Development of a Genome-Wide Oligonucleotide Microarray Platform for Detection of DNA Copy Number Aberrations in Feline Cancers
Abstract
1. Introduction
2. Materials and Methods
2.1. Design and Construction of the Feline Oligonucleotide CGH Array
2.2. Experimental Validation of the Feline oaCGH Array Design
2.3. DNA Copy Number Profiling of a Feline Injection-Site Sarcoma
3. Results
3.1. Guiding Development of a Feline oaCGH Array Design Using a Canine Template
3.2. Experimental Evaluation Using Sex-Mismatched Reference DNA
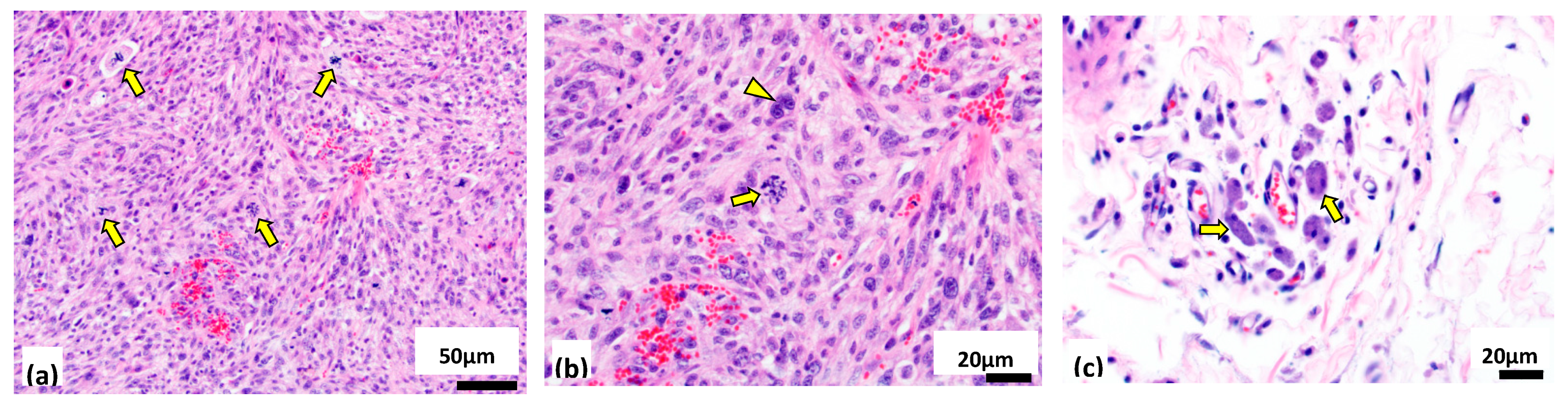
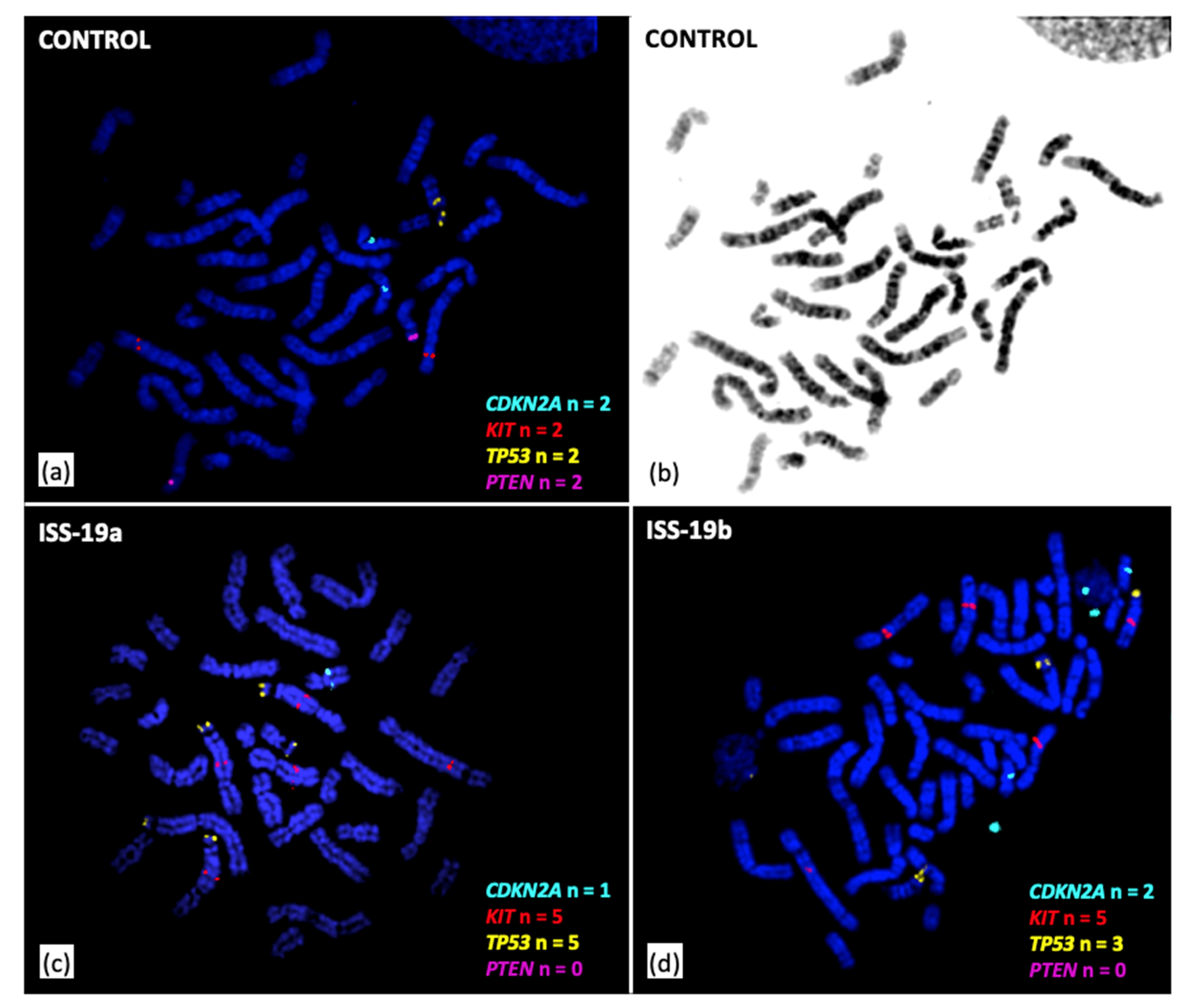
3.3. Cytogenomic Evaluation of Tumor Specimens
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Murphy, W.J. The feline genome. Genome. Dyn. 2006, 2, 60–68. [Google Scholar] [PubMed]
- Online Mendelian Inheritance in Animals. Available online: https://omia.org/ (accessed on 23 June 2020).
- Lindblad-Toh, K.; Wade, C.M.; Mikkelsen, T.S.; Karlsson, E.K.; Jaffe, D.B.; Kamal, M.; Clamp, M.; Chang, J.L.; Kulbokas, E.J.; Zody, M.C.; et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 2005, 438, 803–819. [Google Scholar] [CrossRef] [PubMed]
- Pontius, J.U.; Mullikin, J.C.; Smith, D.R.; Lindblad-Toh, K.; Gnerre, S.; Clamp, M.; Chang, J.; Stephens, R.; Neelam, B.; Volfovsky, N.; et al. Initial sequence and comparative analysis of the cat genome. Genome. Res. 2007, 17, 1675–1689. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, A.K.; Breen, M.; Choyke, P.; Dewhirst, M.; Fan, T.M.; Gustafson, D.L.; Helman, L.J.; Kastan, M.B.; Knapp, D.W.; Levin, W.J.; et al. Perspectives from man’s best friend: National Academy of Medicine’s Workshop on Comparative Oncology. Sci. Transl. Med. 2016, 8, 324–325. [Google Scholar] [CrossRef]
- Schiffman, J.D.; Breen, M. Comparative oncology: What dogs and other species can teach us about humans with cancer. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2015, 370. [Google Scholar] [CrossRef]
- Thomas, R.; Valli, V.E.; Ellis, P.; Bell, J.; Karlsson, E.K.; Cullen, J.; Lindblad-Toh, K.; Langford, C.F.; Breen, M. Microarray-based cytogenetic profiling reveals recurrent and subtype-associated genomic copy number aberrations in feline sarcomas. Chromosome. Res. 2009, 17, 987–1000. [Google Scholar] [CrossRef]
- Buckley, R.M.; Davis, B.W.; Brashear, W.A.; Farias, F.H.G.; Kuroki, K.; Graves, T.; Hillier, L.W.; Kremitzki, M.; Li, G.; Middleton, R.; et al. A new domestic cat genome assembly based on long sequence reads empowers feline genomic medicine and identifies a novel gene for dwarfism. BioRxiv 2020. [Google Scholar] [CrossRef]
- Poorman, K.; Borst, L.; Moroff, S.; Roy, S.; Labelle, P.; Motsinger-Reif, A.; Breen, M. Comparative cytogenetic characterization of primary canine melanocytic lesions using array CGH and fluorescence in situ hybridization. Chromosome. Res. 2015, 23, 171–186. [Google Scholar] [CrossRef]
- Shapiro, S.G.; Raghunath, S.; Williams, C.; Motsinger-Reif, A.A.; Cullen, J.M.; Liu, T.; Albertson, D.; Ruvolo, M.; Bergstrom Lucas, A.; Jin, J.; et al. Canine urothelial carcinoma: Genomically aberrant and comparatively relevant. Chromosome. Res. 2015, 23, 311–331. [Google Scholar] [CrossRef]
- Thomas, R.; Borst, L.; Rotroff, D.; Motsinger-Reif, A.; Lindblad-Toh, K.; Modiano, J.F.; Breen, M. Genomic profiling reveals extensive heterogeneity in somatic DNA copy number aberrations of canine hemangiosarcoma. Chromosome. Res. 2014, 22, 305–319. [Google Scholar] [CrossRef]
- Thomas, R.; Demeter, Z.; Kennedy, K.A.; Borst, L.; Singh, K.; Valli, V.E.; Le Boedec, K.; Breen, M. Integrated immunohistochemical and DNA copy number profiling analysis provides insight into the molecular pathogenesis of canine follicular lymphoma. Vet. Comp. Oncol. 2017, 15, 852–867. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J. BLAT--the BLAST-like alignment tool. Genome. Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Seiser, E.L.; Motsinger-Reif, A.; Borst, L.; Valli, V.E.; Kelley, K.; Suter, S.E.; Argyle, D.; Burgess, K.; Bell, J.; et al. Refining tumor-associated aneuploidy through ’genomic recoding’ of recurrent DNA copy number aberrations in 150 canine non-Hodgkin lymphomas. Leuk. Lymphoma. 2011, 52, 1321–1335. [Google Scholar] [CrossRef]
- Cheung, S.W.; Bi, W. Novel applications of array comparative genomic hybridization in molecular diagnostics. Expert. Rev. Mol. Diagn. 2018, 18, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Murphy, W.J.; Davis, B.; David, V.A.; Agarwala, R.; Schaffer, A.A.; Pearks Wilkerson, A.J.; Neelam, B.; O’Brien, S.J.; Menotti-Raymond, M. A 1.5-Mb-resolution radiation hybrid map of the cat genome and comparative analysis with the canine and human genomes. Genomics 2007, 89, 189–196. [Google Scholar] [CrossRef][Green Version]
- Young, A.C.; Kirkness, E.F.; Breen, M. Tackling the characterization of canine chromosomal breakpoints with an integrated in-situ/in-silico approach: The canine PAR and PAB. Chromosome. Res. 2008, 16, 1193–1202. [Google Scholar] [CrossRef]
- Raudsepp, T.; Chowdhary, B.P. The Eutherian Pseudoautosomal Region. Cytogenet. Genome. Res. 2015, 147, 81–94. [Google Scholar] [CrossRef]
- Murphy, W.J.; Pearks Wilkerson, A.J.; Raudsepp, T.; Agarwala, R.; Schaffer, A.A.; Stanyon, R.; Chowdhary, B.P. Novel gene acquisition on carnivore Y chromosomes. PLoS. Genet. 2006, 2. [Google Scholar] [CrossRef]
- Zhang, C.Z.; Leibowitz, M.L.; Pellman, D. Chromothripsis and beyond: Rapid genome evolution from complex chromosomal rearrangements. Genes. Dev. 2013, 27, 2513–2530. [Google Scholar] [CrossRef] [PubMed]
- Storchova, Z.; Kloosterman, W.P. The genomic characteristics and cellular origin of chromothripsis. Curr. Opin. Cell. Biol. 2016, 40, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Cathcart, J.; Pulkoski-Gross, A.; Cao, J. Targeting Matrix Metalloproteinases in Cancer: Bringing New Life to Old Ideas. Genes. Dis. 2015, 2, 26–34. [Google Scholar] [CrossRef]
- Sorensen, K.C.; Kitchell, B.E.; Schaeffer, D.J.; Mardis, P.E. Expression of matrix metalloproteinases in feline vaccine site-associated sarcomas. Am. J. Vet. Res. 2004, 65, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Durrbaum, M.; Storchova, Z. Effects of aneuploidy on gene expression: Implications for cancer. FEBS. J. 2016, 283, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Santos, S.; Chaves, R.; Adega, F.; Bastos, E.; Guedes-Pinto, H. Amplification of the major satellite DNA family (FA-SAT) in a cat fibrosarcoma might be related to chromosomal instability. J. Hered. 2006, 97, 114–118. [Google Scholar] [CrossRef]
- von Erichsen, J.; Hecht, W.; Lohberg-Gruene, C.; Reinacher, M. Cell lines derived from feline fibrosarcoma display unstable chromosomal aneuploidy and additionally centrosome number aberrations. Vet. Pathol. 2012, 49, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Dufresne, A.; Cassier, P.; Heudel, P.; Pissaloux, D.; Wang, Q.; Blay, J.Y.; Ray-Coquard, I. Molecular biology of sarcoma and therapeutic choices. Bull. Cancer. 2015, 102, 6–16. [Google Scholar] [CrossRef]
- Brashear, W.A.; Raudsepp, T.; Murphy, W.J. Evolutionary conservation of Y Chromosome ampliconic gene families despite extensive structural variation. Genome. Res. 2018, 28, 1841–1851. [Google Scholar] [CrossRef]
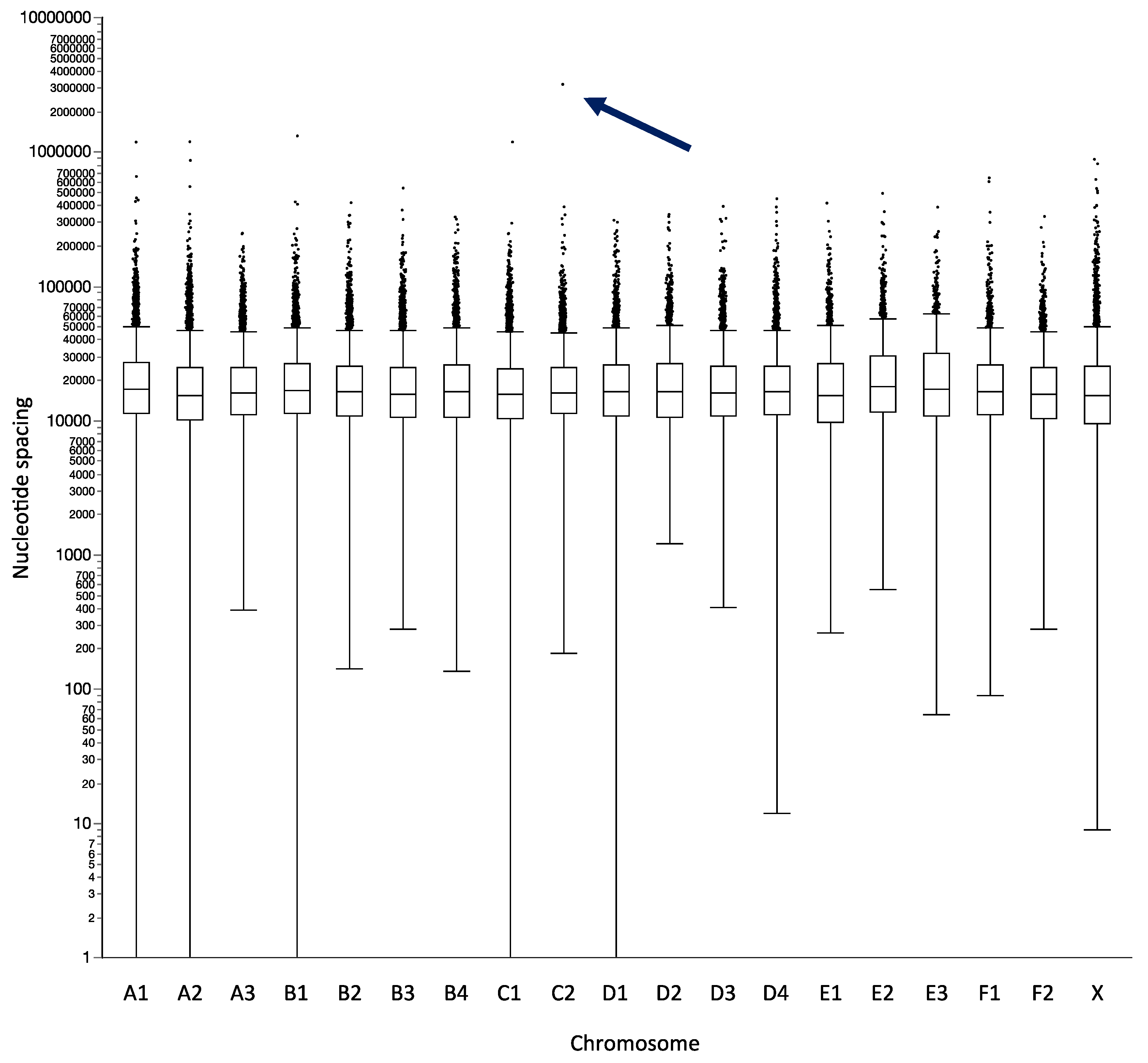


| Chromosome | Chromosome Size (bp) | Total Number of Probes | Mean Spacing (bp) | Median Spacing (bp) | Maximum Spacing (bp) | Probes/Mb |
|---|---|---|---|---|---|---|
| A1 | 242,100,913 | 10,648 | 22,664 | 17,125 | 2,635,625 | 44 |
| A2 | 171,471,747 | 7944 | 21,469 | 15,552 | 2,272,323 | 46 |
| A3 | 143,202,405 | 6653 | 21,434 | 16,188 | 2,630,398 | 46 |
| B1 | 208,212,889 | 9420 | 22,001 | 16,824 | 2,362,299 | 45 |
| B2 | 155,302,638 | 7160 | 21,603 | 16,296 | 2,303,124 | 46 |
| B3 | 149,751,809 | 7026 | 21,173 | 15,704 | 2,516,731 | 47 |
| B4 | 144,528,695 | 6559 | 21,942 | 16,343 | 2,103,152 | 45 |
| C1 | 222,790,142 | 10,724 | 20,646 | 15,770 | 2,108,648 | 48 |
| C2 | 161,193,150 | 7462 | 21,511 | 16,288 | 3,159,135 | 46 |
| D1 | 117,648,028 | 5174 | 22,610 | 16,463 | 2,184,516 | 44 |
| D2 | 90,186,660 | 3863 | 23,132 | 16,397 | 2,155,220 | 43 |
| D3 | 96,884,206 | 4275 | 22,564 | 16,227 | 2,223,652 | 44 |
| D4 | 96,521,652 | 4249 | 22,622 | 16,361 | 2,213,074 | 44 |
| E1 | 63,494,689 | 2712 | 22,955 | 15,407 | 2,301,848 | 43 |
| E2 | 64,340,295 | 2352 | 27,026 | 18,082 | 2,536,363 | 37 |
| E3 | 44,648,284 | 1561 | 28,241 | 17,413 | 3,394,005 | 35 |
| F1 | 71,664,243 | 3082 | 22,498 | 16,523 | 650,963 | 43 |
| F2 | 85,752,456 | 4137 | 20,102 | 15,601 | 333,037 | 48 |
| X | 130,557,009 | 5455 | 23,800 | 15,598 | 2,487,257 | 42 |
| Total | 2,460,251,910 | 110,456 | 22,631 | - | - | 44 (mean) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, R.; Pontius, J.U.; Borst, L.B.; Breen, M. Development of a Genome-Wide Oligonucleotide Microarray Platform for Detection of DNA Copy Number Aberrations in Feline Cancers. Vet. Sci. 2020, 7, 88. https://doi.org/10.3390/vetsci7030088
Thomas R, Pontius JU, Borst LB, Breen M. Development of a Genome-Wide Oligonucleotide Microarray Platform for Detection of DNA Copy Number Aberrations in Feline Cancers. Veterinary Sciences. 2020; 7(3):88. https://doi.org/10.3390/vetsci7030088
Chicago/Turabian StyleThomas, Rachael, Joan U Pontius, Luke B Borst, and Matthew Breen. 2020. "Development of a Genome-Wide Oligonucleotide Microarray Platform for Detection of DNA Copy Number Aberrations in Feline Cancers" Veterinary Sciences 7, no. 3: 88. https://doi.org/10.3390/vetsci7030088
APA StyleThomas, R., Pontius, J. U., Borst, L. B., & Breen, M. (2020). Development of a Genome-Wide Oligonucleotide Microarray Platform for Detection of DNA Copy Number Aberrations in Feline Cancers. Veterinary Sciences, 7(3), 88. https://doi.org/10.3390/vetsci7030088





