, D.G.; Bryan, M.A.; Vermunt, J.J.; Weir, A.M. Clinical and Bacteriological Response to Treatment of Clinical Mastitis with One of Three Intramammary Antibiotics. N. Z. Vet. J. 2007, 55, 161–170. [Google Scholar] [CrossRef]
- Petrovski, K.R.; Laven, R.A.; Lopez-Villalobos, N. A Descriptive Analysis of the Antimicrobial Susceptibility of Mastitis-Causing Bacteria Isolated from Samples Submitted to Commercial Diagnostic Laboratories in New Zealand (2003–2006). N. Z. Vet. J. 2011, 59, 59–66. [Google Scholar] [CrossRef]
- Bonsaglia, E.C.R.; Silva, N.C.C.; Rossi, B.F.; Camargo, C.H.; Dantas, S.T.A.; Langoni, H.; Guimarães, F.F.; Lima, F.S.; Fitzgerald, J.R.; Fernandes, A.; et al. Molecular Epidemiology of Methicillin-Susceptible Staphylococcus aureus (MSSA) Isolated from Milk of Cows with Subclinical Mastitis. Microb. Pathog. 2018, 124, 130–135. [Google Scholar] [CrossRef]
- Rossi, B.F.; Bonsaglia, E.C.R.; Castilho, I.G.; Dantas, S.T.A.; Salina, A.; Langoni, H.; Pantoja, J.C.F.; Budri, P.E.; Fitzgerald-Hughes, D.; Júnior, A.F.; et al. Genotyping of Long-Term Persistent Staphylococcus aureus in Bovine Subclinical Mastitis. Microb. Pathog. 2019, 132, 45–50. [Google Scholar] [CrossRef]
- McDougall, S.; Hussein, H.; Petrovski, K. Antimicrobial Resistance in Staphylococcus aureus, Streptococcus uberis and Streptococcus dysgalactiae from Dairy Cows with Mastitis. N. Z. Vet. J. 2014, 62, 68–76. [Google Scholar] [CrossRef]
- Petrovski, K.R.; Grinberg, A.; Williamson, N.B.; Abdalla, M.E.; Lopez-Villalobos, N.; Parkinson, T.J.; Tucker, I.G.; Rapnicki, P. Susceptibility to Antimicrobials of Mastitis-Causing Staphylococcus aureus, Streptococcus uberis and Str. dysgalactiae from New Zealand and the USA as Assessed by the Disk Diffusion Test. Aust. Vet. J. 2015, 93, 227–233. [Google Scholar] [CrossRef]
- Mehndiratta, P.L.; Bhalla, P. Use of Antibiotics in Animal Agriculture & Emergence of Methicillin-Resistant Staphylococcus aureus (MRSA) Clones: Need to Assess the Impact on Public Health. Indian J. Med. Res. 2014, 140, 339–344. [Google Scholar] [PubMed]
- Cuny, C.; Wieler, L.; Witte, W. Livestock-Associated MRSA: The Impact on Humans. Antibiotics 2015, 4, 521–543. [Google Scholar] [CrossRef] [PubMed]
- Grinberg, A.; Kingsbury, D.D.; Gibson, I.R.; Kirby, B.M.; Mack, H.J.; Morrison, D. Clinically Overt Infections with Methicillin-Resistant Staphylococcus aureus in Animals in New Zealand: A Pilot Study. N. Z. Vet. J. 2008, 56, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Heffernan, H.; Bakker, S. 2017 Survey of Methicillin-Resistant Staphylococcus Aureus (MRSA); Nosocomial Infections Laboratory, Institute of Environmental Science and Research Ltd.: Porirua, New Zealand, 2018. [Google Scholar]
- NZVA AMR Guidelines. Dairy. NZVA Dairy Cattle Vets Newsl. 2016, 34, 26–27. Available online: https://www.nzva.org.nz/resource/general/amr/ (accessed on 18 June 2021).
- National Mastitis Council (U.S.) Research Committee. Microbiological Procedures for the Diagnosis of Bovine Udder Infection and Determination of Milk Quality, 4th ed.; National Mastitis Council: New Prague, MN, USA, 2004. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Souvorov, A.; Agarwala, R.; Lipman, D.J. SKESA: Strategic k-Mer Extension for Scrupulous Assemblies. Genome Biol. 2018, 19, 153. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinform. Oxf. Engl. 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Croucher, N.J.; Page, A.J.; Connor, T.R.; Delaney, A.J.; Keane, J.A.; Bentley, S.D.; Parkhill, J.; Harris, S.R. Rapid Phylogenetic Analysis of Large Samples of Recombinant Bacterial Whole Genome Sequences Using Gubbins. Nucleic Acids Res. 2015, 43, e15. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-Sites: Rapid Efficient Extraction of SNPs from Multi-FASTA Alignments. Microb. Genom. 2016, 2, e56. [Google Scholar] [CrossRef]
- Zhang, J.; Halkilahti, J.; Hänninen, M.-L.; Rossi, M. Refinement of Whole-Genome Multilocus Sequence Typing Analysis by Addressing Gene Paralogy. J. Clin. Microbiol. 2015, 53, 1765–1767. [Google Scholar] [CrossRef]
- R-Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A Reference Database for Bacterial Virulence Factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, M.P.; Patel, J.B.; Campeau, S.; Eliopoulos, G.M.; Galas, M.F.; Humphries, R.M.; Jenkins, S.G.; Limbago, B.; Mathers, A.J.; Mazzulli, T.; et al. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing Supplement M100, 26th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Brynildsrud, O.; Bohlin, J.; Scheffer, L.; Eldholm, V. Rapid Scoring of Genes in Microbial Pan-Genome-Wide Association Studies with Scoary. Genome Biol. 2016, 17, e238. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Maddison, W.P. Testing Character Correlation Using Pairwise Comparisons on a Phylogeny. J. Theor. Biol. 2000, 202, 195–204. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Dimmer, E.C.; Huntley, R.P.; Alam-Faruque, Y.; Sawford, T.; O’Donovan, C.; Martin, M.J.; Bely, B.; Browne, P.; Mun Chan, W.; Eberhardt, R.; et al. The UniProt-GO Annotation Database in 2011. Nucleic Acids Res. 2012, 40, D565–D570. [Google Scholar] [CrossRef]
- Argimón, S.; Abudahab, K.; Goater, R.J.E.; Fedosejev, A.; Bhai, J.; Glasner, C.; Feil, E.J.; Holden, M.T.G.; Yeats, C.A.; Grundmann, H.; et al. Microreact: Visualizing and Sharing Data for Genomic Epidemiology and Phylogeography. Microb. Genom. 2016, 2. [Google Scholar] [CrossRef] [PubMed]
- Christensen, G.J.M.; Scholz, C.F.P.; Enghild, J.; Rohde, H.; Kilian, M.; Thürmer, A.; Brzuszkiewicz, E.; Lomholt, H.B.; Brüggemann, H. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and Its Genomic Basis. BMC Genom. 2016, 17, e152. [Google Scholar] [CrossRef] [PubMed]
- Stapleton, P.; Pike, R.; Mullany, P.; Lucas, V.; Roberts, G.; Rowbury, R.; Wilson, M.; Richards, H. Mercuric Resistance Genes in Gram-Positive Oral Bacteria. FEMS Microbiol. Lett. 2004, 236, 213–220. [Google Scholar] [CrossRef][Green Version]
- Ojo, K.K.; Tung, D.; Luis, H.; Bernardo, M.; Leitao, J.; Roberts, M.C. Gram-Positive MerA Gene in Gram-Negative Oral and Urine Bacteria. FEMS Microbiol. Lett. 2004, 238, 411–416. [Google Scholar] [CrossRef]
- Kwak, Y.G.; Truong-Bolduc, Q.C.; Bin Kim, H.; Song, K.-H.; Kim, E.S.; Hooper, D.C. Association of NorB Overexpression and Fluoroquinolone Resistance in Clinical Isolates of Staphylococcus aureus from Korea. J. Antimicrob. Chemother. 2013, 68, 2766–2772. [Google Scholar] [CrossRef] [PubMed]
- Hata, E.; Katsuda, K.; Kobayashi, H.; Uchida, I.; Tanaka, K.; Eguchi, M. Genetic Variation among Staphylococcus aureus Strains from Bovine Milk and Their Relevance to Methicillin-Resistant Isolates from Humans. J. Clin. Microbiol. 2010, 48, 2130–2139. [Google Scholar] [CrossRef]
- Leijon, M.; Atkins, E.; Persson Waller, K.; Artursson, K. Longitudinal Study of Staphylococcus aureus Genotypes Isolated from Bovine Clinical Mastitis. J. Dairy Sci. 2021, 104, 11945–11954. [Google Scholar] [CrossRef]
- Boss, R.; Cosandey, A.; Luini, M.; Artursson, K.; Bardiau, M.; Breitenwieser, F.; Hehenberger, E.; Lam, T.; Mansfeld, M.; Michel, A.; et al. Bovine Staphylococcus aureus: Subtyping, Evolution, and Zoonotic Transfer. J. Dairy Sci. 2016, 99, 515–528. [Google Scholar] [CrossRef]
- Sakwinska, O.; Giddey, M.; Moreillon, M.; Morisset, D.; Waldvogel, A.; Moreillon, P. Staphylococcus Aureus Host Range and Human-Bovine Host Shift. Appl. Environ. Microbiol. 2011, 77, 5908–5915. [Google Scholar] [CrossRef]
- Bar-Gal, G.K.; Blum, S.E.; Hadas, L.; Ehricht, R.; Monecke, S.; Leitner, G. Host-Specificity of Staphylococcus aureus Causing Intramammary Infections in Dairy Animals Assessed by Genotyping and Virulence Genes. Vet. Microbiol. 2015, 176, 143–154. [Google Scholar] [CrossRef]
- Aires-de-Sousa, M. Methicillin-Resistant Staphylococcus aureus among Animals: Current Overview. Clin. Microbiol. Infect. 2017, 23, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Courvalin, P. Antimicrobial Drug Resistance: Prediction Is Very Difficult, Especially about the Future. Emerg. Infect. Dis. 2005, 11, 1503–1506. [Google Scholar] [CrossRef]
- Gordon, N.C.; Price, J.R.; Cole, K.; Everitt, R.; Morgan, M.; Finney, J.; Kearns, A.M.; Pichon, B.; Young, B.; Wilson, D.J.; et al. Prediction of Staphylococcus aureus Antimicrobial Resistance by Whole-Genome Sequencing. J. Clin. Microbiol. 2014, 52, 1182–1191. [Google Scholar] [CrossRef]
- Van den Borne, B.H.P.; Nielen, M.; van Schaik, G.; Melchior, M.B.; Lam, T.J.G.M.; Zadoks, R.N. Host Adaptation of Bovine Staphylococcus aureus Seems Associated with Bacteriological Cure after Lactational Antimicrobial Treatment. J. Dairy Sci. 2010, 93, 2550–2558. [Google Scholar] [CrossRef] [PubMed]
- Steele, N.; McDougall, S. Effect of Prolonged Duration Therapy of Subclinical Mastitis in Lactating Dairy Cows Using Penethamate Hydriodide. N. Z. Vet. J. 2014, 62, 38–46. [Google Scholar] [CrossRef]
- OIE. OIE List of Antimicrobial Agents of Veterinary Importance; World Organisation for Animal Health: Paris, France, 2018. [Google Scholar]
- Hennekinne, J.-A.; De Buyser, M.-L.; Dragacci, S. Staphylococcus aureus and Its Food Poisoning Toxins: Characterization and Outbreak Investigation. FEMS Microbiol. Rev. 2012, 36, 815–836. [Google Scholar] [CrossRef]
- Argudín, M.A.; Mendoza, M.C.; González-Hevia, M.A.; Bances, M.; Guerra, B.; Rodicio, M.R. Genotypes, Exotoxin Gene Content, and Antimicrobial Resistance of Staphylococcus aureus Strains Recovered from Foods and Food Handlers. Appl. Environ. Microbiol. 2012, 78, 2930–2935. [Google Scholar] [CrossRef] [PubMed]
- Nazari, R.; Godarzi, H.; Rahimi Baghi, F.; Moeinrad, M. Enterotoxin Gene Profiles among Staphylococcus aureus Isolated from Raw Milk. Iran. J. Vet. Res. 2014, 15, 409–412. [Google Scholar]
- Carfora, V.; Caprioli, A.; Marri, N.; Sagrafoli, D.; Boselli, C.; Giacinti, G.; Giangolini, G.; Sorbara, L.; Dottarelli, S.; Battisti, A.; et al. Enterotoxin Genes, Enterotoxin Production, and Methicillin Resistance in Staphylococcus aureus Isolated from Milk and Dairy Products in Central Italy. Int. Dairy J. 2015, 42, 12–15. [Google Scholar] [CrossRef]
- Mason, A.; Foster, D.; Bradley, P.; Golubchik, T.; Doumith, M.; Gordon, N.C.; Pichon, B.; Iqbal, Z.; Staves, P.; Crook, D.; et al. Accuracy of Different Bioinformatics Methods in Detecting Antibiotic Resistance and Virulence Factors from Staphylococcus aureus Whole-Genome Sequences. J. Clin. Microbiol. 2018, 56, e1815–e1817. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, D.; Peton, V.; Almeida, S.; Le Maréchal, C.; Miyoshi, A.; Azevedo, V.; Berkova, N.; Rault, L.; François, P.; Schrenzel, J.; et al. Genome Sequence of Staphylococcus aureus Newbould 305, a Strain Associated with Mild Bovine Mastitis. J. Bacteriol. 2012, 194, 6292–6293. [Google Scholar] [CrossRef] [PubMed]
- Maréchal, C.L.; Seyffert, N.; Jardin, J.; Hernandez, D.; Jan, G.; Rault, L.; Azevedo, V.; François, P.; Schrenzel, J.; van de Guchte, M.; et al. Molecular Basis of Virulence in Staphylococcus aureus Mastitis. PLoS ONE 2011, 6, e27354. [Google Scholar] [CrossRef] [PubMed]
- Peton, V.; Le Loir, Y. Staphylococcus aureus in Veterinary Medicine. Infect. Genet. Evol. 2014, 21, 602–615. [Google Scholar] [CrossRef] [PubMed]
| Sample Level | Multilocus Sequence Type (ST) | |||||||
|---|---|---|---|---|---|---|---|---|
| ST-1 | ST-188 | ST-5 | ST-705 | ST-1247 | ST-97 | ST-151 | ST-425 | |
| Isolates (%) | ||||||||
| Clinical | 12 (63.2) | 0 | 0 | 1 (5.3) | 0 | 2 (10.5) | 1 (5.3) | 0 |
| Sub-clinical | 23 (60.5) | 5 (13.2) | 1 (2.6) | 1 (2.6) | 2 (5.3) | 3 (7.9) | 0 | 1 (2.6) |
| Total | 35 (61.4) | 5 (8.8) | 1 (1.8) | 2 (3.5) | 2 (3.5) | 5 (8.8) | 1 (1.8) | 1 (1.8) |
| Farms (%) | 11 (64.7) | 1 (5.9) | 1 (5.9) | 2 (11.8) | 2 (11.8) | 3 (17.6) | 1 (5.9) | 1 (5.9) |
| Cows (%) | 32 (61.5) | 4 (7.7) | 1 (1.9) | 2 (3.8) | 2 (3.8) | 5 (9.6) | 1 (1.9) | 1 (1.9) |
| Gene(s) | Drug Class | Common Use in the New Zealand Dairy Industry |
|---|---|---|
| ant(9)-Ia | Aminoglycosides | Intra-mammary antimicrobials for the treatment of mastitis in lactating cows e.g., neomycin and streptomycin. |
| blaI blaPC1 blaR1 blaZ mecA mecR1 | ß-lactams | Broad range antimicrobials used to treat a range of intra-mammary, intra-uterine and systemic infections e.g., penicillin, amoxicillin and cloxacillin. |
| dfrC | Diaminopyrimidines | Limited use in cattle, with the exception of Trimethoprim, which is commonly used in combination with sulpha drugs to treat enteric or respiratory tract diseases. |
| erm(A) | Macrolides | Antimicrobials used in the treatment of various systemic and localised bacterial infections including mastitis, respiratory infection, and foot-rot although tilmicosin and tulathromycin have a very long milk withholding period, so are not used in lactating cattle, and rarely on dairy farms whilst erythromycin is also no longer used in cattle in New Zealand. |
| fosD | Fosfomycin | Used to treat a broad variety of bacterial infections in humans, particularly urinary tract infections but it is not registered for animal use in New Zealand. |
| fusC | Fusidic acid | Fusidic acid is not registered for cattle use in New Zealand but has registration for use in dogs. |
| qacA qacB | Fluoroquinolones | Injectable antimicrobials used in a range of treatments including Escherichia coli and Pseudomonas mastitis, osteomyelitis, and respiratory infections, but with very limited usage in the dairy industry. |
| tet(38) | Tetracyclines | Antimicrobial used in the broad-spectrum treatment of local and systemic infections particularly uterine infections and other soft tissue infections in cattle. |
| Gene(s) | GO Terms | No. Isolates Gene Present (%) | |
|---|---|---|---|
| ST-1 (n = 35) | Other STs (n = 22) | ||
| agrB | Quorum sensing, pathogenesis, and peptidase activity | 35 (100) | 0 |
| entH | Virulence, metal ion binding, and toxin activity | 35 (100) | 0 |
| flr | Pathogenesis and signal peptide | 35 (100) | 0 |
| catE-2 | Transcription regulation and DNA-binding | 35 (100) | 1 (4.5) |
| gltR | 35 (100) | 1 (4.5) | |
| yofA | 35 (100) | 0 | |
| gdmA | Cytolysis and signalling receptor binding | 35 (100) | 0 |
| nisC | Maturation of the lantibiotic | 35 (100) | 0 |
| repE/N | DNA replication initiation and binding | 35 (100) | 0 |
| group-2156/7 | Signal peptide | 35 (100) | 0 |
| ssbA-1 | DNA replication, repair and recombination, and single-stranded DNA binding | 23 (65.7) | 0 |
| ssbA-2 | 14 (40.0) | 1 (4.5) | |
| dnaC-2 | DNA replication, helicase activity, synthesis of RNA primers, and ATP binding | 21 (60.0) | 0 |
| brnQ-3 | Branched-chain amino acid transmembrane transporter | 16 (45.7) | 0 |
| dut | dUMP biosynthetic process, dUTP activity, and Mg binding | 14 (40.0) | 0 |
| bcgIA/B | DNA modification and N-methyltransferase activity | 14 (40.0) | 0 |
| hin | DNA integration, DNA-binding, and recombinase activity | 10 (28.6) | 0 |
| cna | Pathogenesis, cell adhesion, and collagen binding | 9 (25.7) | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greening, S.S.; Zhang, J.; Midwinter, A.C.; Wilkinson, D.A.; McDougall, S.; Gates, M.C.; French, N.P. The Genetic Relatedness and Antimicrobial Resistance Patterns of Mastitis-Causing Staphylococcus aureus Strains Isolated from New Zealand Dairy Cattle. Vet. Sci. 2021, 8, 287. https://doi.org/10.3390/vetsci8110287
Greening SS, Zhang J, Midwinter AC, Wilkinson DA, McDougall S, Gates MC, French NP. The Genetic Relatedness and Antimicrobial Resistance Patterns of Mastitis-Causing Staphylococcus aureus Strains Isolated from New Zealand Dairy Cattle. Veterinary Sciences. 2021; 8(11):287. https://doi.org/10.3390/vetsci8110287
Chicago/Turabian StyleGreening, Sabrina S., Ji Zhang, Anne C. Midwinter, David A. Wilkinson, Scott McDougall, M. Carolyn Gates, and Nigel P. French. 2021. "The Genetic Relatedness and Antimicrobial Resistance Patterns of Mastitis-Causing Staphylococcus aureus Strains Isolated from New Zealand Dairy Cattle" Veterinary Sciences 8, no. 11: 287. https://doi.org/10.3390/vetsci8110287
APA StyleGreening, S. S., Zhang, J., Midwinter, A. C., Wilkinson, D. A., McDougall, S., Gates, M. C., & French, N. P. (2021). The Genetic Relatedness and Antimicrobial Resistance Patterns of Mastitis-Causing Staphylococcus aureus Strains Isolated from New Zealand Dairy Cattle. Veterinary Sciences, 8(11), 287. https://doi.org/10.3390/vetsci8110287






