Efficient Transduction and Expansion of Ovine Macrophages for Gene Therapy Implementations
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Ovine Monocytes
2.2. Macrophage Phagocytosis Assay
2.3. Lentiviral Vector Production and Titration
2.4. Macrophage Transduction
2.5. Vector Copy Number Analysis
2.6. Statistics
3. Results
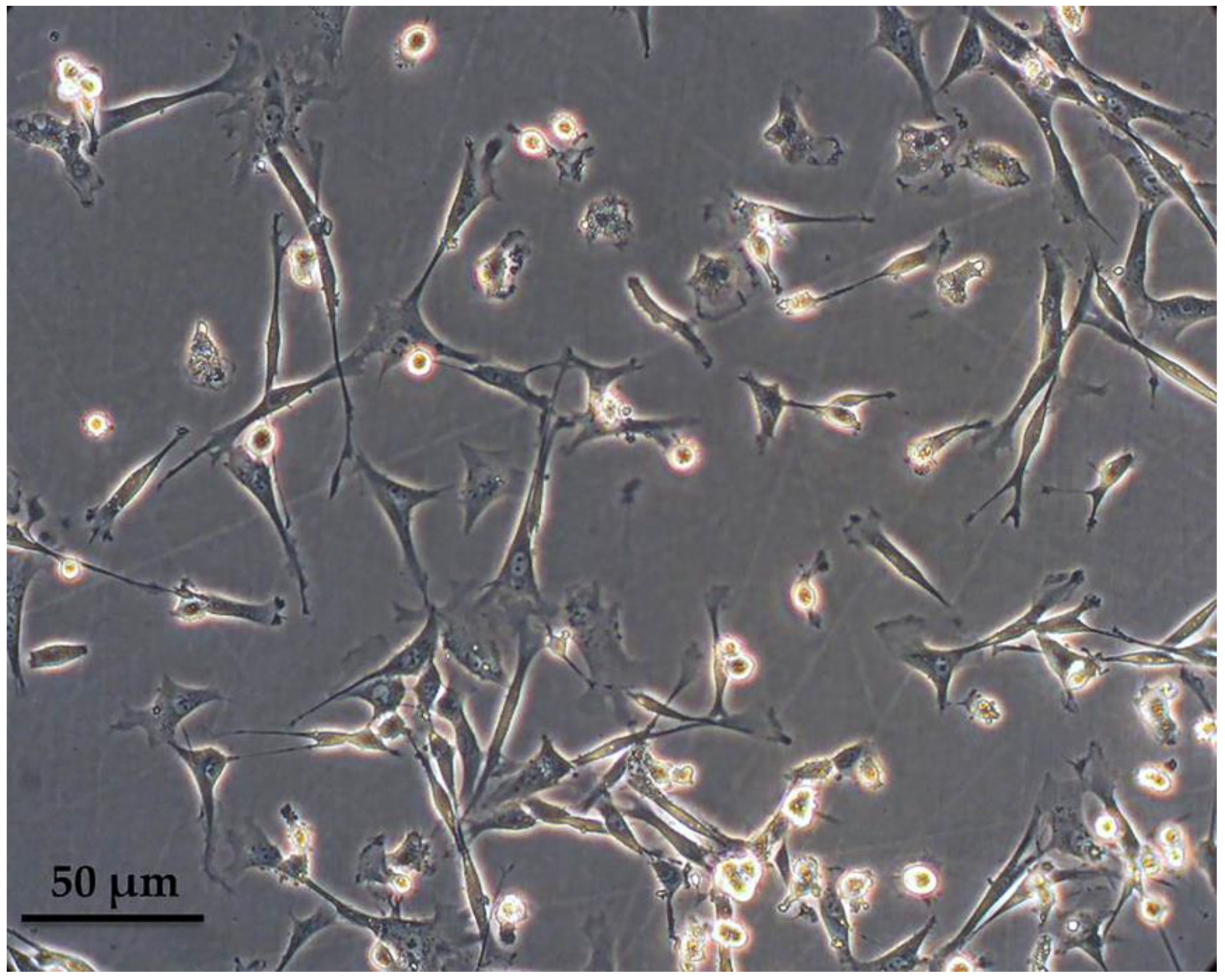
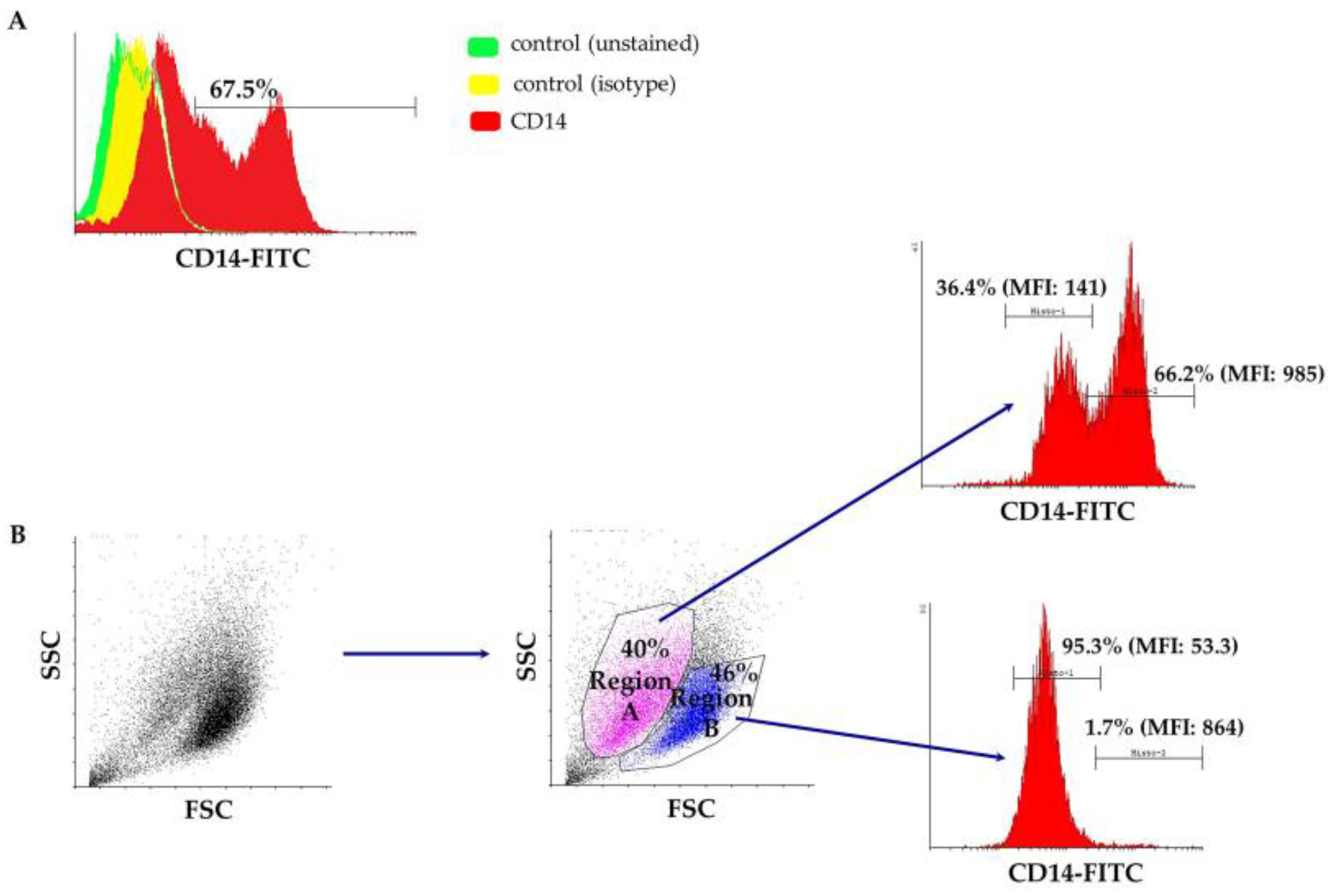
3.1. Characterization of Peripheral Blood-Derived Ovine Macrophages
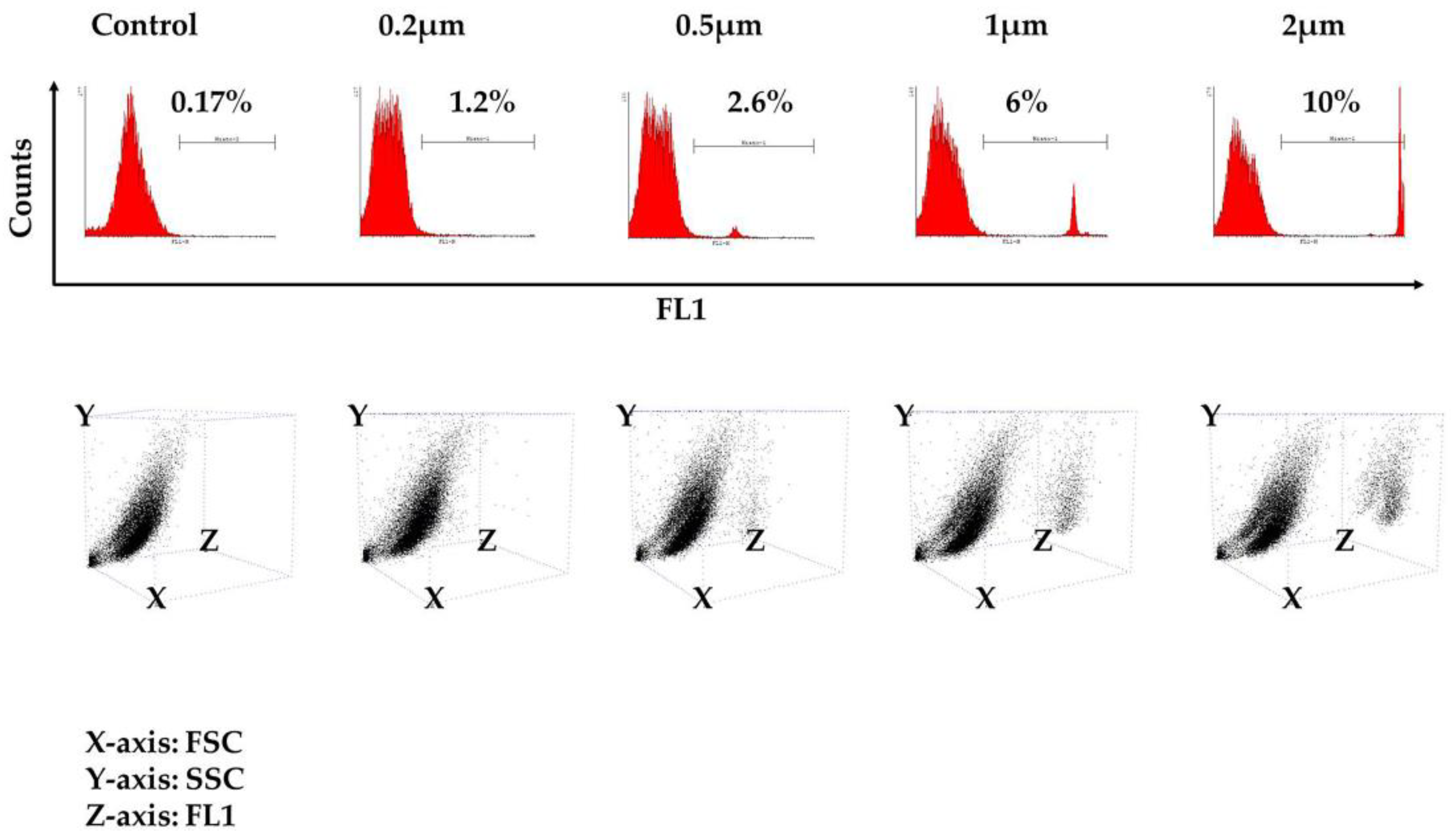
3.2. Phagocytosis Assay
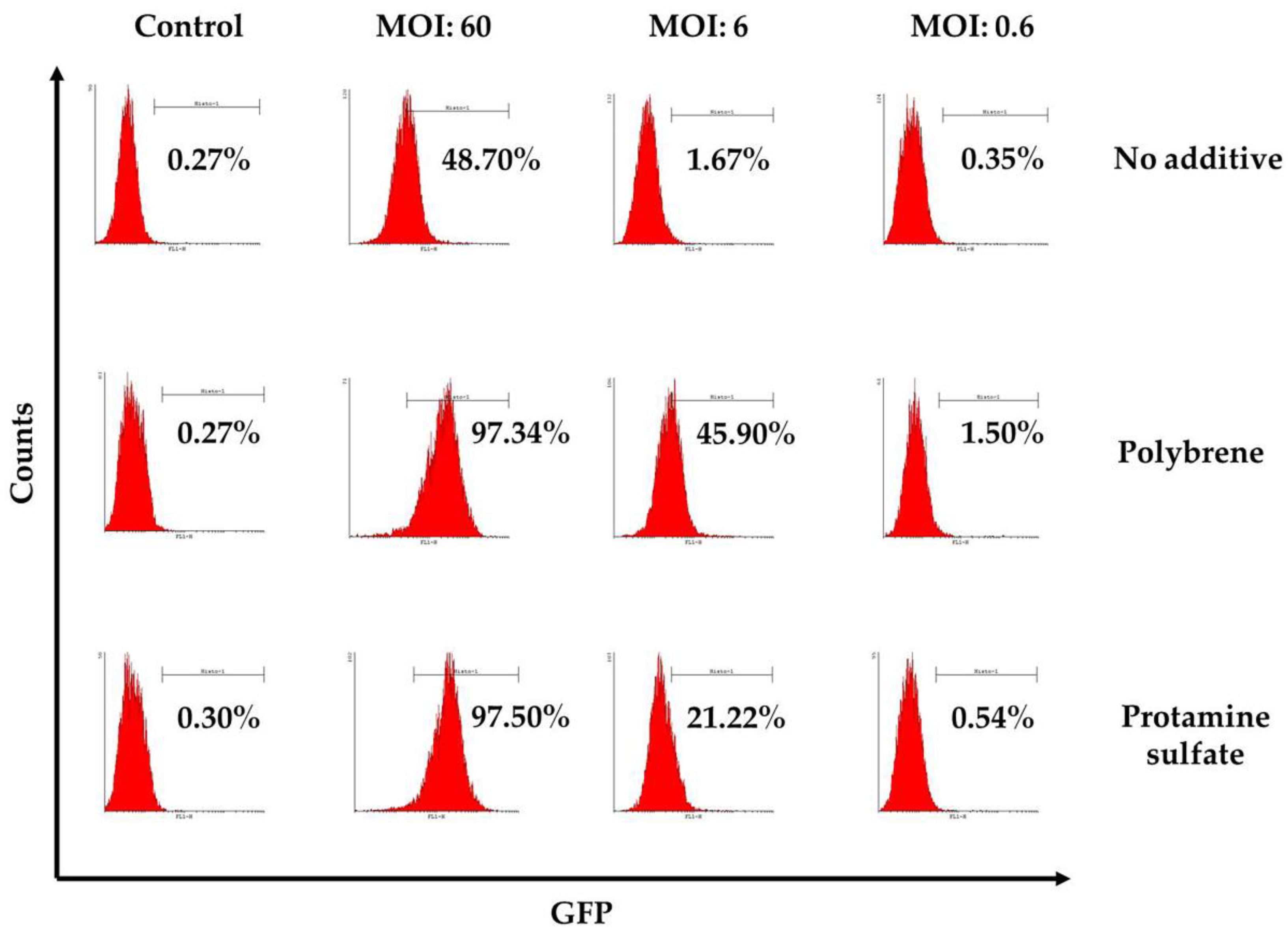
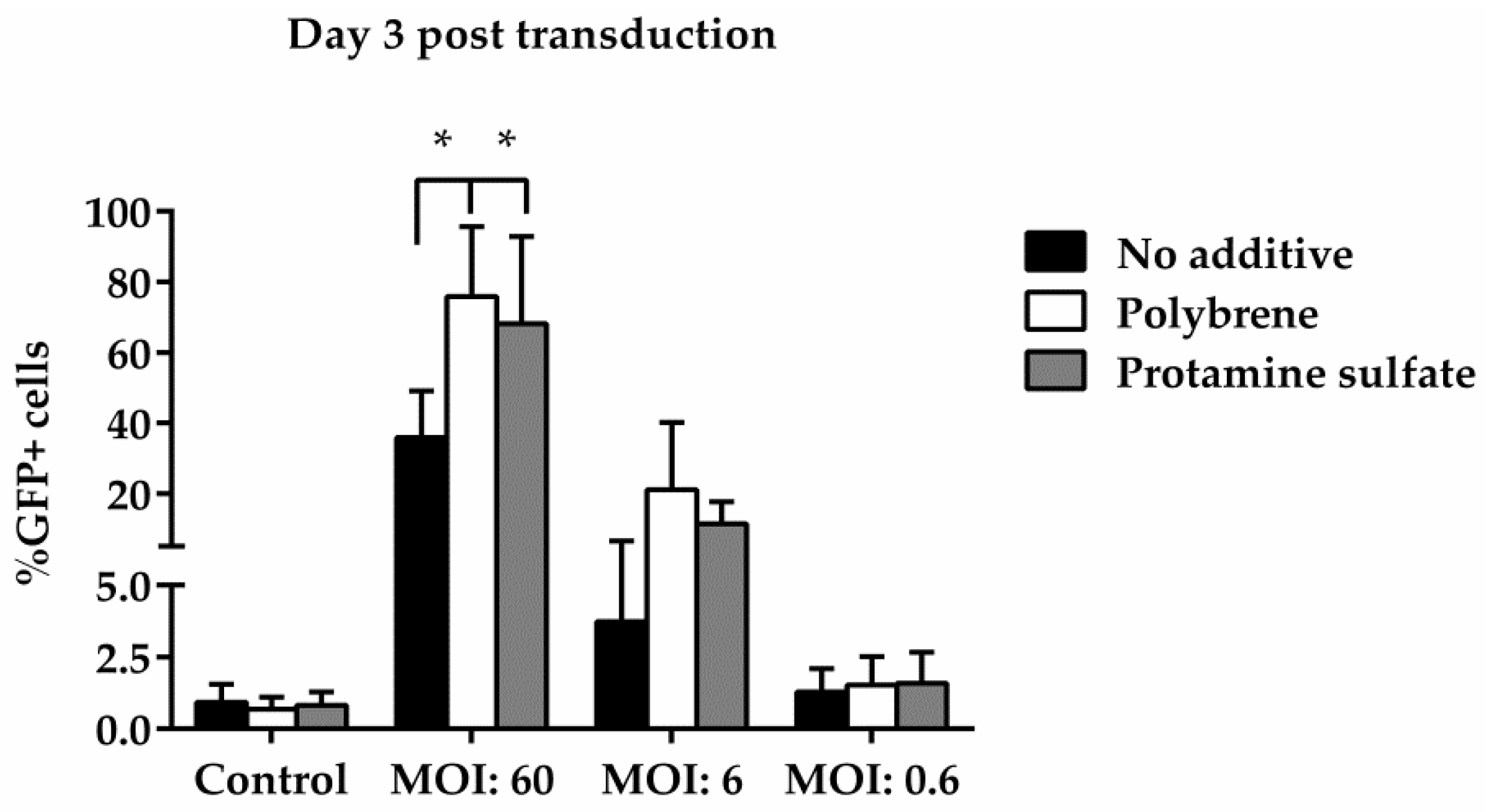
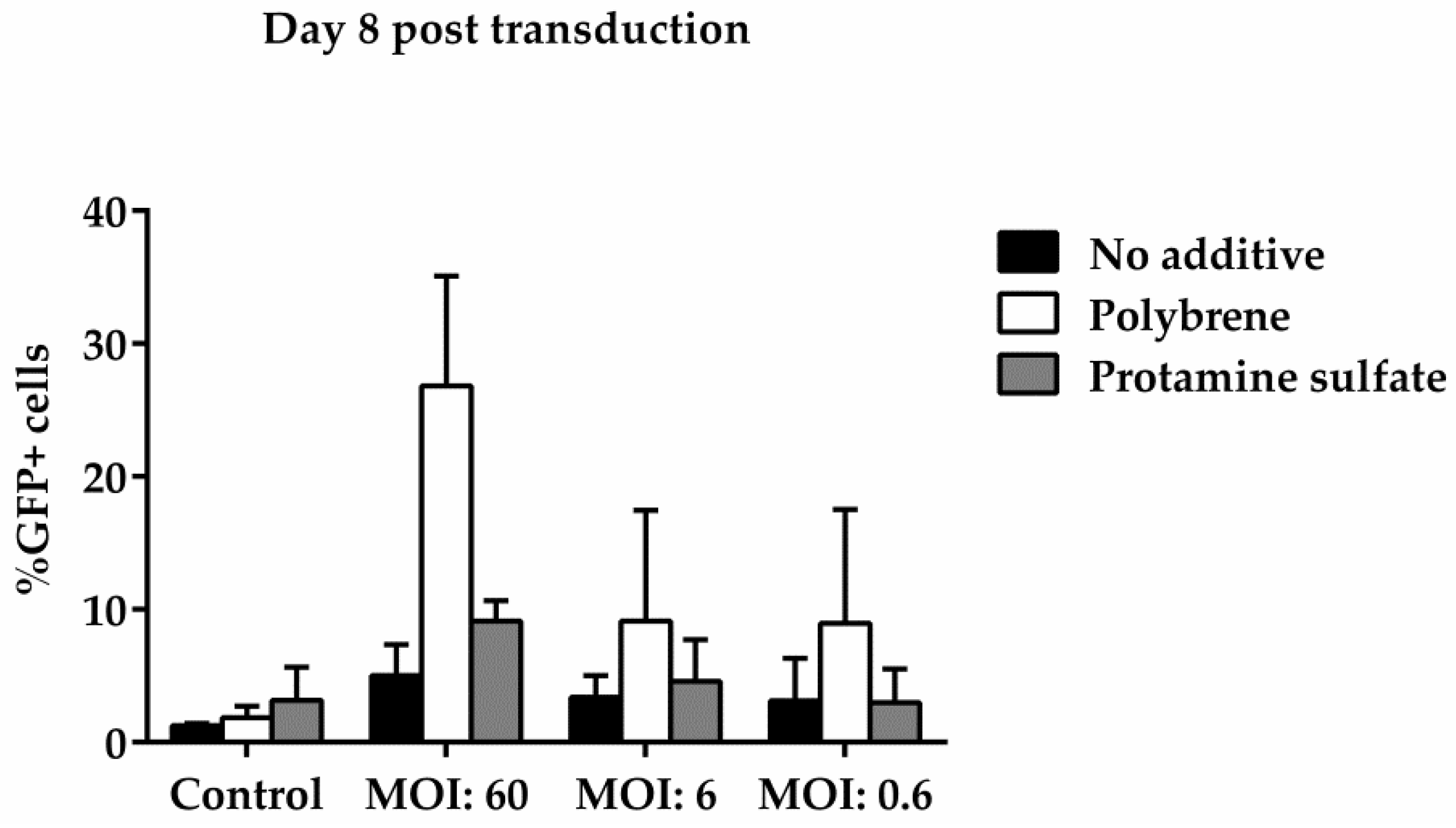
3.3. Ovine Macrophages’ Resistance to Transduction May Be Averted with Transduction Enhancers
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Von Bargen, K.; Gorvel, J.P.; Salcedo, S.P. Internal affairs: Investigating the Brucella intracellular lifestyle. FEMS Microbiol. Rev. 2012, 36, 533–562. [Google Scholar] [CrossRef] [PubMed]
- Blasco, J.M. A review of the use of B. melitensis Rev 1 vaccine in adult sheep and goats. Prev. Vet. Med. 1997, 31, 275–283. [Google Scholar] [CrossRef]
- Minas, A.; Minas, M.; Stournara, A.; Tselepidis, S. The "effects" of Rev-1 vaccination of sheep and goats on human brucellosis in Greece. Prev. Vet. Med. 2004, 64, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Kay, M.A. State-of-the-art gene-based therapies: The road ahead. Nat. Rev. Genet. 2011, 12, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Cavazzana-Calvo, M.; Hacein-Bey, S.; de Saint Basile, G.; Gross, F.; Yvon, E.; Nusbaum, P.; Selz, F.; Hue, C.; Certain, S.; Casanova, J.L.; et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science 2000, 288, 669–672. [Google Scholar] [CrossRef] [PubMed]
- Cartier, N.; Hacein-Bey-Abina, S.; Bartholomae, C.C.; Veres, G.; Schmidt, M.; Kutschera, I.; Vidaud, M.; Abel, U.; Dal-Cortivo, L.; Caccavelli, L.; et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 2009, 326, 818–823. [Google Scholar] [CrossRef] [PubMed]
- Cavazzana-Calvo, M.; Payen, E.; Negre, O.; Wang, G.; Hehir, K.; Fusil, F.; Down, J.; Denaro, M.; Brady, T.; Westerman, K.; et al. Transfusion independence and HMGA2 activation after gene therapy of human b-thalassaemia. Nature 2010, 467, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Aiuti, A.; Biasco, L.; Scaramuzza, S.; Ferrua, F.; Cicalese, M.P.; Baricordi, C.; Dionisio, F.; Calabria, A.; Giannelli, S.; Castiello, M.C.; et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science 2013, 341. [Google Scholar] [CrossRef] [PubMed]
- Biffi, A.; Montini, E.; Lorioli, L.; Cesani, M.; Fumagalli, F.; Plati, T.; Baldoli, C.; Martino, S.; Calabria, A.; Canale, S.; et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 2013, 341. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.A.; Walters, M.C.; Kwiatkowski, J.; Rasko, J.E.J.; Ribeil, J.A.; Hongeng, S.; Magrin, E.; Schiller, G.J.; Payen, E.; Semeraro, M.; et al. Gene therapy in patients with transfusion-dependent β-thalassemia. N. Engl. J. Med. 2018, 378, 1479–1493. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.R.; Yang, H.C.; Kuo, Y.T.; Liu, C.J.; Yang, T.Y.; Sung, K.C.; Lin, Y.Y.; Wang, H.Y.; Wang, C.C.; Shen, Y.C.; et al. The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo. Mol. Ther. Nucleic Acids 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Kaminski, R.; Yang, F.; Zhang, Y.; Cosentino, L.; Li, F.; Luo, B.; Alvarez-Carbonell, D.; Garcia-Mesa, Y.; Karn, J.; et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl. Acad. Sci. USA 2014, 111, 11461–11466. [Google Scholar] [CrossRef] [PubMed]
- Jarrosson-Wuilleme, L.; Goujon, C.; Bernaud, J.; Rigal, D.; Darlix, J.L.; Cimarelli, A. Transduction of nondividing human macrophages with gammaretrovirus-derived vectors. J. Virol. 2006, 80, 1152–1159. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Planelles, V.; Sui, Z.; Gartner, S.; Maggirwar, S.B.; Dewhurst, S.; Ye, L.; Nerurkar, V.R.; Yanagihara, R.; Lu, Y. HIV-1-based defective lentiviral vectors efficiently transduce human monocytes-derived macrophages and suppress replication of wild-type HIV-1. J. Gene Med. 2006, 8, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Yang, S.; Wu, C.; Ye, L.; Lu, Y. Effective transduction of primary mouse blood- and bone marrow-derived monocytes/macrophages by HIV-based defective lentiviral vectors. J. Virol. Methods 2006, 134, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Linterman, K.S.; Palmer, D.N.; Kay, G.W.; Barry, L.A.; Mitchell, N.L.; McFarlane, R.G.; Black, M.A.; Sands, M.S.; Hughes, S.M. Lentiviral-mediated gene transfer to the sheep brain: Implications for gene therapy in Batten disease. Hum. Gene Ther. 2011, 22, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Gray-Edwards, H.L.; Randle, A.N.; Maitland, S.A.; Benatti, H.R.; Hubbard, S.M.; Canning, P.F.; Vogel, M.B.; Brunson, B.L.; Hwang, M.; Ellis, L.E.; et al. Adeno-associated virus gene therapy in a sheep model of Tay-Sachs disease. Hum. Gene Ther. 2018, 29, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wong, E.; Miller, D.; Smith, G.; Anson, D.; Parsons, D. Lentiviral airway gene transfer in lungs of mice and sheep: Successes and challenges. J. Gene Med. 2010, 12, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Banin, E.; Gootwine, E.; Obolensky, A.; Ezra-Elia, R.; Ejzenberg, A.; Zelinger, L.; Honig, H.; Rosov, A.; Yamin, E.; Sharon, D.; et al. Gene augmentation therapy restores retinal function and visual behavior in a sheep model of CNGA3 achromatopsia. Mol. Ther. 2015, 23, 1423–1433. [Google Scholar] [CrossRef] [PubMed]
- Porada, C.D.; Park, P.; Almeida-Porada, G.; Zanjani, E.D. The sheep model of in utero gene therapy. Fetal Diagn. Ther. 2004, 19, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Crespo, H.; Bertolotti, L.; Juganaru, M.; Glaria, I.; de Andrés, D.; Amorena, B.; Rosati, S.; Reina, R. Small ruminant macrophage polarization may play a pivotal role on lentiviral infection. Vet. Res. 2013, 44. [Google Scholar] [CrossRef] [PubMed]
- Jarczak, J.; Kaba, J.; Reczynska, D.; Bagnicka, E. Impaired expression of cytokines as a result of viral infections with an emphasis on small ruminant lentivirus infection in goats. Viruses 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- De Pablo-Maiso, L.; Glaria, I.; Crespo, H.; Nistal-Villán, E.; Andrésdóttir, V.; de Andrés, D.; Amorena, B.; Reina, R. Characterization of ovine A3Z1 restriction properties against small ruminant lentiviruses (SRLVs). Viruses 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.T.; Griffin, F.T. A comparison of ovine monocyte-derived macrophage function following infection with Mycobacterium avium ssp. avium and Mycobacterium avium ssp. paratuberculosis. Immunol. Cell Biol. 2006, 84, 349–356. [Google Scholar] [PubMed]
- Papanikolaou, E.; Georgomanoli, M.; Stamateris, E.; Panetsos, F.; Karagiorga, M.; Tsaftaridis, P.; Graphakos, S.; Anagnou, N.P. The new self-inactivating lentiviral vector for thalassemia gene therapy combining two HPFH activating elements corrects human thalassemic hematopoietic stem cells. Hum. Gene Ther. 2012, 23, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Papanikolaou, E.; Kontostathi, G.; Drakopoulou, E.; Georgomanoli, M.; Stamateris, E.; Vougas, K.; Vlahou, A.; Maloy, A.; Ware, M.; Anagnou, N.P. Characterization and comparative performance of lentiviral vector preparations concentrated by either one-step ultrafiltration or ultracentrifugation. Virus Res. 2013, 175, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.Y.; Westaway, D.; Smit, A.F.A.; Wang, K.; Seto, J.; Chen, L.; Acharya, C.; Ankener, M.; Baskin, D.; Cooper, C.; et al. Complete genomic sequence and analysis of the prion protein gene region from three mammalian species. Genome Res. 1998, 8, 1022–1037. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Crespo, D.; Juste, R.A.; Hurtado, A. Selection of ovine housekeeping genes for normalisation by real-time RT-PCR; analysis of PrP gene expression and genetic susceptibility to scrapie. BMC Vet. Res. 2005, 1, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Donev, D.M. Brucellosis as priority public health challenge in South Eastern European countries. Croat. Med. J. 2010, 51, 283–284. [Google Scholar] [CrossRef] [PubMed]
- Avila-Calderón, E.; Lopez-Merino, A.; Sriranganathan, N.; Boyle, S.; Contreras-Rodríguez, A. A history of the development of Brucella vaccines. Biomed. Res. Int. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Pappas, G.; Papadimitriou, P.; Akritidis, N.; Christou, L.; Tsianos, E.V. The new global map of human brucellosis. Lancet Infect. Dis. 2006, 6, 91–99. [Google Scholar] [CrossRef]
- Gupta, V.K.; McConnell, I.; Dalziel, R.G.; Hopkins, J. Identification of the sheep homologue of the monocyte cell surface molecule - CD14. Vet. Immunol. Immunopathol. 1996, 51, 89–99. [Google Scholar] [CrossRef]
- Berthon, P.; Hopkins, J. Ruminant cluster CD14. Vet. Immunol. Immunopathol. 1996, 52, 245–248. [Google Scholar] [CrossRef]
- May, K.F.; Jinushi, M.; Dranoff, G. Immunosurveillance: Innate and adaptive antitumor immunity. In Cancer Immunotherapy: Immune Suppression and Tumor Growth, 2nd ed.; Prendergast, G.C., Jaffee, E.M., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 101–113. ISBN 9780123942968. [Google Scholar]
- Gordon, S.; Martinez, F.O. Alternative activation of macrophages: Mechanism and functions. Immunity 2010, 32, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Nagl, M.; Kacani, L.; Mullauer, B.; Lemberger, E.M.; Stoiber, H.; Sprinzl, G.M.; Schennach, H.; Dierich, M.P. Phagocytosis and killing of bacteria by professional phagocytes and dendritic cells. Clin. Vaccine Immunol. 2002, 9, 1165–1168. [Google Scholar] [CrossRef]
- Paul, D.; Achouri, S.; Yoon, Y.Z.; Herre, J.; Bryant, C.E.; Cicuta, P. Phagocytosis dynamics depends on target shape. Biophys. J. 2013, 105, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Edwards, J.P.; Mosser, D.M. The expression of exogenous genes in macrophages: Obstacles and opportunities. Methods Mol. Biol. 2009, 531, 123–143. [Google Scholar] [PubMed]
- Bobadilla, S.; Sunseri, N.; Landau, N.R. Efficient transduction of myeloid cells by an HIV-1-derived lentiviral vector that packages the Vpx accessory protein. Gene Ther. 2013, 20, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Cronin, J.; Zhang, X.Y.; Reiser, J. Altering the tropism of lentiviral vectors through pseudotyping. Curr. Gene Ther. 2005, 5, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.E.; Rosinski, M.; Morgan, J.R.; Yarmush, M.L. Charged polymers modulate retrovirus transduction via membrane charge neutralization and virus aggregation. Biophys. J. 2004, 86, 1234–1242. [Google Scholar] [CrossRef]
- Denning, W.; Das, S.; Guo, S.; Xu, J.; Kappes, J.C.; Hel, Z. Optimization of the transductional efficiency of lentiviral vectors: Effect of sera and polycations. Mol. Biotechnol. 2013, 53, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.E.; Morgan, J.R.; Yarmush, M.L. Polybrene increases retrovirus gene transfer efficiency by enhancing receptor-independent virus adsorption on target cell membranes. Biophys. Chem. 2002, 97, 159–172. [Google Scholar] [CrossRef]
| 1st Experiment | 2nd Experiment | 3rd Experiment | |
|---|---|---|---|
| Control | 0.1 | 0.17 | 0.13 |
| 0.2 μm | 1.15 | 1.21 | 1 |
| 0.5 μm | 1.7 | 2.6 | 2.2 |
| 1 μm | 3.6 | 6 | 5.75 |
| 2 μm | 8.9 | 8.6 | 10 |
| 1st Experiment | 2nd Experiment | 3rd Experiment | 4th Experiment | 5th Experiment | ||
|---|---|---|---|---|---|---|
| No additive | control | 0.27 | 0.09 | 1.4 | 0.9 | 1.54 |
| MOI *:60 | 48.7 | 48.1 | 43.7 | 34.2 | 18.3 | |
| MOI:6 | 1.67 | 1.66 | 8.8 | 2.7 | 2.37 | |
| MOI:0.6 | 0.35 | 0.16 | 1.7 | 1.9 | 1.47 | |
| Polybrene | control | 0.27 | 0.09 | 0.86 | 0.7 | 0.95 |
| MOI:60 | 97.34 | 97.46 | 78 | 75.6 | 51.4 | |
| MOI:6 | 45.9 | 42.7 | 14.55 | 14.7 | 2.75 | |
| MOI:0.6 | 1.5 | 0.71 | 1.34 | 1.25 | 0.91 | |
| Protamine sulfate | control | 0.3 | 0.15 | 1.04 | 1.05 | 1.14 |
| MOI:60 | 97.5 | 94.7 | 70 | 64.7 | 40.3 | |
| MOI:6 | 21.2 | 16.8 | 6.8 | 10.8 | 7.55 | |
| MOI:0.6 | 0.54 | 0.41 | 1.06 | 2.8 | 1.76 |
| Days Post Transduction | MOI: 60 | |
|---|---|---|
| No additive | 3 | 10.46 |
| 8 | 0.13 | |
| Polybrene | 3 | 11.33 |
| 8 | 0.29 | |
| Protamine sulfate | 3 | 10.94 |
| 8 | 0.21 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karponi, G.; Kritas, S.; Petridou, E.; Papanikolaou, E. Efficient Transduction and Expansion of Ovine Macrophages for Gene Therapy Implementations. Vet. Sci. 2018, 5, 57. https://doi.org/10.3390/vetsci5020057
Karponi G, Kritas S, Petridou E, Papanikolaou E. Efficient Transduction and Expansion of Ovine Macrophages for Gene Therapy Implementations. Veterinary Sciences. 2018; 5(2):57. https://doi.org/10.3390/vetsci5020057
Chicago/Turabian StyleKarponi, Garyfalia, Spyridon Kritas, Evanthia Petridou, and Eleni Papanikolaou. 2018. "Efficient Transduction and Expansion of Ovine Macrophages for Gene Therapy Implementations" Veterinary Sciences 5, no. 2: 57. https://doi.org/10.3390/vetsci5020057
APA StyleKarponi, G., Kritas, S., Petridou, E., & Papanikolaou, E. (2018). Efficient Transduction and Expansion of Ovine Macrophages for Gene Therapy Implementations. Veterinary Sciences, 5(2), 57. https://doi.org/10.3390/vetsci5020057






