Fluorescent Protein Expressing Rickettsia buchneri and Rickettsia peacockii for Tracking Symbiont-Tick Cell Interactions
Abstract
1. Introduction
2. Materials and Methods
2.1. Tick Cell Lines
2.2. Rickettsiae
2.3. Plasmid Constructs
2.4. Plasmid Transformation of R. buchneri and R. peacockii
2.5. Transposon Mutagenesis (HIMAR1 A7) of R. peacockii
2.6. Cloning and Sequencing of Transposon Integration Sites
2.7. Microscopy
2.8. Preparation of Genomic DNA for PFGE and Southern Blot Analysis
3. Results
3.1. Plasmid Transformation of R. buchneri and R. peacockii
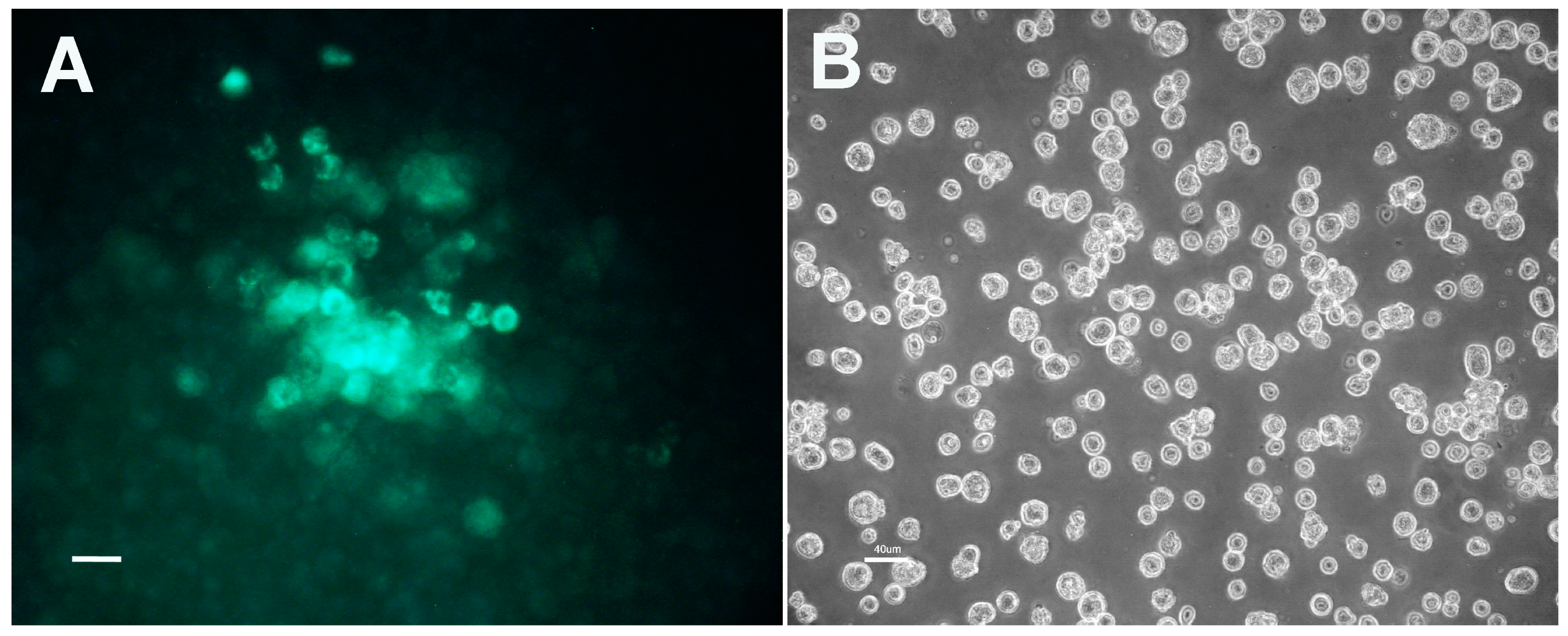
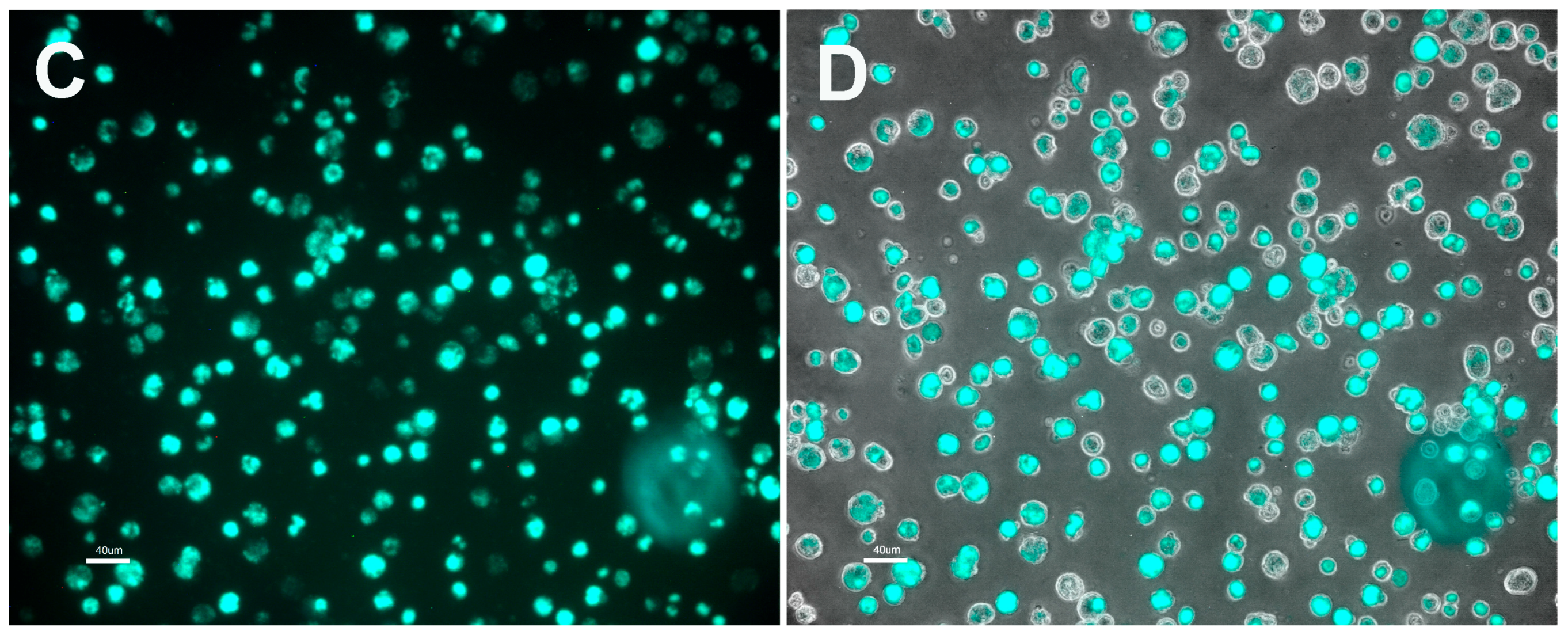
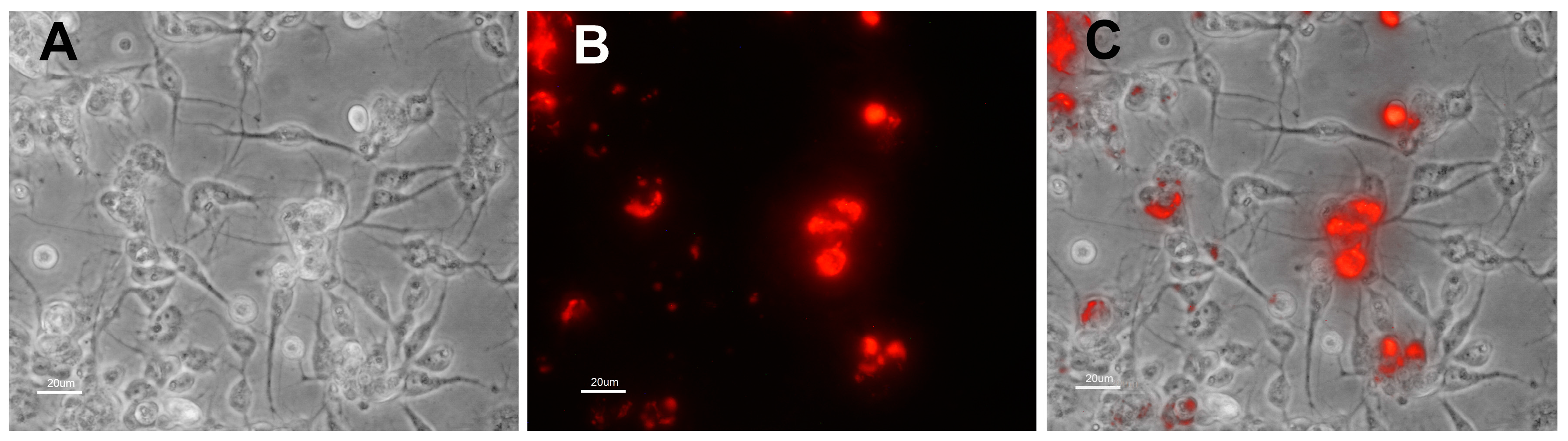
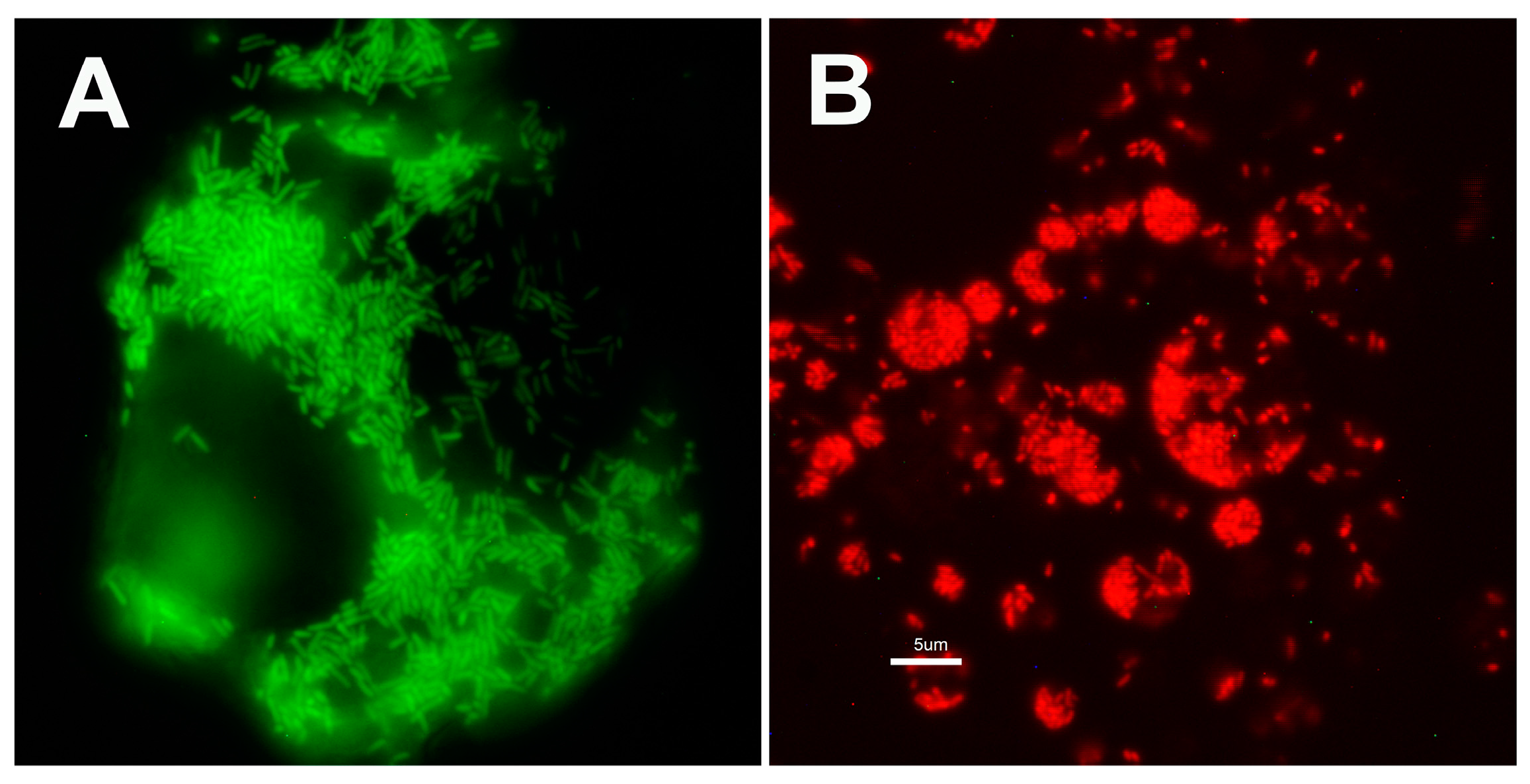
3.2. Characteristics of Transformed R. buchneri and R. peacockii in Tick Cell Culture
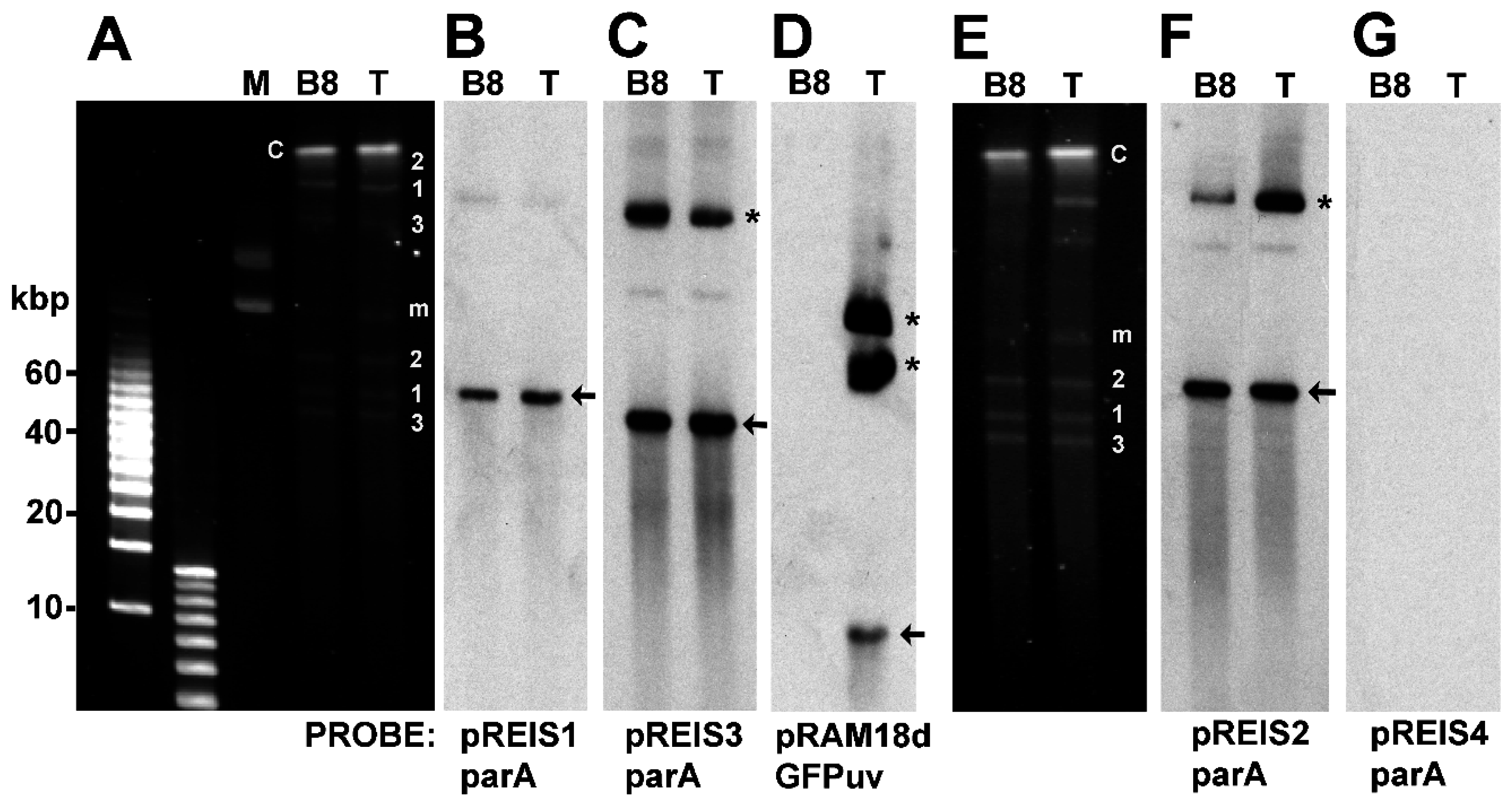
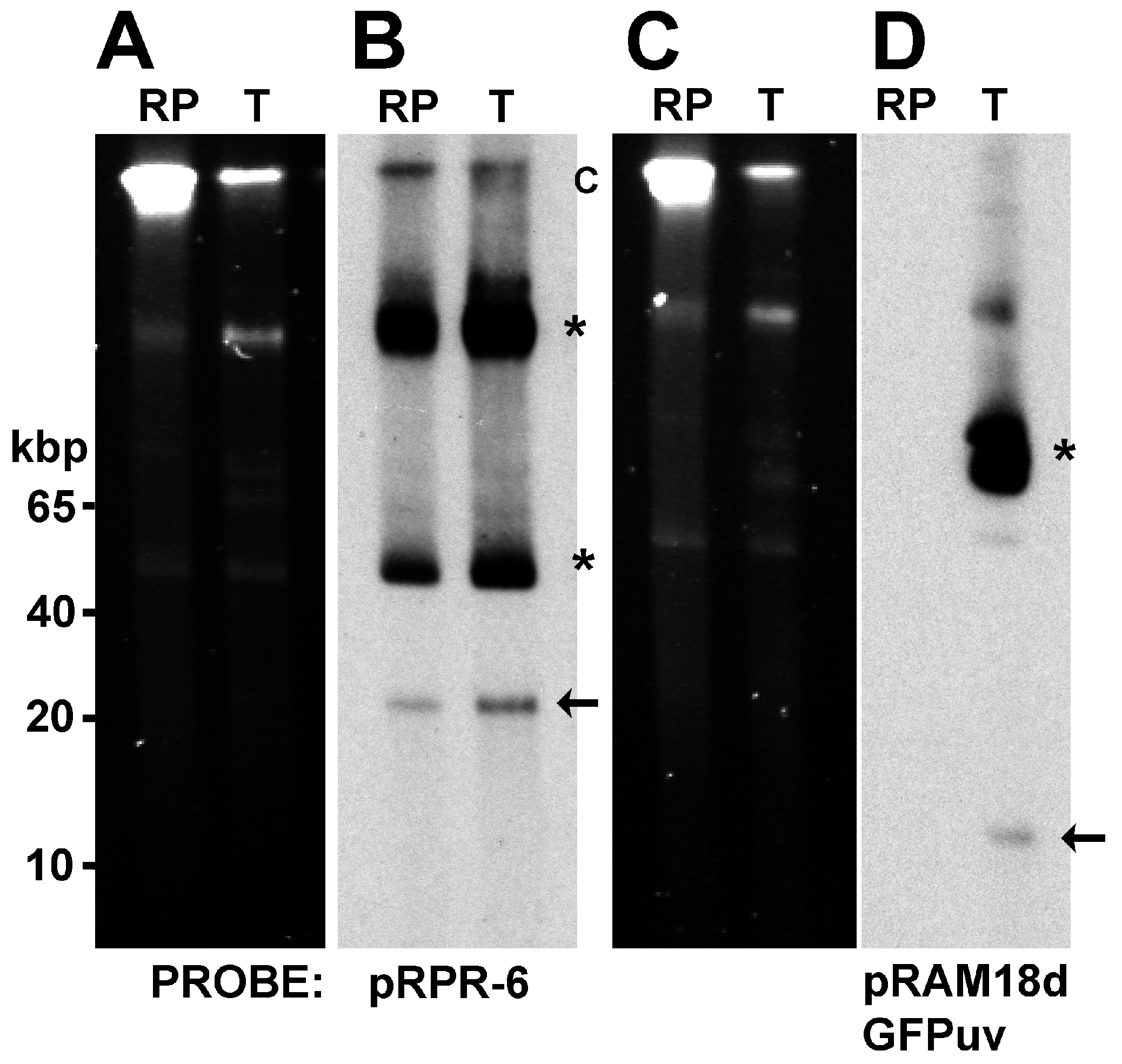
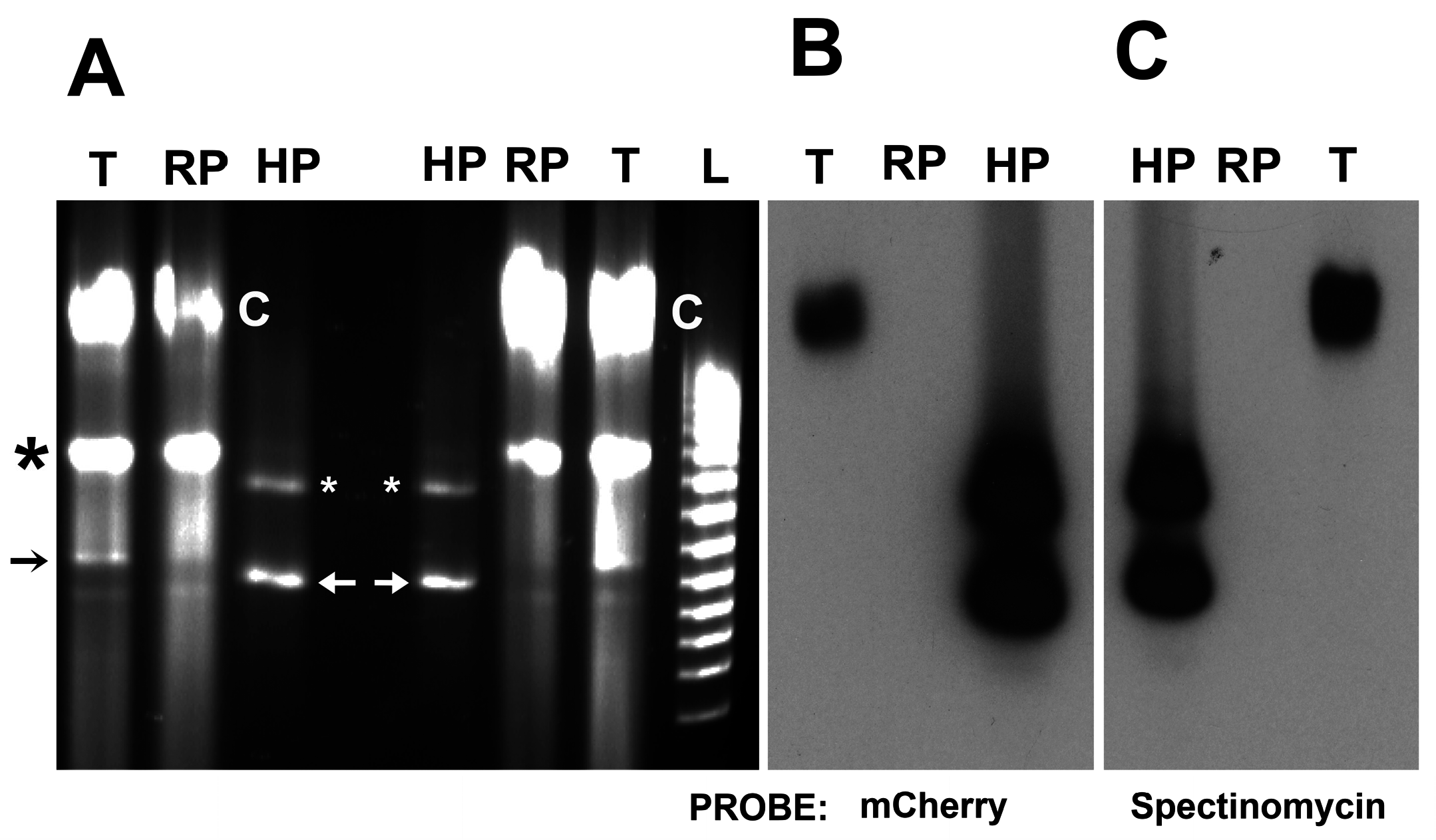
3.3. Identification of Shuttle Vector pRAM18dRGA and Native Plasmids pREIS1-3 in R. buchneri Transformant and Clone B8
3.4. Identification of pRPR and pRAM18dSGA in R. peacockii and Transformants
3.5. Transposon Mutagenesis (HIMAR1 A7) of R. peacockii
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Niebylski, M.L.; Schrumpf, M.E.; Burgdorfer, W.; Fischer, E.R.; Gage, K.L.; Schwan, T. Rickettsia peacockii sp. nov., a new species infecting wood ticks, Dermacentor andersoni, in western Montana. Int. J. Syst. Bacteriol. 1997, 47, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, J.J.; Joardar, V.; Williams, K.P.; Driscoll, T.; Hostetler, J.B.; Nordberg, E.; Shukla, M.; Walenz, B.; Hill, C.A.; Nene, V.M.; et al. A rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J. Bacteriol. 2012, 194, 376–394. [Google Scholar] [CrossRef] [PubMed]
- Kurtti, T.J.; Felsheim, R.F.; Burkhardt, N.Y.; Oliver, J.D.; Heu, C.C.; Munderloh, U.G. Rickettsia buchneri sp. nov., a rickettsial endosymbiont of the blacklegged tick Ixodes scapularis. Int. J. Syst. Evol. Microbiol. 2015, 65, 965–970. [Google Scholar] [CrossRef] [PubMed]
- Hunter, D.J.; Torkelson, J.L.; Bodnar, J.; Mortazavi, B.; Laurent, T.; Deason, J.; Thephavongsa, K.; Zhong, J. The Rickettsia endosymbiont of Ixodes pacificus contains all the genes of de novo folate biosynthesis. PLoS ONE 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Apperson, C.S.; Engber, B.; Nicholson, W.L.; Mead, D.G.; Engel, J.; Yabsley, M.J.; Dail, K.; Johnson, J.; Watson, D.W. Tick-borne disease in North Carolina: Is “Rickettsia amblyommii” a possible cause of rickettsiosis reported as Rocky Mountain spotted fever? Vector Borne Zoonotic Dis. 2008, 8, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Dahlgren, F.S.; Paddock, C.D.; Springer, Y.P.; Eisen, R.J.; Behravesh, C.B. Expanding range of Amblyomma americanum and simultaneous changes in the epidemiology of spotted fever group rickettsiosis in the United States. Am. J. Trop. Med. Hyg. 2016, 94, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, J.J.; Driscoll, T.P.; Verhoeve, V.I.; Utsuki, T.; Husseneder, C.; Chouljenko, V.N.; Azad, A.F.; Macaluso, K.R. Genomic diversification in strains of Rickettsia felis isolated from different arthropods. Genome Biol. Evol. 2015, 7, 35–56. [Google Scholar] [CrossRef] [PubMed]
- Sahni, S.K.; Narra, H.P.; Sahni, A.; Walker, D.H. Recent molecular insights into rickettsial pathogenesis and immunity. Future Microbiol. 2013, 8, 1265–1288. [Google Scholar] [CrossRef] [PubMed]
- Simser, A.; Palmer, A.T.; Kurtti, T.J.; Munderloh, U.G. Isolation of a spotted fever group rickettsia, Rickettsia peacockii, in a Rocky Mountain wood tick, Dermacentor andersoni, cell line. Appl. Environ. Microbiol. 2001, 67, 546–552. [Google Scholar] [CrossRef] [PubMed]
- Simser, J.A.; Palmer, A.T.; Fingerle, V.; Wilske, B.; Kurtti, T.J.; Munderloh, U.G. Rickettsia monacensis sp. nov., a spotted fever group rickettsia from ticks (Ixodes ricinus) collected in a European city park. Appl. Environ. Microbiol. 2002, 68, 4559–4566. [Google Scholar] [CrossRef] [PubMed]
- Felsheim, R.F.; Kurtti, T.J.; Munderloh, U.G. Genome sequence of the endosymbiont Rickettsia peacockii and comparison with virulent Rickettsia rickettsii: Identification of virulence factors. PLoS ONE 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Munderloh, U.G.; Jauron, S.D.; Kurtti, T.J. The tick: A different kind of host for human pathogens. In Tick-Borne Diseases of Humans; Goodman, J.L., Dennis, D.T., Sonenshine, D.E., Eds.; ASM Press: Washington, DC, USA, 2005; pp. 37–64. [Google Scholar]
- Kurlovs, A.H.; Li, J.; Cheng, D.; Zhong, J. Ixodes pacificus ticks maintain embryogenesis and egg hatching after antibiotic treatment of Rickettsia endosymbiont. PLoS ONE 2009, 9. [Google Scholar] [CrossRef] [PubMed]
- Mattila, J.T.; Burkhardt, N.Y.; Hutcheson, J.H.; Munderloh, U.G.; Kurtti, T.J. Isolation of cell lines and a rickettsial endosymbiont from the soft tick Carios capensis (Acari: Argasidae: Ornithodorinae). J. Med. Entomol. 2007, 44, 1091–1101. [Google Scholar] [CrossRef]
- Burgdorfer, W.; Brinton, L.P. Mechanisms of transovarial infection of spotted fever rickettsiae in ticks. Ann. N. Y. Acad. Sci. 1975, 266, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Niebylski, M.L.; Peacock, M.G.; Schwan, T.G. Lethal effect of Rickettsia rickettsii on its tick vector (Dermacentor andersoni). Appl. Environ. Microbiol. 1999, 65, 773–778. [Google Scholar] [PubMed]
- Santos, A.S.; Bacellar, F.; Santos-Silva, M.; Formosinho, P.; Grácio, A.J.; Franca, S. Ultrastructural study of the infection process of Rickettsia conorii in the salivary glands of the vector tick Rhipicephalus sanguineus. Vector Borne Zoonotic Dis. 2002, 2, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Sahni, S.K.; Rydkina, E. Host-cell interactions with pathogenic Rickettsia species. Future Microbiol. 2009, 4, 323–339. [Google Scholar] [CrossRef] [PubMed]
- Ellison, D.W.; Clark, T.R.; Sturdevant, D.E.; Virtaneva, K.; Porcella, S.F.; Hackstadt, T. Genomic comparison of virulent Rickettsia rickettsii Sheila Smith and avirulent Rickettsia rickettsii Iowa. Infect. Immun. 2008, 76, 542–550. [Google Scholar] [CrossRef] [PubMed]
- Munderloh, U.G.; Felsheim, R.F.; Burkhardt, N.Y.; Herron, M.J.; Oliva Chávez, A.S.; Nelson, C.M.; Kurtti, T.J. The way forward: Improving genetic systems. In Intracellular Pathogens II. Rickettsiales, 1st ed.; Palmer, G.H., Azad, A.F., Eds.; ASM Press: Washington, DC, USA, 2012; pp. 416–432. [Google Scholar]
- Burkhardt, N.Y.; Baldridge, G.D.; Williamson, P.C.; Billingsley, P.M.; Heu, C.C.; Felsheim, R.F.; Kurtti, T.J.; Munderloh, U.G. Development of shuttle vectors for transformation of diverse Rickettsia species. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Felsheim, R.F.; Herron, M.J.; Nelson, C.M.; Burkhardt, N.Y.; Barbet, A.F.; Kurtti, T.J.; Munderloh, U.G. Transformation of Anaplasma phagocytophilum. BMC Biotechnol. 2006, 6. [Google Scholar] [CrossRef] [PubMed]
- Munderloh, U.G.; Liu, Y.; Wang, M.; Chen, C.; Kurtti, T.J. Establishment, maintenance and description of cell lines from the tick Ixodes scapularis. J. Parasitol. 1994, 80, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.D.; Burkhardt, N.Y.; Felsheim, R.F.; Kurtti, T.J.; Munderloh, U.G. Motility characteristics are altered for Rickettsia bellii transformed to overexpress a heterologous rickA gene. Appl. Environ. Microbiol. 2014, 80, 1170–1176. [Google Scholar] [CrossRef] [PubMed]
- Crameri, A.; Whitehorn, E.A.; Tate, E.; Stemmer, W.P.C. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 1996, 14, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Shcherbo, D.; Merzlyak, E.M.; Chepurnykh, T.V.; Fradkov, A.; Ermakova, G.V.; Solovieva, E.A.; Lukyanov, K.A.; Bogdanova, E.A.; Zaraisky, A.G.; Lukyanov, S.; et al. Bright far-red fluorescent protein for whole-body imaging. Nat. Methods 2007, 4, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Steinbach, P.A.; Tsien, R.Y. A guide to choosing fluorescent proteins. Nat. Methods 2005, 2, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Barbet, A.F.; Agnes, J.T.; Moreland, A.L.; Lundgren, A.M.; Alleman, A.R.; Noh, S.M.; Brayton, K.A.; Munderloh, U.G.; Palmer, G.H. Identification of functional promoters in the msp2 expression loci of Anaplasma marginale and Anaplasma phogocytophilum. Gene 2005, 353, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Lampe, D.J.; Akerley, B.J.; Rubin, E.J.; Mekalanos, J.J.; Robertson, H.M. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci. USA 1999, 96, 11428–11433. [Google Scholar] [CrossRef] [PubMed]
- Baldridge, G.D.; Burkhardt, N.; Felsheim, R.F.; Kurtti, T.J.; Munderloh, U.G. Transposon insertion reveals pRM, a plasmid of Rickettsia monacensis. Appl. Environ. Microbiol. 2007, 73, 4984–4995. [Google Scholar] [CrossRef] [PubMed]
- Baldridge, G.D.; Burkhardt, N.Y.; Labruna, M.B.; Pacheco, R.C.; Paddock, C.D.; Williamson, P.C.; Billingsley, P.M.; Felsheim, R.F.; Kurtti, T.J.; Munderloh, U.G. Wide dispersal and possible multiple origins of low-copy-number plasmids in Rickettsia species associated with blood feeding arthropods. Appl. Environ. Microbiol. 2010, 76, 1718–1731. [Google Scholar] [CrossRef] [PubMed]
- Baldridge, G.D.; Burkhardt, N.Y.; Felsheim, R.F.; Kurtti, T.J.; Munderloh, U.G. Plasmids of the pRM/pRF family occur in diverse Rickettsia species. Appl. Environ. Microbiol. 2008, 74, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Kurtti, T.J.; Simser, J.A.; Baldridge, G.D.; Palmer, A.T.; Munderloh, U.G. Factors influencing in vitro infectivity and growth of Rickettsia peacockii (Rickettsiales: Rickettsiaceae), an endosymbiont of the Rocky Mountain wood tick, Dermacentor andersoni (Acari, Ixodidae). J. Invertbr. Pathol. 2005, 90, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Driskell, L.O.; Yu, X.; Zhang, L.; Liu, Y.; Popov, V.L.; Walker, D.H.; Tucker, A.M.; Wood, D.O. Directed mutagenesis of the Rickettsia prowazekii pld gene encoding phospholipase D. Infect. Immun. 2009, 77, 3244–3248. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Nair, A.D.S.; Indukuri, V.V.; Gong, S.; Felsheim, R.F.; Jaworski, D.; Munderloh, U.G.; Ganta, R.R. Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for in vivo infection. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef]
- Noriea, N.F.; Clark, T.R.; Hackstadt, T. Targeted knockout of the Rickettsia rickettsii ompA surface antigen does not diminish virulence in a mammalian model system. mBio 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Driskell, L.O.; Tucker, A.M.; Woodard, A.; Wood, R.R.; Wood, D.O. Fluorescence activated cell sorting of Rickettsia prowazekii-infected host cells based on bacterial burden and early detection of fluorescent rickettsial transformants. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Perotti, A.A.; Clarke, H.K.; Turner, B.D.; Braig, H.R. Rickettsia as obligate and mycetomic bacteria. FASEB J. 2006, 20, E1646–E1656. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, M.; Turner, B. Characterization of Wolbachia-like bacteria isolated from the parthenogenetic stored-product pest psocid Liposcelis bostrychophila (Badonnel) (Psocoptera). J. Stored Prod. Res. 2004, 40, 207–225. [Google Scholar] [CrossRef]
| Construct | MW (bp) | Fluoro-Chrome | Antibiotic Selection | Symbiont |
|---|---|---|---|---|
| Shuttle Vector Plasmids | ||||
| pRAM18dRGA | 10,248 | GFPuv | Rifampin | R. buchneri |
| pRAM18dRGA[MCS] | 10,309 | GFPuv | Rifampin | R. buchneri |
| pRAM18dSGA[MCS] | 10,736 | GFPuv | Spectinomycin | R. peacockii |
| pRAM18dSGK(23)[MCS] | 11,525 | GFPuv | Spectinomycin | R. peacockii |
| pRAM18dSFA[MCS] | 10,829 | mKate | Spectinomycin | R. peacockii |
| Himar I Transposase-Transposon Plasmid | ||||
| pCis mCherry-SS HIMAR1 A7 | 8423 | mCherry Spec. | Spectinomycin | R. peacockii |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurtti, T.J.; Burkhardt, N.Y.; Heu, C.C.; Munderloh, U.G. Fluorescent Protein Expressing Rickettsia buchneri and Rickettsia peacockii for Tracking Symbiont-Tick Cell Interactions. Vet. Sci. 2016, 3, 34. https://doi.org/10.3390/vetsci3040034
Kurtti TJ, Burkhardt NY, Heu CC, Munderloh UG. Fluorescent Protein Expressing Rickettsia buchneri and Rickettsia peacockii for Tracking Symbiont-Tick Cell Interactions. Veterinary Sciences. 2016; 3(4):34. https://doi.org/10.3390/vetsci3040034
Chicago/Turabian StyleKurtti, Timothy J., Nicole Y. Burkhardt, Chan C. Heu, and Ulrike G. Munderloh. 2016. "Fluorescent Protein Expressing Rickettsia buchneri and Rickettsia peacockii for Tracking Symbiont-Tick Cell Interactions" Veterinary Sciences 3, no. 4: 34. https://doi.org/10.3390/vetsci3040034
APA StyleKurtti, T. J., Burkhardt, N. Y., Heu, C. C., & Munderloh, U. G. (2016). Fluorescent Protein Expressing Rickettsia buchneri and Rickettsia peacockii for Tracking Symbiont-Tick Cell Interactions. Veterinary Sciences, 3(4), 34. https://doi.org/10.3390/vetsci3040034





