Genetic Characterization of Indubrasil Cattle Breed Population
Abstract
1. Introduction
2. Material and Methods
- (1)
- The percentage of homozygous genotypes in relation to the total number of genotypes (FSNP).
- (2)
- The inbreeding coefficient (Fx) using the frequency of the observed homozygous genotypes (HO) in relation to the expected homozygous genotypes (HE): Fx = [(HO − HE)/(HE)]).
- (3)
- Runs of homozygosis (ROH). For the verification of FROH, the ROH were detected using the PLINK program with a sliding window of 50 SNPs with minimum length of 1000 kb, with a possibility to have 1 heterozygous SNP and 1 missing SNP within each window. For this, we used the following formula:FROH = Σk Length (ROHk)/L
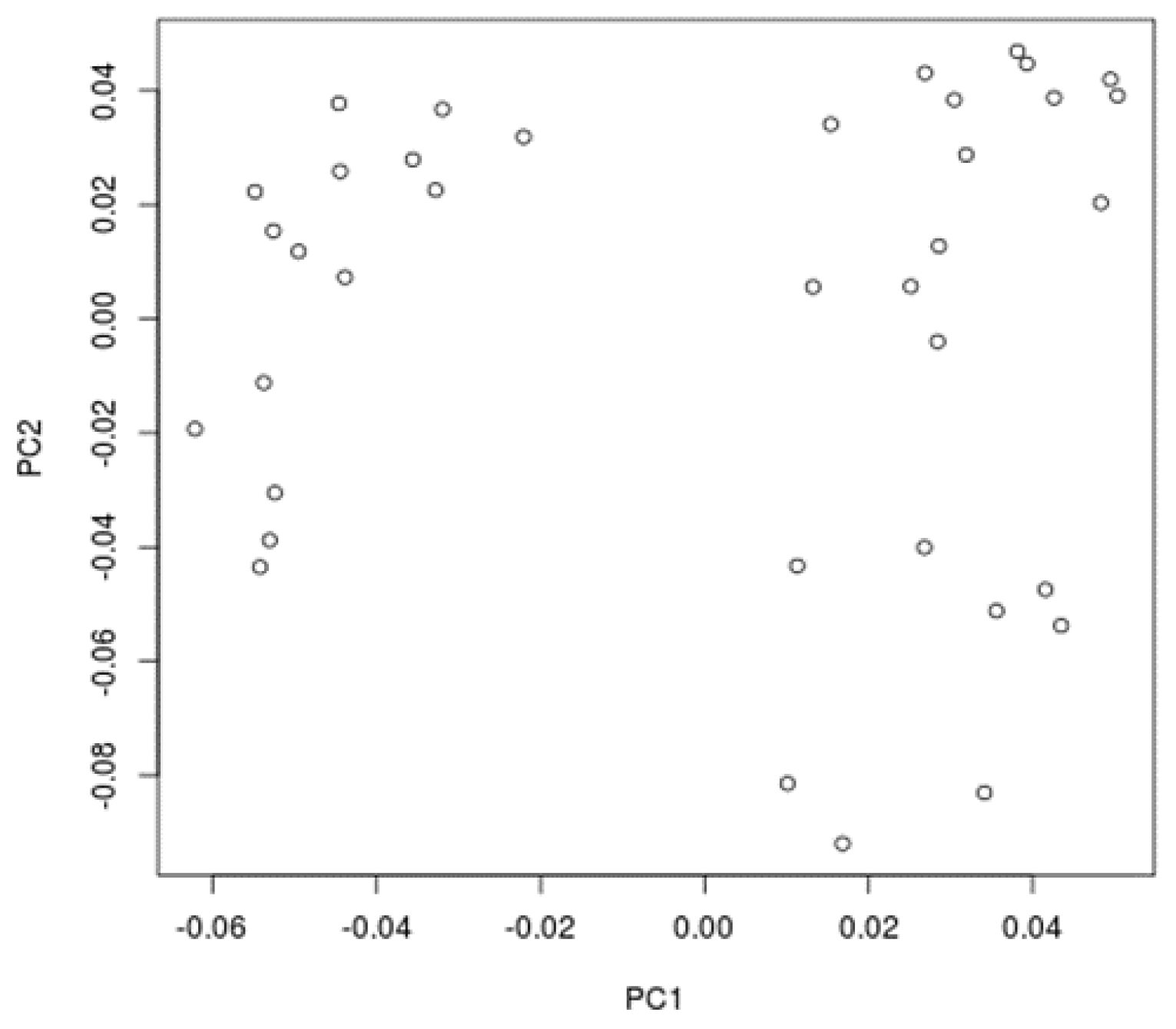
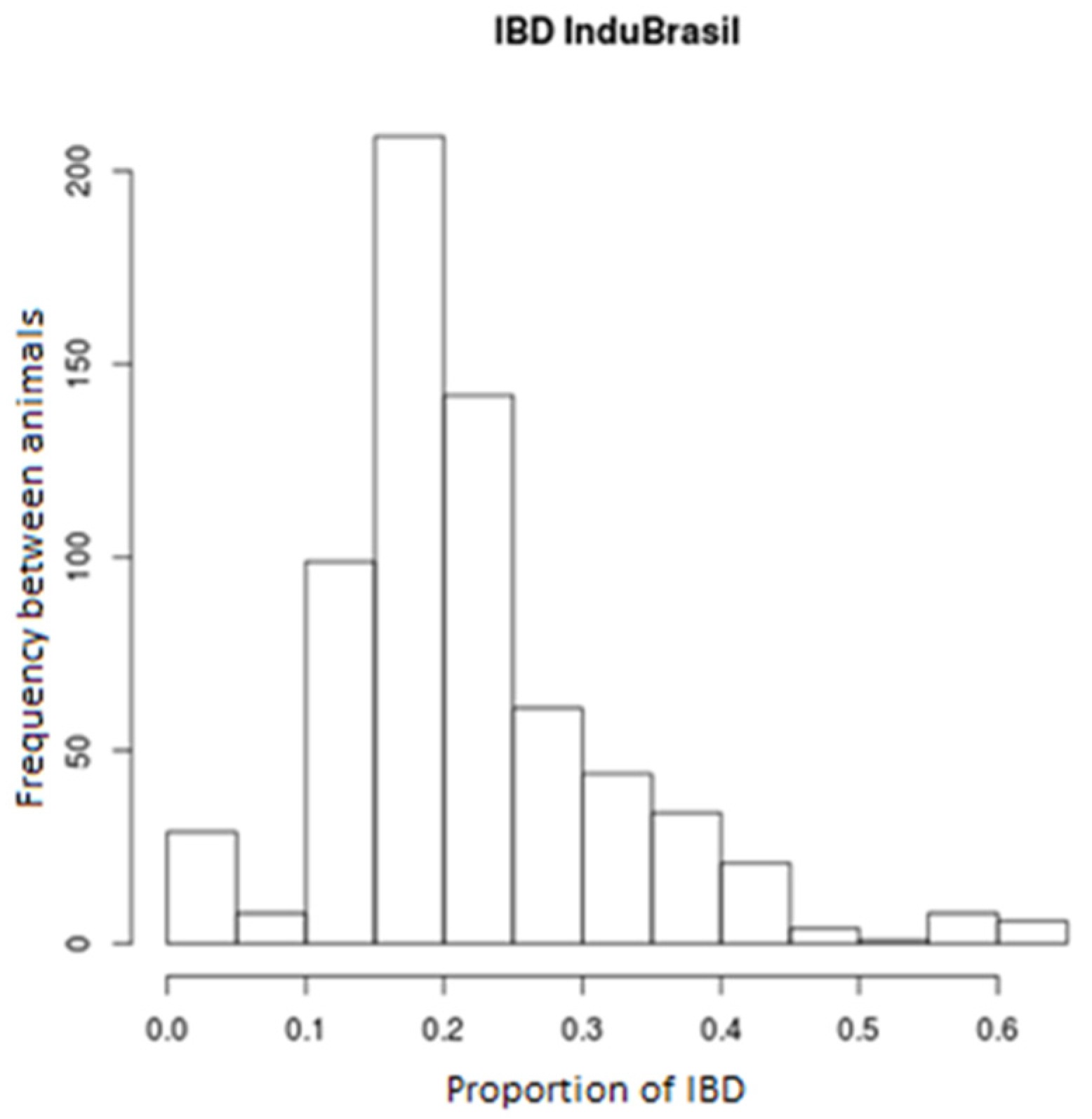
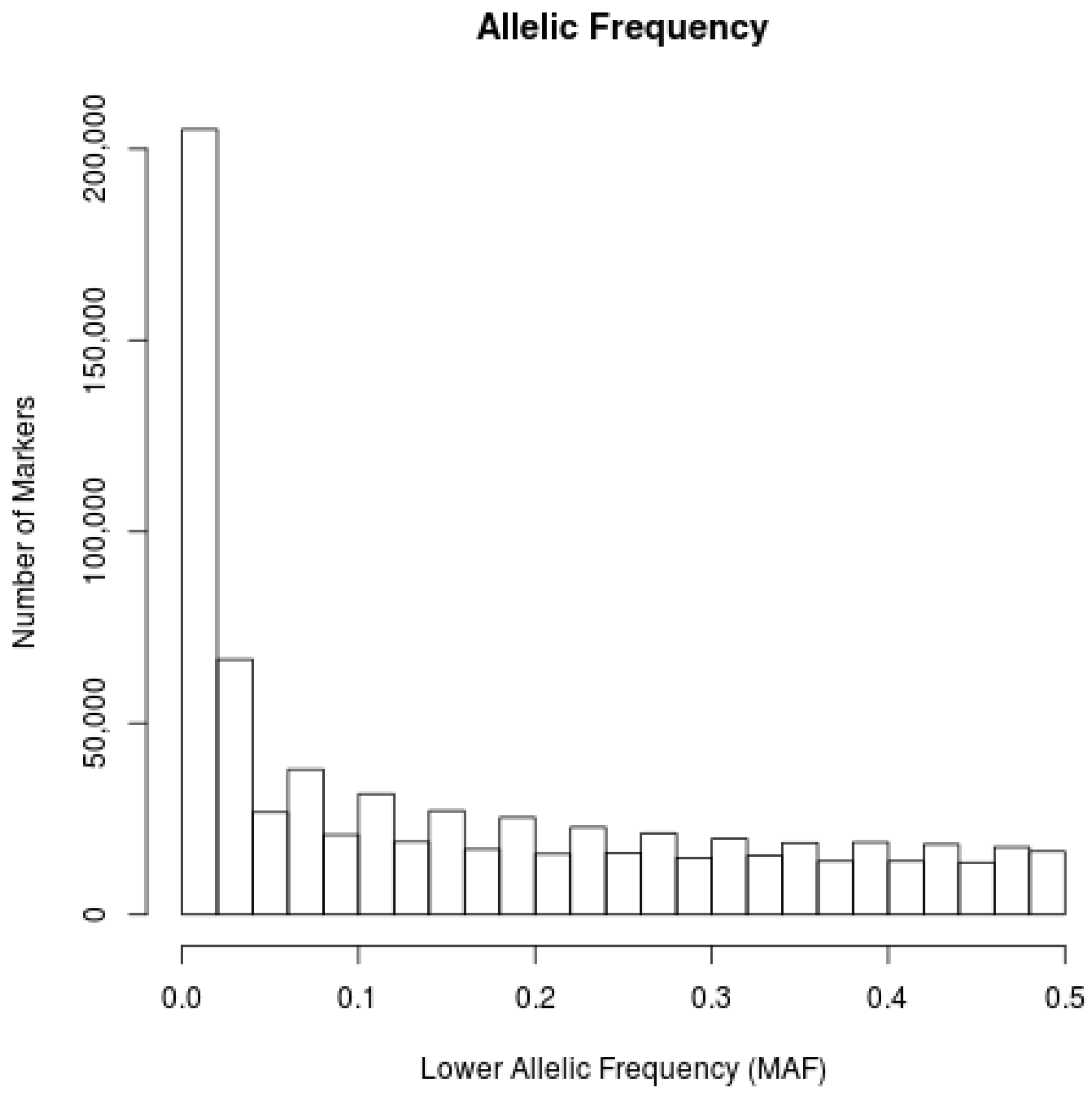
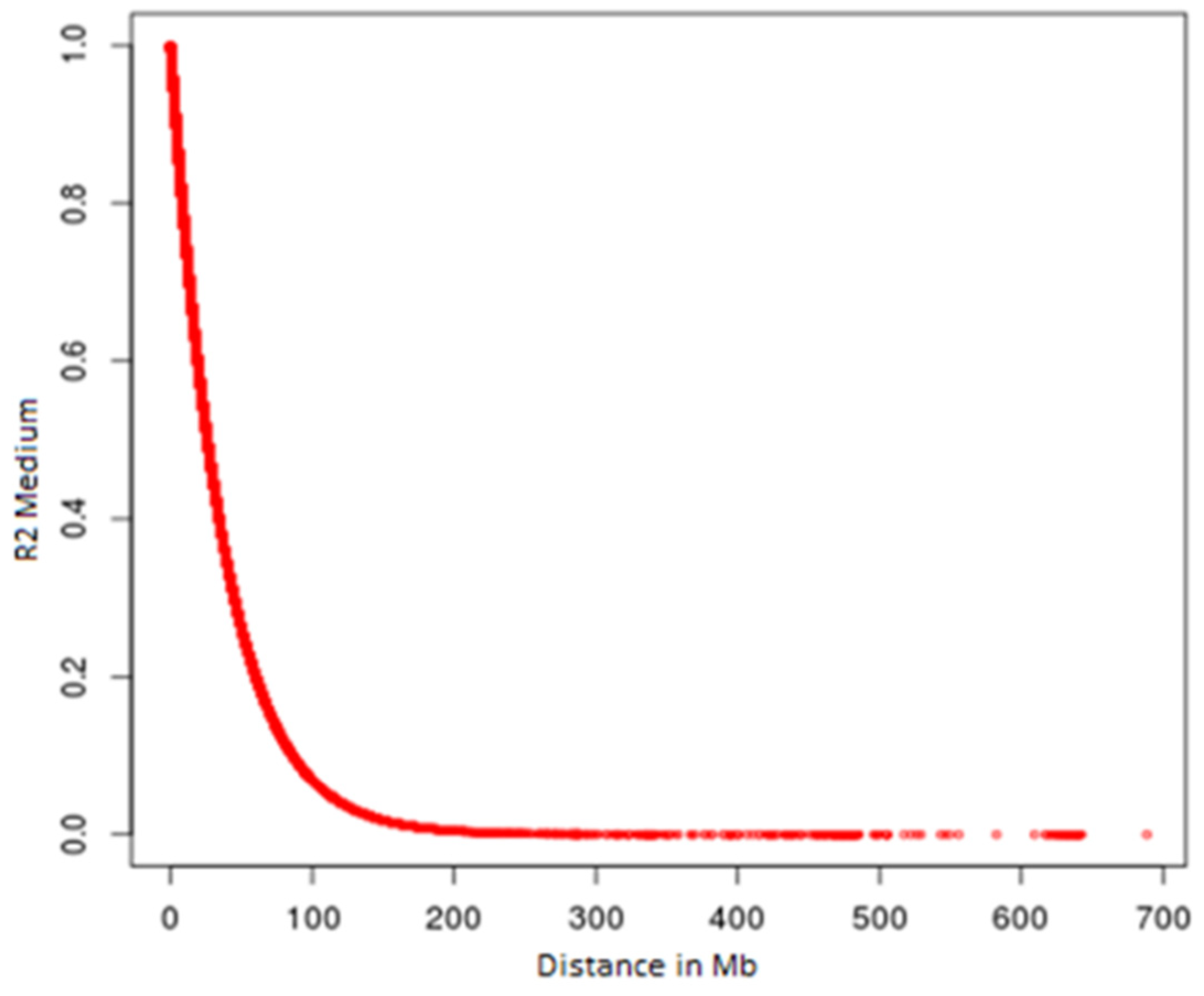
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgements
Conflicts of Interest
References
- Santiago, A.A. O Zebu na Índia, no Brasil e No Mundo; Instituto Campineiro de Ensino Agrícola: Campinas, Brazil, 1986; 744p. [Google Scholar]
- Toral, F.L.B.; Pereira, J.C.C.; Bergmann, J.A.G.; Josahkian, L.A. Parâmetros genéticos do peso desde o nascimento até 730 dias de idade na raça Indubrasil. Pesqui. Agropecu. Bras. 2014, 49, 595–603. [Google Scholar] [CrossRef]
- Carneiro, L.S.C.; Malhado, C.H.M.; Filho, R.M.; Carneiro, A.P.S.; Silva, F.F.; Torres, R.A. A Raça Indubrasil No Nordeste Brasileiro: Melhoramento e Estrutura Populacional. Rev. Bras. Zootec. 2009, 38, 2327–2334. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E.; Woolliams, J. Effective sizes of livestock populations to prevent a decline in fitness. Theor. Appl. Genet. 1994, 89, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- Frankham, R. Conservation genetics. Ann. Rev. Genet. 1995, 29, 305–327. [Google Scholar] [CrossRef] [PubMed]
- Scraggs, E.; Zanella, R.; Wojtowicz, A.; Taylor, J.F.; Gaskins, C.T.; Reeves, J.J.; de Avila, J.M.; Neibergs, H.L. Estimation of inbreeding and effective population size of full-blood wagyu cattle registered with the American Wagyu Cattle Association. J. Anim. Breed. Genet. 2014, 131, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Zanella, R.; Peixoto, J.O.; Cardoso, F.F.; Cardoso, L.L.; Biegelmeyer, P.; Cantão, M.E.; Otaviano, A.; Freitas, M.S.; Caetano, A.R.; Ledur, M.C. Genetic diversity analysis of two commercial breeds of pigs using genomic and pedigree data. Genet. Sel. Evol. GSE 2016, 48, 24. [Google Scholar] [CrossRef] [PubMed]
- Burrow, H.M. The effects of inbreeding in beef cattle. Anim. Breed. Abstr. 1993, 61, 737–751. [Google Scholar]
- Queiroz, S.A.; Albuquerque, L.G.; Lanzoni, N.A. Efeito da endogamia sobre características de crescimento de bovinos da raça Gir no Brasil. Rev. Bras. Zootec. 2000, 29, 1014–1019. [Google Scholar] [CrossRef]
- Schenkel, F.S.; Lagioia, D.R.; Riboldi, J. Níveis de Endogamia e Depressão Endogâmica no Ganho de Peso de Raças Zebuínas no Brasil. Anais do IV Simpósio de Melhoramento Animal. 2002. Available online: http://www.sbmaonline.org.br/anais/iv/trabalhos/pdfs/ivt06bc.pdf (accessed on 5 March 2018).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser. Genome Res. 2002, 6, 996–1006. [Google Scholar] [CrossRef]
- Cohen-Zinder, M.; Seroussi, E.; Larkin, D.M.; Loor, J.J.; Everts-van der Wind, A.; Lee, J.H.; Drackley, J.K.; Band, M.R.; Hernandez, A.G.; Shani, M.; et al. Identification of a missense mutation in the bovine ABCG2 gene with a major effect on the QTL on chromosome 6 affecting milk yield and composition in Holstein cattle. Genome Res. 2005, 15, 936–944. [Google Scholar] [CrossRef] [PubMed]
- Ron, M.; Blanc, Y.; Band, M.; Ezra, E.; Weller, J.I. Misidentification rate in the Israeli dairy cattle population and its implications for genetic improvement. J. Dairy Sci. 1996, 79, 676–681. [Google Scholar] [CrossRef]
- Ferencakovic, M.; Hamzić, E.; Gredler, B.; Solberg, T.R.; Klemetsdal, G.; Curik, I.; Sölkner, J. Estimates of autozygosity derived from runs of homozygosity: Empirical evidence from selected cattle populations. J. Anim. Breed. Genet. 2013, 130, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Ferencakovic, M.; Hamzic, E.; Gredler, B.; Curik, I.; Sölkner, J. Runs of homozygosity reveal genome-wide autozygosity in the Austrian fleckvieh cattle. Agric. Conspec. Sci. 2011, 76, 325–328. [Google Scholar]
- Purfield, D.C.; Berry, D.P.; McParland, S.; Bradley, D.G. Runs of homozygosity and population history in cattle. BMC Genet. 2012, 13, 70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Guldbrandtsen, B.; Bosse, M.; Lund, M.S.; Sahana, G. Runs of homozygosity and distribution of functional variants in the cattle genome. BMC Genomics 2015, 16, 542. [Google Scholar] [CrossRef]
- Zavarez, L.B.; Utsunomiya, Y.T.; Carmo, A.S.; Neves, H.H.; Carvalheiro, R.; Ferenčaković, M.; Pérez O’Brien, A.M.; Curik, I.; Cole, J.B.; Van Tassell, C.P.; et al. Assessment of autozygosity in Nellore cows (Bos indicus) through high-density SNP genotypes. Front. Genet. 2015, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Howrigan, D.P.; Simonson, M.A.; Keller, M.C. Detecting autozygosity through runs of homozygosity: A comparison of three autozygosity detection algorithms. BMC Genomics 2011, 12, 460. [Google Scholar] [CrossRef]
- Vercesi Filho, A.E.; Faria, F.J.C.; Madalena, F.E.; Josahkian, L.A. Estrutura populacional do rebanho Indubrasil registrado no Brasil. Arch. Latinoam. Prod. Anim. 2002, 10, 86–92. [Google Scholar]
- Wright, S. Evolution in Mendelian genetics. Genetics 1931, 16, 97–159. [Google Scholar]
- Oliveira, J.A.; Bastos, J.F.P.; Tonhati, H. Endogamia em um rebanho da raça Guzerá. Rev. Bras. Zootec. 1999, 28, 721–728. [Google Scholar] [CrossRef]
- Muniz, L.M.S.; Souza, L.A.; Barbosa, A.C.B.; Ambrosini, D.P.; Oliveira, A.P.; Carneiro, P.L.S.; Malhado, C.H.M.; Martins Filho, R.; Duarte, R.A.B. A raça Gir Mocha na região Nordeste do Brasil: Estrutura genética populacional via análise de pedigree. Arq. Bras. Med. Vet. Zootec. 2012, 64, 1656–1664. [Google Scholar] [CrossRef]
- Nothnagel, M.; Lu, T.; Kayser, M.; Krawczak, M. Genomic and geographic distribution of SNP defined runs of homozygosity in Europeans. Hum. Mol. Genet. 2009, 19, 2927–2935. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Duan, X.; Chen, S.; He, H.; Liu, X. NCAPG is differentially expressed during longissimus muscle development and is associated with growth traits in Chinese Qinchuan beef cattle. Genet. Mol. Biol. 2015, 38, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Setoguchi, K.; Furuta, M.; Hirano, T.; Nagao, T.; Watanabe, T.; Sugimoto, Y.; Takasuga, A. Cross-breed comparisons identified a critical 591-kb region for bovine carcass weight QTL (CW-2) on chromosome 6 and the Ile-442-Met substitution in NCAPG as a positional candidate. BMC Genet. 2009, 10, 43. [Google Scholar] [CrossRef] [PubMed]
- Bongiorni, S.; Mancini, G.; Chillemi, G.; Pariset, L.; Valentini, A. Identification of a Short Region on Chromosome 6 Affecting Direct Calving Ease in Piedmontese Cattle Breed. PLoS ONE 2012, 7, e50137. [Google Scholar] [CrossRef] [PubMed]
- Olson, K.M.; Cassell, B.G.; McAllister, A.J.; Washburn, S.P. Dystocia, stillbirth, gestation length, and birth weight in Holstein, Jersey, and reciprocal crosses from a planned experiment. J. Dairy Sci. 2009, 92, 6167–6175. [Google Scholar] [CrossRef] [PubMed]
- Sahana, G.; Hoglund, J.K.; Guldbrandtsen, B.; Lund, M.S. Loci associated with adult stature also affect calf birth survival in cattle. BMC Genet. 2015, 16, 47. [Google Scholar] [CrossRef]
- Lindholm-Perry, A.K.; Sexten, A.K.; Kuehn, L.A.; Smith, T.P.; King, D.A.; Shackelford, S.D.; Wheeler, T.L.; Ferrell, C.L.; Jenkins, T.G.; Snelling, W.M.; et al. Association, effects and validation of polymorphisms within the NCAPG—LCORL locus located on BTA6 with feed intake, gain, meat and carcass traits in beef cattle. BMC Genet. 2011, 12, 103. [Google Scholar] [CrossRef]
- Silva, C.R.; Neves, H.; Queiroz, S.; Sena, J.A.D.; Pimentel, E. Extent of linkage disequilibrium in Brazilian Gyr dairy cattle based on genotypes of AI sires for dense SNP markers. In Proceedings of the 9th World Congress on Genetics Applied to Livestock Production, Leipzig, Germany, 1–6 August 2010; pp. 1–29. [Google Scholar]
| Chromosome (BTA) | Markers (SNPs) |
|---|---|
| 1 | 46,495 |
| 2 | 40,056 |
| 3 | 35,579 |
| 4 | 34,980 |
| 5 | 34,842 |
| 6 | 35,519 |
| 7 | 33,168 |
| 8 | 33,529 |
| 9 | 31,060 |
| 10 | 30,449 |
| 11 | 32,015 |
| 12 | 26,127 |
| 13 | 23,594 |
| 14 | 24,780 |
| 15 | 24,755 |
| 16 | 24,178 |
| 17 | 22,266 |
| 18 | 19,386 |
| 19 | 18,908 |
| 20 | 21,490 |
| 21 | 21,175 |
| 22 | 18,034 |
| 23 | 15,215 |
| 24 | 18,620 |
| 25 | 12,931 |
| 26 | 15,242 |
| 27 | 13,152 |
| 28 | 13,038 |
| 29 | 14,710 |
| Total | 735,293 |
| Chromosome (BTA) | Number of Shared Regions (ROH) |
|---|---|
| 1 | 43 |
| 2 | 33 |
| 3 | 23 |
| 4 | 19 |
| 5 | 53 |
| 6 | 51 |
| 7 | 28 |
| 8 | 25 |
| 9 | 20 |
| 10 | 34 |
| 11 | 28 |
| 12 | 32 |
| 13 | 28 |
| 14 | 22 |
| 15 | 17 |
| 16 | 19 |
| 17 | 17 |
| 18 | 23 |
| 19 | 20 |
| 20 | 19 |
| 21 | 21 |
| 22 | 20 |
| 23 | 7 |
| 24 | 19 |
| 25 | 15 |
| 26 | 17 |
| 27 | 11 |
| 28 | 5 |
| 29 | 18 |
| Total | 687 |
| % of Animals | Cromosome (BTA) | Initial Position (bp) | Final Position (bp) | Genes |
|---|---|---|---|---|
| 51% | 6 | 34,496,622 | 34,671,490 | -None- |
| 54% | 6 | 36,717,524 | 36,738,110 | -None- |
| 59% | 6 | 35,211,888 | 35,992,229 | CCSER1 |
| 62% | 6 | 37,380,548 | 38,188,436 | FAM13A, HERC3, NAP1L5, PIGY, PYURF, HERC5, HERC6, PPM1K, ABCG2, PKD2, SPP1 |
| 65% | 6 | 40,186,267 | 41,452,236 | SLIT2 |
| 86% | 6 | 38,698,886 | 39,581,936 | DCAF16, NCAPG, LCORL |
| Average | Standard Deviation | Minimun | Maximum | |
|---|---|---|---|---|
| Fx | −0.032 | 0.075 | −0.181 | 0.135 |
| FROH | 0.046 | 0.035 | 0.012 | 0.180 |
| FSNP | 0.713 | 0.020 | 0.673 | 0.761 |
| r | Fx | FROH | FSNP |
|---|---|---|---|
| Fx | 1 | 0.752 | 0.999 |
| FROH | 0.752 | 1 | 0.752 |
| FSNP | 0.999 | 0.752 | 1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zanella, R.; Lago, L.V.; Da Silva, A.N.; Pértille, F.; De Carvalho, N.S.; Do Carmo Panetto, J.C.; Zanella, G.C.; Facioli, F.L.; Da Silva, M.V.G.B. Genetic Characterization of Indubrasil Cattle Breed Population. Vet. Sci. 2018, 5, 98. https://doi.org/10.3390/vetsci5040098
Zanella R, Lago LV, Da Silva AN, Pértille F, De Carvalho NS, Do Carmo Panetto JC, Zanella GC, Facioli FL, Da Silva MVGB. Genetic Characterization of Indubrasil Cattle Breed Population. Veterinary Sciences. 2018; 5(4):98. https://doi.org/10.3390/vetsci5040098
Chicago/Turabian StyleZanella, Ricardo, Luísa V. Lago, Arthur N. Da Silva, Fábio Pértille, Nathã S. De Carvalho, João Cláudio Do Carmo Panetto, Giovana C. Zanella, Fernanda L. Facioli, and Marcos Vinicius G.B. Da Silva. 2018. "Genetic Characterization of Indubrasil Cattle Breed Population" Veterinary Sciences 5, no. 4: 98. https://doi.org/10.3390/vetsci5040098
APA StyleZanella, R., Lago, L. V., Da Silva, A. N., Pértille, F., De Carvalho, N. S., Do Carmo Panetto, J. C., Zanella, G. C., Facioli, F. L., & Da Silva, M. V. G. B. (2018). Genetic Characterization of Indubrasil Cattle Breed Population. Veterinary Sciences, 5(4), 98. https://doi.org/10.3390/vetsci5040098





