Sheep Pox Susceptibility: Role of Genetic Variants, Gene Expression, and Immune-Oxidative Markers
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Study Design
2.2. Sampling
2.2.1. Sample Selection and Exposure
2.2.2. Blood Samples
2.2.3. Scab Sample Collections and Processing
2.3. Extraction of Genomic DNA and PCR Amplification
2.3.1. Molecular Detection of SPV
2.3.2. PCR Conditions
2.3.3. Gel Electrophoresis of PCR Products
2.4. Gene Sequencing and Phylogenetic Analyses
2.5. Total RNA Extraction, Reverse Transcription and Quantitative Real-Time PCR
2.6. DNA Sequencing of Real-Time PCR Products (RT-PCR) and Polymorphism Detection
2.7. Biochemical, Antioxidant and Immunological Parameters
2.8. Statistical Analysis
3. Results
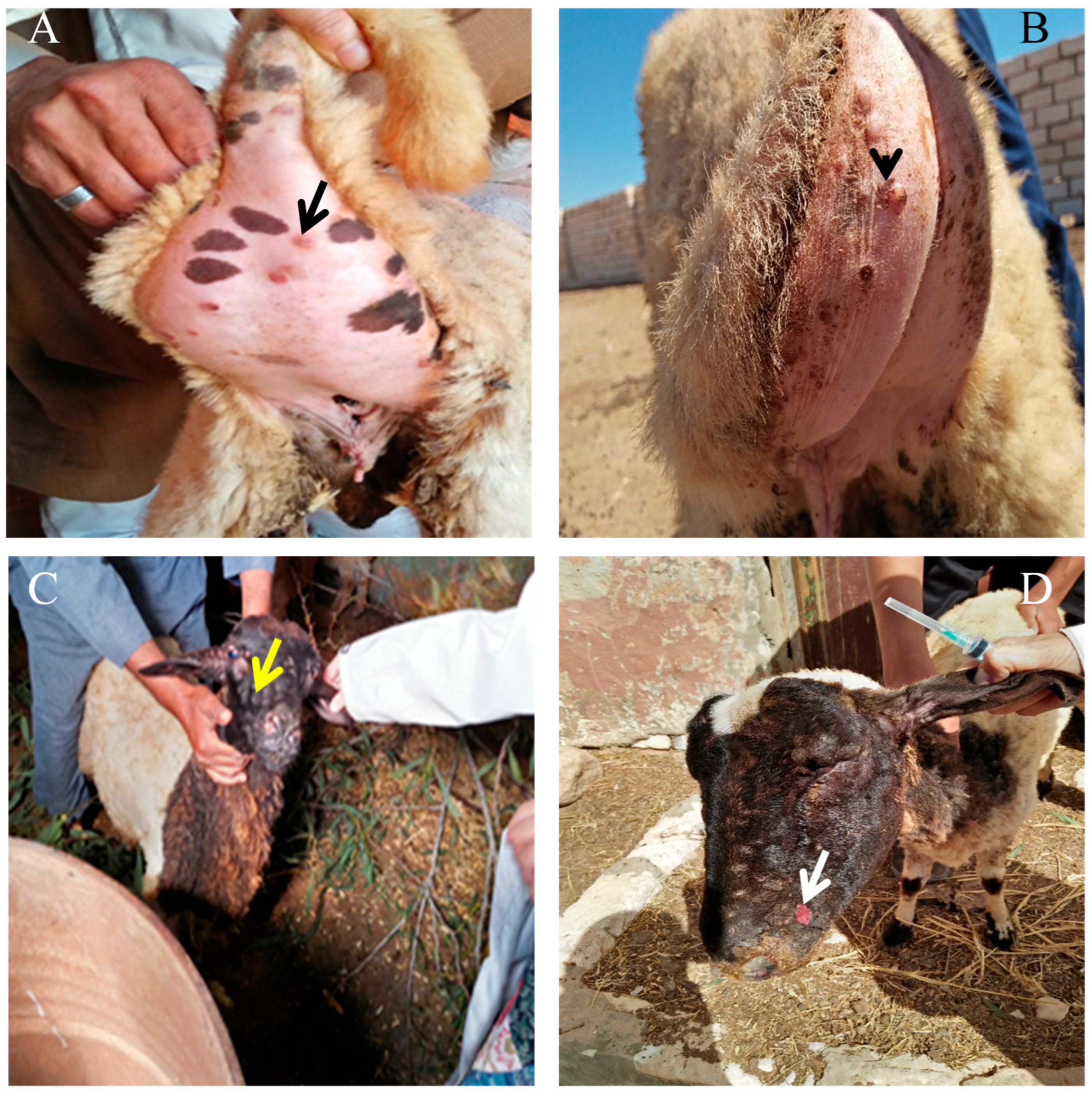
3.1. Pathological Evolution of Sheep Pox Lesions in Naturally Infected Sheep
3.2. Genetic Variants and SNP Analysis
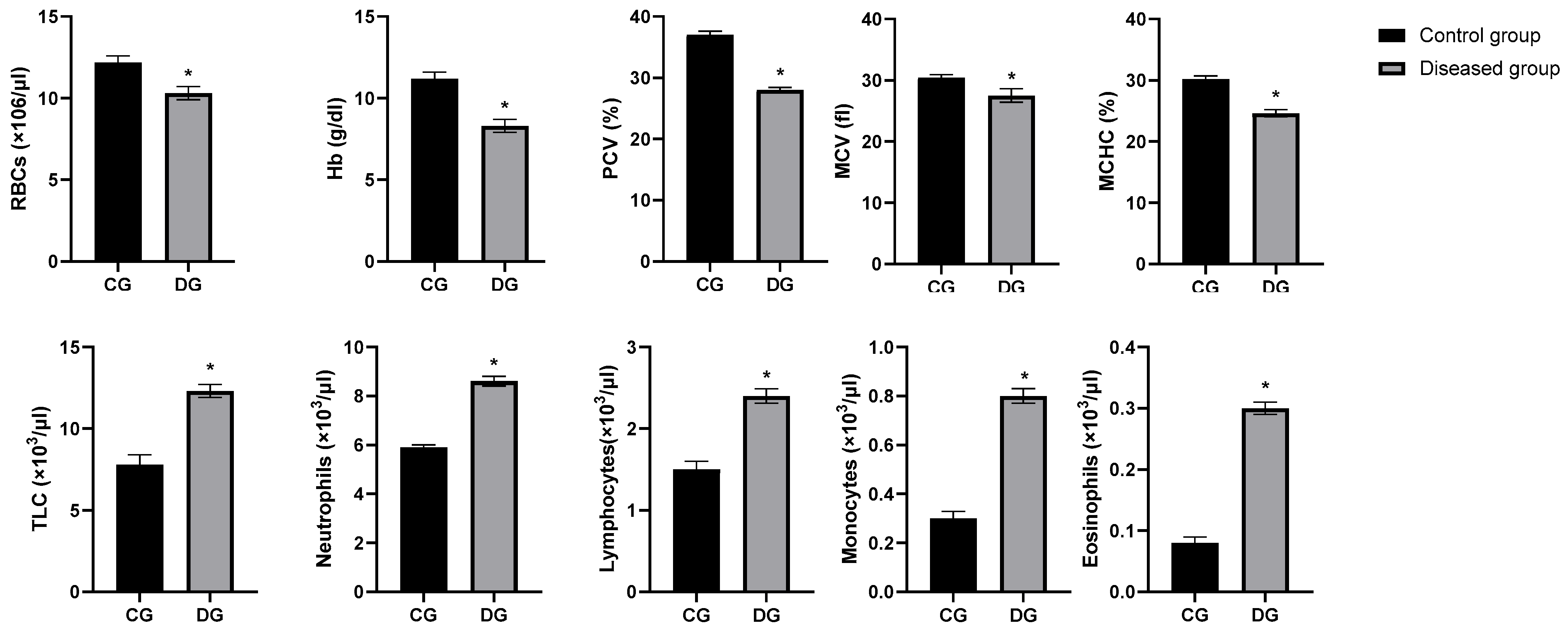
3.3. Hematological Parameters
3.4. PCR Detection Rate Among Symptomatic Submissions
3.5. Homology and Genetic Evolution Analysis of ORF 103Gene
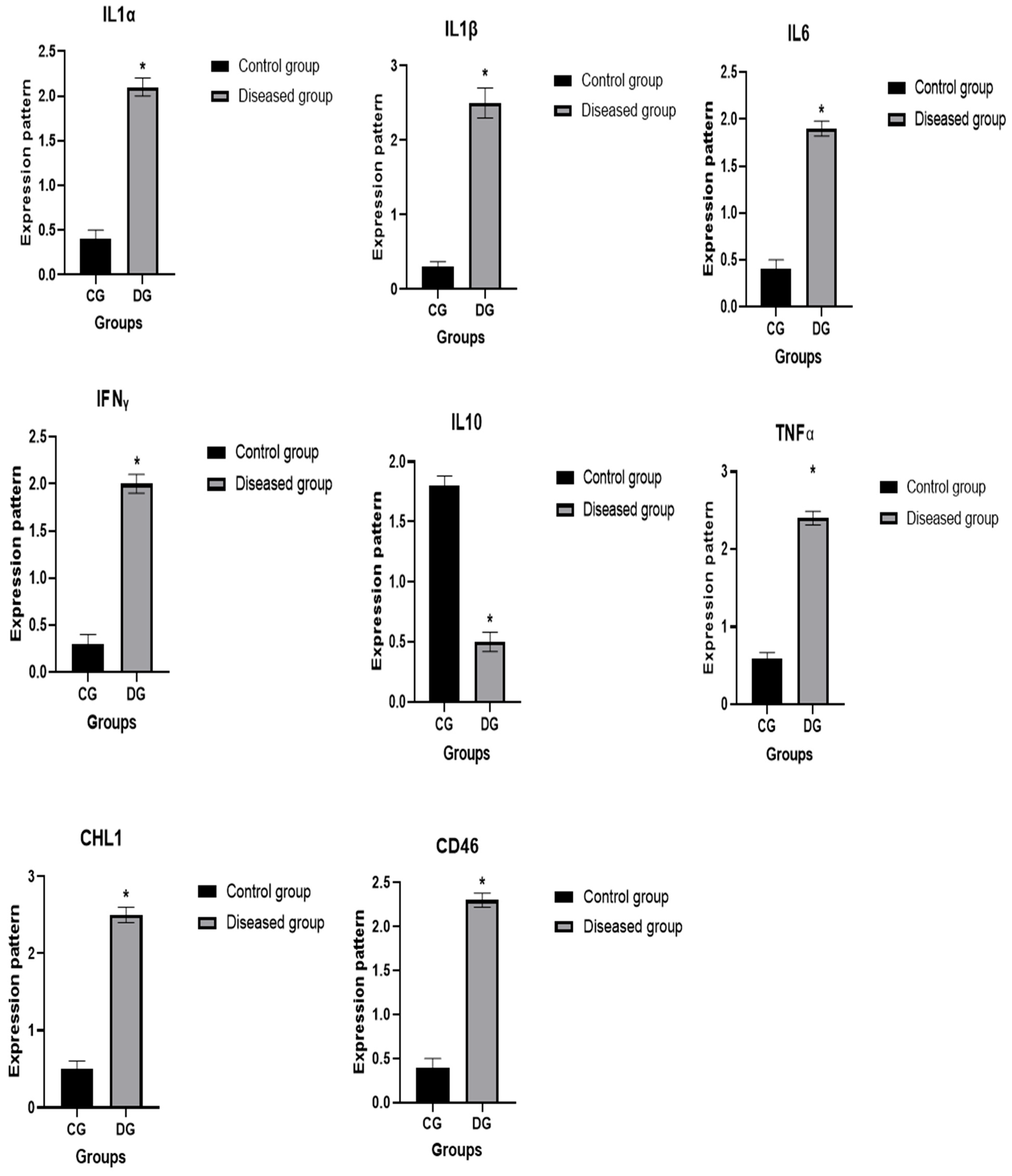
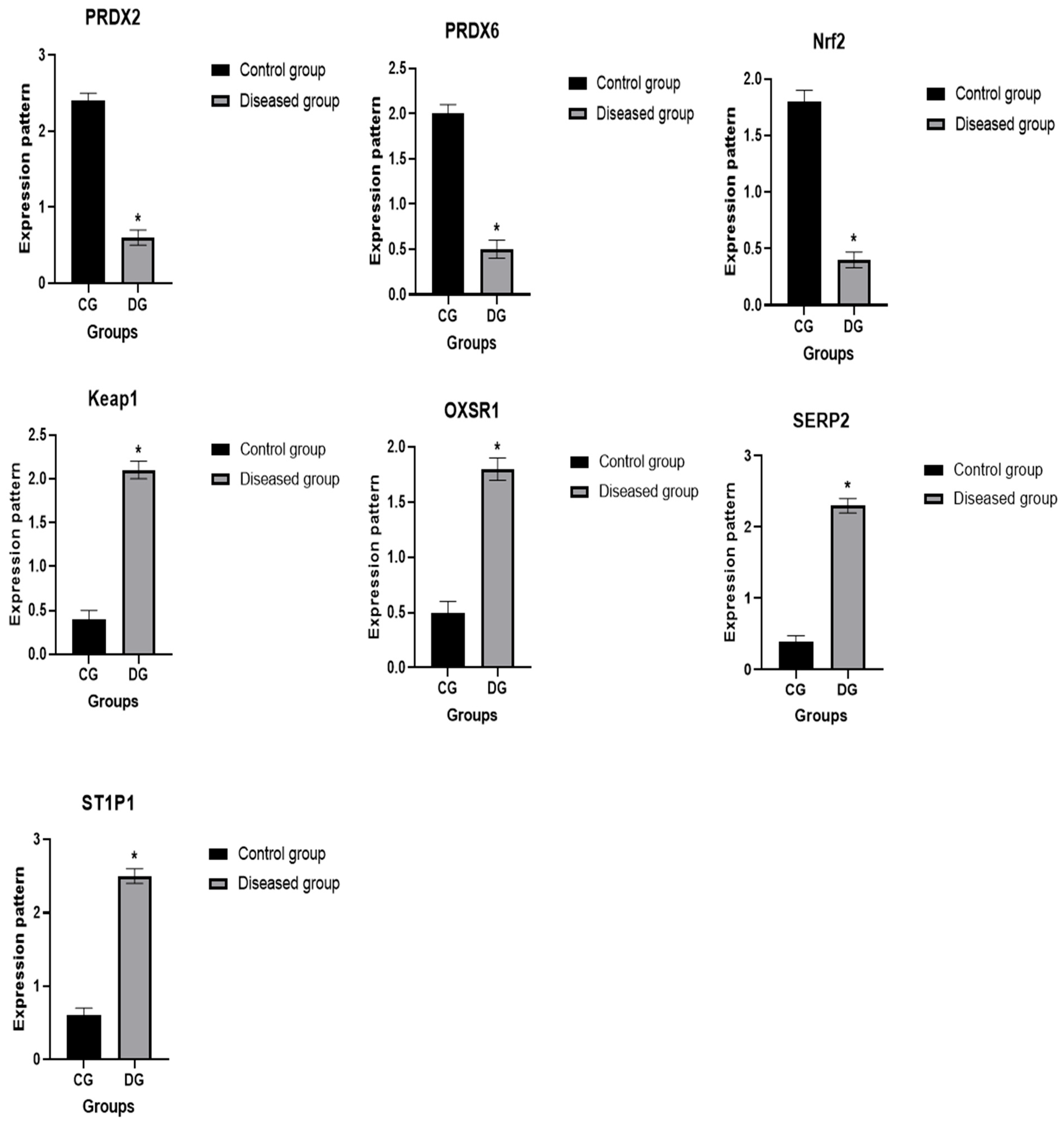
3.6. Patterns for Transcript Levels of Immune and Antioxidant Indicators
3.7. Genetic Polymorphisms of Immune and Antioxidant Genes
3.8. Biochemical Parameters
3.9. Diagnostic Performance of Biomarkers
4. Discussion
4.1. Clinical and Pathological Findings
4.2. Genetic Susceptibility
4.3. Hematological Alterations and Inflammatory Response
4.4. The Prevalence and Molecular Detection of Sheep Pox Virus
4.5. Genetic Associations and SPV Susceptibility
4.6. Patterns for Transcript Levels and Genetic Polymorphisms of Immune and Antioxidant Genes
4.7. Biochemical Indicators of Organ Involvement
4.8. Electrolyte and Metabolic Disturbances
4.9. Lipid Profile and Trace Element Depletion
4.10. Oxidative Stress and Antioxidant Deficiency
4.11. Immunological Dysregulation and Cytokine Profiles
4.12. Acute Phase Proteins and Endocrine Changes
4.13. Iron Homeostasis and Anemia of Inflammation
4.14. Diagnostic Utility of Biomarkers
4.15. Diagnostic Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sallam, A.M. Risk factors and genetic analysis of pre-weaning mortality in Barki lambs. Livest. Sci. 2019, 230, 103818. [Google Scholar] [CrossRef]
- Abousoliman, I.; Reyer, H.; Oster, M.; Murani, E.; Mohamed, I.; Wimmers, K. Genome-wide analysis for early growth-related traits of the locally adapted Egyptian Barki sheep. Genes 2021, 12, 1243. [Google Scholar] [CrossRef]
- Elbagory, G.; Aboul Soud, E.A.S.; Hassan, N. Isolation, identification and characterization of Capripox virus among clinically infected cases of small ruminants in different Egyptian governorates. Benha Vet. Med. J. 2021, 40, 174–178. [Google Scholar] [CrossRef]
- OIE. Internationale des Epizooties (World Health Organization for Animals) Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Sheep Pox Goat Pox 2017, 7, 1–12. [Google Scholar]
- Bukar, B.A.; Mabu, I.M. Common diseases of goats, treatment and preventive measures. In Goat Science—From Keeping to Precision Production; IntechOpen: London, UK, 2023. [Google Scholar]
- Dawle, Y. Review on Sheep and Goat Pox. Eur. J. Med. Res. Clin. Trials 2021, 1, 1–11. [Google Scholar] [CrossRef]
- Hassan, A.; Uyigue, P.; Akinleye, A.; Oyeromi, O. The role of common housefly as a mechanical vector of pathogenic microorganisms. Funksec Here 2021, 3, 261–269. [Google Scholar]
- Paslaru, A.I.; Verhulst, N.O.; Maurer, L.M.; Brendle, A.; Pauli, N.; Vögtlin, A.; Renzullo, S.; Ruedin, Y.; Hoffmann, B.; Torgerson, P.R. Potential mechanical transmission of Lumpy skin disease virus (LSDV) by the stable fly (Stomoxys calcitrans) through regurgitation and defecation. Curr. Res. Insect Sci. 2021, 1, 100007. [Google Scholar] [CrossRef]
- Fares, M.; Fawzy, M.; Shahin, M.; Elshahidy, M. A Comprehensive Review of Sheep and Goat Pox Viruses: Perspective of Their Epidemiology and Economic Importance in Egypt. Suez Canal Vet. Med. J. SCVMJ 2019, 24, 313–328. [Google Scholar] [CrossRef][Green Version]
- Hamdi, J.; Munyanduki, H.; Omari Tadlaoui, K.; El Harrak, M.; Fassi Fihri, O. Capripoxvirus infections in ruminants: A review. Microorganisms 2021, 9, 902. [Google Scholar] [CrossRef]
- Hunter, P.A. Common and Reportable Infectious Diseases of Small Ruminants. Prof. Agric. Work. J. (PAWJ) 2020, 6, 47–61. [Google Scholar][Green Version]
- Zewdie, G.; Derese, G.; Getachew, B.; Belay, H.; Akalu, M. Review of sheep and goat pox disease: Current updates on epidemiology, diagnosis, prevention and control measures in Ethiopia. Anim. Dis. 2021, 1, 28. [Google Scholar] [CrossRef]
- Arbaga, A.; Hassan, H.; Anis, A.; Othman, N.; Kamr, A. Clinicopathological and Electrophoretic Pattern of Serum Protein Alterations in Acute Pneumonic Sheep. Pak. Vet. J. 2023, 43, 303–308. [Google Scholar]
- Camini, F.C.; da Silva Caetano, C.C.; Almeida, L.T.; de Brito Magalhães, C.L. Implications of oxidative stress on viral pathogenesis. Arch. Virol. 2017, 162, 907–917. [Google Scholar] [CrossRef]
- Rehman, Z.U.; Meng, C.; Sun, Y.; Safdar, A.; Pasha, R.H.; Munir, M.; Ding, C. Oxidative stress in poultry: Lessons from the viral infections. Oxidative Med. Cell. Longev. 2018, 2018, 5123147. [Google Scholar] [CrossRef]
- Ntyonga-Pono, M.-P. COVID-19 infection and oxidative stress: An under-explored approach for prevention and treatment? Pan Afr. Med. J. 2020, 35 (Suppl. 2), 12. [Google Scholar] [CrossRef]
- Berry, D.P.; Bermingham, M.L.; Good, M.; More, S.J. Genetics of animal health and disease in cattle. Ir. Vet. J. 2011, 64, 5. [Google Scholar] [CrossRef]
- Pierce, C.F.; Brown, V.R.; Olsen, S.C.; Boggiatto, P.; Pedersen, K.; Miller, R.S.; Speidel, S.E.; Smyser, T.J. Loci associated with antibody response in feral swine (Sus scrofa) infected with Brucella suis. Front. Vet. Sci. 2020, 7, 554674. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Lu, T.; Niu, L.; Wang, L.; Zhan, S.; Guo, J.; Cao, J.; Li, L.; Zhang, H. Nucleotide variants in TheTLR5 gene and promoter methylation with A susceptibility to brucellosis in Chinese goats. Folia Biol. 2022, 70, 55–66. [Google Scholar] [CrossRef]
- Sallam, A.M.; Abou-Souliman, I.; Reyer, H.; Wimmers, K.; Rabee, A.E. New insights into the genetic predisposition of brucellosis and its effect on the gut and vaginal microbiota in goats. Sci. Rep. 2023, 13, 20086. [Google Scholar] [CrossRef]
- Adams, L.; Schutta, C.J. Natural resistance against brucellosis: A review. Open Vet. Sci. J. 2010, 4, 61–71. [Google Scholar] [CrossRef]
- Pugh, D. Sheep and Goat Medicine, 2nd ed.; Pugh, D.G., Baird, A.N., Eds.; Elsevier: Amsterdam, The Netherlands, 2012; Available online: https://www.elsevier.com (accessed on 24 December 2024).
- Feldman, M.S. Organizational routines as a source of continuous change. Organ. Sci. 2000, 11, 611–629. [Google Scholar] [CrossRef]
- Wondimu, A.; Tassew, H.; Gelaye, E.; Hagos, Y.; Belay, A.; Teshome, Y.; Laiju, S.; Asebe, G. Outbreak investigation and molecular detection of pox virus circulating in sheep and goats in selected districts of west Gojjam and Awi zones northwest, Ethiopia. Vet. Med. Res. Rep. 2021, 12, 303–315. [Google Scholar] [CrossRef]
- Hassanien, R.T.; Afify, A.F.; Abdelmegeed, H.K.; Danial, N.M. Isolation, antigenic and molecular characterization of sheeppox virus from clinical cases in Egypt. Adv. Anim. Vet. Sci 2021, 9, 416–421. [Google Scholar] [CrossRef]
- Zhu, X.; Yang, F.; Li, H.; Dou, Y.; Meng, X.; Li, H.; Luo, X.; Cai, X. Identification and phylogenetic analysis of a sheep pox virus isolated from the Ningxia Hui Autonomous Region of China. Genet. Mol. Res. 2013, 12, 1670–1678. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Boom, R.; Sol, C.; Salimans, M.; Jansen, C.; Wertheim-van Dillen, P.; Van der Noordaa, J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990, 28, 495–503. [Google Scholar] [CrossRef]
- Boesenberg-Smith, K.A.; Pessarakli, M.M.; Wolk, D.M. Assessment of DNA yield and purity: An overlooked detail of PCR troubleshooting. Clin. Microbiol. Newsl. 2012, 34, 1, 3–6. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Jalali, S.; Rasooli, A.; Abad-Shapouri, M.; Daneshi, M. Clinical, hematologic, and biochemical findings in cattle infected with lumpy skin disease during an outbreak in southwest Iran. Arch. Razi Inst. 2017, 72, 255. [Google Scholar] [PubMed]
- Altan, E.; JR, G.S.-S.; DELWART, E. The first discovery of Chaphamaparvovirus in sheep with encephalitis and anemia. Turk. Bull. Hyg. Exp. Biol. 2021, 78, 343–350. [Google Scholar] [CrossRef]
- Farhan, M.; Iqbal, Q.; Jamil, T.; Fayaz, M.; Kabir, A.; Nawaz, E.; Ali, M.; Manzoor, S. An outbreak of parasitic infection due to nutritional deficiency in goats. Res J. Vet. Pract. 2022, 10, 19–23. [Google Scholar]
- Karakurt, E.; Coskun, N.; Beytut, E.; Dag, S.; Yilmaz, V.; Nuhoglu, H.; Yildiz, A.; Kurtbas, E. Cytokine profile in lambs naturally infected with sheeppox virus. Trop. Anim. Health Prod. 2023, 55, 401. [Google Scholar] [CrossRef]
- Smith, S.A.; Kotwa, G.J. Immune response to poxvirus infections in various animals. Crit. Rev. Microbiol. 2002, 28, 149–185. [Google Scholar] [CrossRef]
- Bhanuprakash, V.; Indrani, B.; Hosamani, M.; Singh, R. The current status of sheep pox disease. Comp. Immunol. Microbiol. Infect. Dis. 2006, 29, 27–60. [Google Scholar] [CrossRef]
- Chanie, M. Clinical and histopathological study of sheep pox in Ethiopia. Int. J. Nat. Sci. 2011, 1, 89–92. [Google Scholar] [CrossRef]
- Yacob, H.; Nesanet, B.; Dinka, A. Part II: Prevalences of major skin diseases in cattle, sheep and goats at Adama Veterinary Clinic, Oromia regional state, Ethiopia. Rev. De Médecine Vétérinaire 2008, 159, 455–461. [Google Scholar]
- Kebede, A.; Dugassa, J. Prevalence of common skin diseases of small ruminants in Dibate district Metekel zone of Benishangul Gumuz regional state, Northwestern Ethiopia. Multidiscip. Adv. Vet. Sci. 2018, 2, 283–292. [Google Scholar]
- Molla, B.; Haile, H.; Alemu, S. Prevalence and risk factors associated to skin diseases in small ruminants in Gamo Gofa zone, south-Western Ethiopia. J. Vet. Med. Anim. Health 2017, 9, 228–234. [Google Scholar]
- Senthilkumar, V.; Thirunavukkarasu, M.; Kathiravan, G. Survival time in sheep affected by sheep pox and enterotoxaemia. J. Anim. Vet. Adv. 2006, 5, 647–650. [Google Scholar]
- Babiuk, S.; Bowden, T.R.; Parkyn, G.; Dalman, B.; Hoa, D.M.; Long, N.T.; Vu, P.P.; Bieu, D.X.; Copps, J.; Boyle, D.B. Yemen and Vietnam capripoxviruses demonstrate a distinct host preference for goats compared with sheep. J. Gen. Virol. 2009, 90, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.-A.M.; Hamad, M.E.; Ali, B.H.; Ali, A.-W.S. Alterations in some epidemiological patterns and virus heterogeneity recently observed in sheeppox outbreaks in the Sudan. Vet. Arh. 2004, 74, 341–350. [Google Scholar]
- Zro, K.; Azelmat, S.; Bendouro, Y.; Kuhn, J.; El Fahime, E.; Ennaji, M. PCR-based assay to detect sheeppox virus in ocular, nasal, and rectal swabs from infected Moroccan sheep. J. Virol. Methods 2014, 204, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, M.; Khafagi, M. Detection, identification, and differentiation of sheep pox virus and goat pox virus from clinical cases in Giza Governorate, Egypt. Vet. World 2016, 9, 1445. [Google Scholar] [CrossRef]
- Manjunatha Reddy, G.B.; Krishnappa, V.K.; Siddalingaiah, C.D.; Rao, S.; Nayakvadi, S.; Harlipura Basavarajappa, C.K.; Gualti, B.R. Epidemiological, Pathological, and Molecular Studies on Sheeppox Disease Outbreaks in Karnataka, India. Microorganisms 2024, 12, 1373. [Google Scholar] [CrossRef]
- Zeedan, G.S.; Mahmoud, A.H.; Abdalhamed, A.M.; Ghazy, A.A.; Abd EL-Razik, K.A. Rapid detection and differentiation between sheep pox and goat pox viruses by real-time QPCR and conventional PCR in sheep and goats in Egypt. World’s Vet. J. 2020, 10, 80–87. [Google Scholar] [CrossRef]
- Tian, H.; Wu, J.; Chen, Y.; Zhang, K.; Shang, Y.; Liu, X. Development of a SYBR green real-time PCR method for rapid detection of sheep pox virus. Virol. J. 2012, 9, 291. [Google Scholar] [CrossRef]
- Salim, T.; Sershen, C.L.; May, E.E. Investigating the role of TNF-α and IFN-γ activation on the dynamics of iNOS gene expression in LPS stimulated macrophages. PLoS ONE 2016, 11, e0153289. [Google Scholar] [CrossRef] [PubMed]
- Asadpour, R.; Zangiband, P.; Nofouzi, K.; Saberivand, A. Differential expression of antioxidant genes during clinical mastitis of cow caused by Staphylococcus aureus and Escherichia coli. Vet. Arh. 2021, 91, 451–458. [Google Scholar] [CrossRef]
- Fonseca, I.; Silva, P.V.; Lange, C.C.; Guimarães, M.F.; Weller, M.M.D.C.A.; Sousa, K.R.S.; Lopes, P.S.; Guimarães, J.D.; Guimarães, S.E. Expression profile of genes associated with mastitis in dairy cattle. Genet. Mol. Biol. 2009, 32, 776–781. [Google Scholar] [CrossRef] [PubMed]
- Benedict, C.A.; Banks, T.A.; Ware, C.F. Death and survival: Viral regulation of TNF signaling pathways. Curr. Opin. Immunol. 2003, 15, 59–65. [Google Scholar] [CrossRef]
- Li, M.O.; Flavell, R.A. Contextual regulation of inflammation: A duet by transforming growth factor-β and interleukin-10. Immunity 2008, 28, 468–476. [Google Scholar] [CrossRef]
- Huang, X.; Sun, J.; Rong, W.; Zhao, T.; Li, D.; Ding, X.; Wu, L.; Wu, K.; Schachner, M.; Xiao, Z.-C. Loss of cell adhesion molecule CHL1 improves homeostatic adaptation and survival in hypoxic stress. Cell Death Dis. 2013, 4, e768. [Google Scholar] [CrossRef]
- Yang, C.; Ning, L.; Zhou, F.; Sun, Q.; Meng, H.; Han, Z.; Liu, Y.; Huang, W.; Liu, S.; Li, X. Downregulation of adhesion molecule CHL1 in B cells but not T cells of patients with major depression and in the brain of mice with chronic stress. Neurotox. Res. 2020, 38, 914–928. [Google Scholar] [CrossRef]
- Wang, X.; Zhong, J.; Gao, Y.; Ju, Z.; Huang, J. A SNP in intron 8 of CD46 causes a novel transcript associated with mastitis in Holsteins. BMC Genom. 2014, 15, 630. [Google Scholar] [CrossRef]
- Wadley, A.J.; Aldred, S.; Coles, S.J. An unexplored role for Peroxiredoxin in exercise-induced redox signalling? Redox Biol. 2016, 8, 51–58. [Google Scholar] [CrossRef]
- Baird, L.; Suzuki, T.; Takahashi, Y.; Hishinuma, E.; Saigusa, D.; Yamamoto, M. Geldanamycin-derived HSP90 inhibitors are synthetic lethal with NRF2. Mol. Cell. Biol. 2020, 40, e00377-20. [Google Scholar] [CrossRef]
- Kobayashi, A.; Kang, M.-I.; Okawa, H.; Ohtsuji, M.; Zenke, Y.; Chiba, T.; Igarashi, K.; Yamamoto, M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004, 24, 7130–7139. [Google Scholar] [CrossRef]
- Ateya, A.; El-Sayed, A.; Mohamed, R. Gene expression and serum profile of antioxidant markers discriminate periparturient period time in dromedary camels. Mammal Res. 2021, 66, 603–613. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Hori, O.; Stern, D.M.; Hartmann, E.; Ogawa, S.; Tohyama, M. Stress-associated endoplasmic reticulum protein 1 (SERP1)/Ribosome-associated membrane protein 4 (RAMP4) stabilizes membrane proteins during stress and facilitates subsequent glycosylation. J. Cell Biol. 1999, 147, 1195–1204. [Google Scholar] [CrossRef]
- Song, Y.; Masison, D.C. Independent regulation of Hsp70 and Hsp90 chaperones by Hsp70/Hsp90-organizing protein Sti1 (Hop1). J. Biol. Chem. 2005, 280, 34178–34185. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.J.; Van der Vies, S.M. Molecular chaperones. Annu. Rev. Biochem. 1991, 60, 321–347. [Google Scholar] [CrossRef]
- Sonowal, J.; Patel, C.L.; Gandham, R.K.; Sajjanar, B.; Khan, R.I.N.; Praharaj, M.R.; Malla, W.A.; Kumar, D.; Dev, K.; Barkathullah, N. Genome-wide expression analysis reveal host genes involved in immediate-early infections of different sheeppox virus strains. Gene 2021, 801, 145850. [Google Scholar] [CrossRef] [PubMed]
- Carter, G.C.; Law, M.; Hollinshead, M.; Smith, G.L. Entry of the vaccinia virus intracellular mature virion and its interactions with glycosaminoglycans. J. Gen. Virol. 2005, 86, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Elgueta, R.; Benson, M.J.; De Vries, V.C.; Wasiuk, A.; Guo, Y.; Noelle, R.J. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol. Rev. 2009, 229, 152–172. [Google Scholar] [CrossRef]
- Ji, W.-T.; Liu, H.J. PI3K-Akt signaling and viral infection. Recent Pat. Biotechnol. 2008, 2, 218–226. [Google Scholar] [CrossRef]
- Bozukluhan, K.; Merhan, O.; Gökçe, H.İ.; Öğün, M.; Atakişi, E.; Kiziltepe, Ş.; Gökçe, G. Determination of some acute phase proteins, biochemical parameters and oxidative stress in sheep with naturally infected sheeppox virus. Kafkas Üniversitesi Vet. Fakültesi Derg. 2018, 24, 437–444. [Google Scholar]
- Królicka, A.L.; Kruczkowska, A.; Krajewska, M.; Kusztal, M.A. Hyponatremia in infectious diseases—A literature review. Int. J. Environ. Res. Public Health 2020, 17, 5320. [Google Scholar] [CrossRef]
- Neamat-Allah, A.N. Immunological, hematological, biochemical, and histopathological studies on cows naturally infected with lumpy skin disease. Vet. World 2015, 8, 1131. [Google Scholar] [CrossRef]
- De, U.; Chander, V.; Akhilesh; Mahajan, S.; Sharma, G.; Nandi, S.; Singh, K.; Gupta, V. Alterations of hemogram, serum biochemistry, oxidative/nitrosative balance, and copper/zinc homeostasis in dromedary camels naturally infected with poxvirus. Trop. Anim. Health Prod. 2020, 52, 2997–3003. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Nashiruddullah, N.; Ahmed, J.A. Pathology of Spontaneous Pox Virus Infection of Sheep and Goat in Jammu Region. Int. J. Curr. Microbiol. App. Sci 2020, 9, 1204–1226. [Google Scholar] [CrossRef]
- Kirmizigul, A.H.; Ogun, M.; Ozen, H.; Erkilic, E.E.; Gokce, E.; Karaman, M.; Kukurt, A. Oxidative stress and total sialic acid levels in sheep naturally infected with pox virus. Pak. Vet. J. 2016, 36, 313–315. [Google Scholar]
- Struzik, J.; Szulc-Dąbrowska, L. NF-κB as an important factor in optimizing poxvirus-based vaccines against viral infections. Pathogens 2020, 9, 1001. [Google Scholar] [CrossRef] [PubMed]
- Kashafi, S. Comparative Evaluation of In-Vitro Immunological Response of Three Ruminant Species to Bluetongue Virus. Ph.D. Dissertation, Indian Veterinary Research Institute, Bareilly, Indian, 2022. [Google Scholar]
- Choudhary, S.; Justin Davis, K.; Chetan Kumar, G.; Sharma, P. Clinical Findings of Diseases of Goats. Princ. Goat Dis. Prev. 2023, 33–48. [Google Scholar]
- Nikpour, H. Influence of Sex and Blood Collection Method on Innate Immune Response Following Lipopolysaccharide Administration in Sheep; West Virginia University: Morgantown, WV, USA, 2017; p. 6320. [Google Scholar]
- Karademir, B. Changes in Serum Mineral Composition During Poxvirus Infection in Sheep and Their Lambs. Turk. J. Agric. Food Sci. Technol. 2021, 9, 106–113. [Google Scholar] [CrossRef]

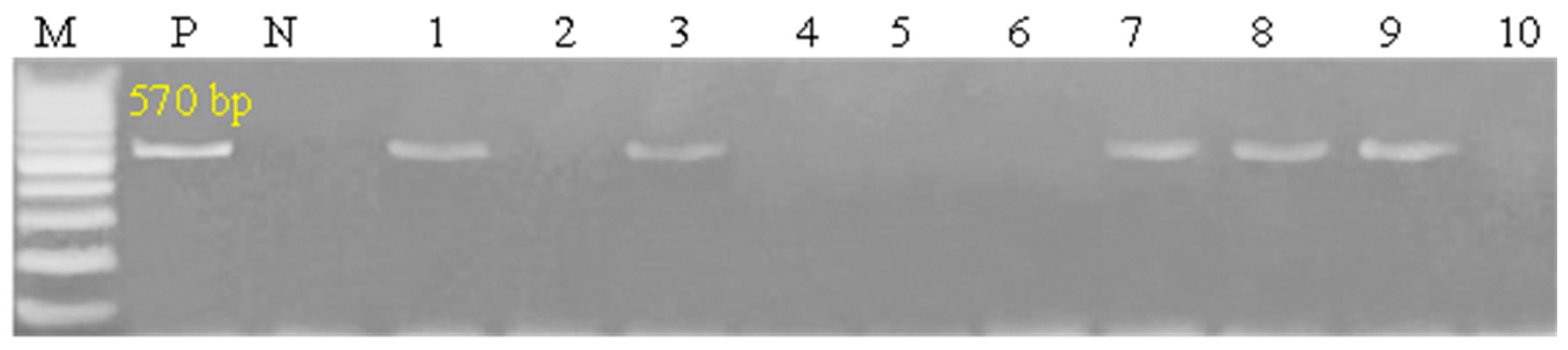

| Target Gene | Primers Sequences | Amplified Segment (bp) | Primary Denaturation | Amplification (35 Cycles) | Final Extension | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Secondary Denaturation | Annealing | Extension | ||||||
| ORF 103 | ATGTCTGATAAAAAATTATCTCG | 570 | 94 °C 5 min | 94 °C 30 s | 52 °C 40 s | 72 °C 45 s | 72 °C 10 min | [26] |
| ATCCATACCATCGTCGATAG | ||||||||
| Investigated Marker | Primer | Product Size (bp) | Annealing Temperature (°C) | GenBank Isolate |
|---|---|---|---|---|
| IL-1α | F5′-AAGACATCCAGGCTTAGCTTC-3′ R5′-GAGATTGGTCTCATCTTTGATG-3′ | 480 | 56 | NM_001009808.1 |
| IL-1β | F5′-GATGAAGAGCTGCACCCAACAC-3′ R5′-CTCTCCTTGTACGAAGCTCATG-3′ | 400 | 58 | NM_001009465.2 |
| IL-6 | F5′-ACAAGCGCCTTCAGTCCACTCGC-3′ R5′-TATATCTGATACTCCAGAAGAC-3′ | 359 | 58 | NM_001009392.1 |
| TNF-α | F5′-ATGTGGAGCTGGCGGAGGAGGT-3′ R5′-TGAAGAGGACCTGCGAGTAGAT-3′ | 399 | 58 | X55152.1 |
| IFN-γ | F5′-ATACACAAGCTCCTTCTTAGCT-3′ R5′-ATTCTGACTTCTCTTCCGCTT-3′ | 466 | 56 | NM_001009803.1 |
| IL-10 | F5′-GTGGCAGCCAGCCGAGATGCCA-3′ R5′-CTTCGTTGTCATGTAGGATTCT-3′ | 480 | 58 | U11421.1 |
| CHL1 | F5′-GAACACATTAAGCAAGATGAGA-3′ R5′-GCTCTTCTACTGTAACATGAA-3′ | 475 | 58 | XM_060402889.1 |
| CD46 | F5′-GGTCAAGTCCTCGCTCTTGTCT-3′ R5′-AAGCCTGCTCTCTCCAATAAGTG-3′ | 405 | 56 | XM_060396230.1 |
| PRDX2 | F5′-GTGGATGGCGCCTTCAAGGAG-3′ R5′-CTGGACCAGCCTCAGAGCCTCA-3′ | 420 | 58 | NM_001166200.1 |
| PRDX6 | F5′-ACTACCATCGGCCGCATCCGT-3′ R5′-GCCAGTGGTAGCTGGGTAGAGG-3′ | 411 | 58 | NM_001280704.1 |
| Nrf2 | F5′-GTAAGTCGAGAGGTATTTGACT-3′ R5′-GCTGCATGCAGTCATCGAAGTA-3′ | 343 | 56 | OR900054.1 |
| Keap1 | F5′-CAGCCGGAACCCAGGCCTAGCG-3′ R5′-GGTGTAGGCGAACTCAATGAGC-3′ | 423 | 58 | OR900055.1 |
| OXSR1 | F5′-GAAGATGGCTCGGTACAGATTG-3′ R5′-TGGTTGGTGCTCTCTGCAATAT-3′ | 461 | 58 | XM_060402087.1 |
| SERP2 | F5′-ATGGTGGCCAAACAGCGGATCC-3′ R5′-TCCTCTTCTGGGTGACTTGTCG-3′ | 304 | 58 | XM_027973536.3 |
| STIP1 | F5′-TACCAGAAGGCTTACGAGGAC-3′ R5′-TCTTCATCCATACTGCCCAGA-3′ | 419 | 56 | XM_042238340.2 |
| ß. actin | F5′-AATTCCATCATGAAGTGTGAC-3′ R5′-GATCTTGATCTTCATCGTGCT-3′ | 150 | 58 | KU365062.1 |
| Gene | SNPs | Healthy n = 50 | Pox n = 50 | Total n = 100 | Chi Square Value X2 | p Value | Kind of Inherited Change | Amino Acid Order and Sort |
|---|---|---|---|---|---|---|---|---|
| IL-1α | T105C | 35/50 | -/50 | 35/100 | 53.8 | 0.001 | Synonymous | 35 D |
| A277C | -/50 | 32/50 | 32/100 | 47 | 0.002 | Non-synonymous | M to L | |
| A431T | -/50 | 25/50 | 25/100 | 33.3 | 0.001 | Non-synonymous | H to L | |
| IL-1β | A74C | -/50 | 40/50 | 40/100 | 66.6 | 0.003 | Non-synonymous | K to Q |
| IL-6 | G216A | 27/50 | -/50 | 27/100 | 36.9 | 0.002 | Synonymous | E |
| TNF-α | C32G | 12/30 | -/50 | 20/100 | 13.6 | 0.001 | Non-synonymous | N to K |
| A287G | -/50 | 35/50 | 35/100 | 53.8 | 0.002 | Synonymous | R | |
| IFN-γ | G345A | 43/50 | -/50 | 43/100 | 75.4 | 0.001 | Non-synonymous | R to K |
| C394A | -/50 | 23/50 | 23/100 | 29.8 | 0.001 | Synonymous | I | |
| IL-10 | A112C | -/50 | 35/50 | 35/100 | 53.8 | 0.001 | Non-synonymous | T to P |
| CHL1 | C167T | 32/50 | -/50 | 32/100 | 47 | 0.002 | Non-synonymous | S to L |
| CD46 | T74A | 35/50 | -/50 | 35/100 | 53.8 | 00.003 | Non-synonymous | L to Q |
| A206G | 22/50 | -/50 | 22/100 | 28.2 | 0.001 | Non-synonymous | D to G | |
| PRDX2 | A128C | 32/50 | -/50 | 32/100 | 47 | 0.002 | Non-synonymous | E to A |
| C288T | -/50 | 30/50 | 30/100 | 42.8 | 0.001 | Synonymous | G | |
| PRDX6 | A234G | 22/50 | -/50 | 22/100 | 28.2 | 0.001 | Synonymous | T |
| Nrf2 | C256G | 28/50 | -/50 | 28/100 | 38.8 | 0.003 | Non-synonymous | P to A |
| Keap1 | A48G | -/50 | 27/50 | 27/100 | 36.9 | 0.002 | Synonymous | L |
| A303G | -/50 | 18/50 | 18/100 | 21.9 | 0.001 | Synonymous | S | |
| OXSR1 | T219A | 23/50 | -/50 | 23/100 | 29.8 | 0.002 | Synonymous | P |
| SERP2 | A114C | -/50 | 40/50 | 40/100 | 66.6 | 0.001 | Synonymous | G |
| STIP1 | T66C | 28/50 | -/50 | 28/100 | 38.8 | 0.001 | Synonymous | Y |
| C177A | 37/50 | -/50 | 37/100 | 58.7 | 0.001 | Non-synonymous | F to L |
| Predicted Group Membership | Total | |||
|---|---|---|---|---|
| Healthy | Diseases | |||
| Count | Healthy | 50 | 0 | 100 |
| Diseased | 0 | 50 | 100 | |
| % | Healthy | 50 | 0.0 | 100.0 |
| Diseased | 0.0 | 50 | 100.0 | |
| Parameter | Unit | CG | DG |
|---|---|---|---|
| Total protein | g/dL | 4.54 ± 0.36 | 7.51 ± 0.51 * |
| Albumin | g/dL | 3.24 ± 0.23 | 2.50 ± 0.49 * |
| Globulin | g/dL | 1.30 ± 0.27 | 5.01 ± 0.64 * |
| A\G | mg/dL | 2.61 ± 0.64 | 0.52 ± 0.15 * |
| Glucose | mg/dL | 113.60 ± 7.78 | 68.42 ± 5.52 * |
| AST | U/L | 26.40 ± 0.26 | 32.93 ± 2.10 * |
| ALT (U/L) | U/L | 30.40 ± 0.26 | 38.14 ± 1.08 * |
| ALP | mg/dL | 27.38 ± 1.23 | 44.51 ± 3.05 * |
| Blood urea | mg/dL | 17.90 ± 1.24 | 41.68 ± 4.58 * |
| Creatinine | mg/dL | 0.75 ± 0.09 | 1.75 ± 0.27 * |
| Total lipids | mg/dL | 419.57 ± 14.10 | 830.75 ± 55.10 |
| Triglycerides | mg/dL | 55.98 ± 2.03 | 147.20 ± 21.83 * |
| Phospholipids | mg/dL | 107.20 ± 3.24 | 397.94 ± 33.95 * |
| T-cholesterol | mg/dL | 195.50 ± 7.60 | 142.81 ± 18.07 * |
| HDL-cholesterol | mg/dL | 88.30 ± 7.82 | 77.20 ± 15.06 * |
| LDL-cholesterol | mg/dL | 107.20 ± 3.24 | 65.60 ± 5.08 * |
| Calcium | mg/dL | 10.36 ± 0.24 | 8.05 ± 0.32 * |
| Phosphorus) | mg/dL | 5.38 ± 0.22 | 2.45 ± 0.47 * |
| Chloride | mmol/L | 97.77 ± 29.94 | 68.65 ± 2.45 * |
| Sodium | mmol/L | 126.02 ± 2.06 | 96.52 ± 7.41 * |
| Potassium | mmol/L | 4.80 ± 0.42 | 2.36 ± 0.60 * |
| Magnesium | mg/dL | 2.37 ± 0.24 | 2.03 ± 0.08 * |
| Copper | μg/dL | 154.27 ± 4.88 | 90.77 ± 3.62 * |
| Zinc | μg/dL | 143.43 ± 4.02 | 98.75 ± 5.84 * |
| MMP-2 | ng/mL | 15.42 ± 0.76 | 35.46 ± 4.64 * |
| MMP-9 | ng/mL | 22.75 ± 1.08 | 42.70 ± 3.98 * |
| Parameter | Unit | CG | DG |
|---|---|---|---|
| IL-1α | (Pg/mL) | 31.60 ± 4.10 | 90.80 ± 4.20 * |
| IL-1β | (Pg/mL) | 28.24 ± 4.34 | 73.60 ± 7.94 * |
| IL-6 | (Pg/mL) | 29.90 ± 1.98 | 54.40 ± 6.54 * |
| TNF-α | (Pg/mL) | 27.22 ± 2.98 | 68.01 ± 2.90 * |
| INF-γ | (pg/mL) | 2.55 ± 0.51 | 8.13 ± 101 * |
| IL-10 | (Pg/mL) | 102.60 ± 4.04 | 60.40 ± 8.55 * |
| Cp | (mg/dL) | 3.39 ± 0.84 | 6.72 ± 0.94 * |
| Hp | (g/L) | 0.15 ± 0.04 | 3.64 ± 0.74 * |
| SAA | (mg/L) | 2.76 ± 0.28 | 7.16 ± 0.80 * |
| TAC | (Mm/L) | 1.62 ± 0.22 | 0.59 ± 0.10 * |
| MDA | (nmol/mL) | 13.27 ± 1.01 | 23.30 ± 1.83 * |
| NO | (μmol/L) | 25.66 ± 1.28 | 41.84 ± 2.01 * |
| CAT | (U/L) | 464.80 ± 30.24 | 301.35 ± 39.80 * |
| GPx | (mU/L) | 1065.60 ± 22.42 | 601.01 ± 82.27 * |
| GSH | (ng/mL) | 21.90 ± 1.11 | 11.20 ± 0.86 * |
| Cortisol | (μg/dL) | 1.77 ± 0.13 | 7.96 ± 0.92 * |
| Insulin | (μIU/mL) | 8.69 ± 0.28 | 5.71 ± 0.93 * |
| SI | (μg/dL) | 104.90 ± 2.08 | 87.40 ± 1.88 * |
| TIBC | (μg/dL) | 263.17 ± 21.49 | 378.01 ± 15.22 * |
| UIBC | (μg/dL) | 158.28 ± 21.28 | 289.62 ± 11.85 * |
| Transferrin | (mg/dL) | 126.30 ± 2.75 | 85.58 ± 2.96 * |
| Tf sat | . % | 40.11 ± 3.36 | 23.21 ± 0.98 * |
| Ferritin | (ng/mL) | 15.85 ± 1.36 | 20.80 ± 2.08 * |
| Cut-Off | Sensitivity | Specificity | LR | PPV | NPV | AR | % of (+,−) | |
|---|---|---|---|---|---|---|---|---|
| IL-1α (Pg/mL) | 37.63 | 100% | 95% | 20 | 95.24% | 100% | 97.50% | 187.34% |
| IL-1β (Pg/mL) | 37.11 | 100% | 90% | 10 | 90.91% | 100% | 95% | 160.62% |
| IL-6 (Pg/mL) | 32.66 | 100% | 95% | 20 | 95.24% | 100% | 97.50% | 81.94% |
| TNF-α (Pg/mL) | 31.00 | 100% | 85% | 6.67 | 86.96% | 100% | 92.50% | 149.85% |
| INF-γ (pg/mL) | 3.10 | 100% | 85% | 6.67 | 86.96% | 100% | 92.50% | 218.82% |
| IL-10 (Pg/mL) | 98.90 | 100% | 85% | 6.67 | 86.96% | 100% | 92.50% | −41.13% |
| Cp (mg/dL) | 4.40 | 100% | 90% | 10 | 90.91% | 100% | 95% | 98.23% |
| Hp (g/L) | 0.18 | 100% | 90% | 10 | 90.91% | 100% | 95% | 2326.67% |
| SAA (mg/L) | 2.96 | 100% | 80% | 5 | 83.33% | 100% | 90% | 159.42% |
| Transferrin (mg/dL) | 121.50 | 100% | 90% | 10 | 90.91% | 100% | 95% | −32.24% |
| Ferritin (ng/mL) | 17.50 | 100% | 85% | 6.67 | 86.96% | 100% | 92.50% | 31.23% |
| MMP-2 (ng/mL) | 15.90 | 100% | 70% | 3.33 | 76.92% | 100% | 85% | 129.96% |
| MMP-9 (ng/mL) | 23.70 | 100% | 75% | 4 | 80% | 100% | 87.50% | 87.69% |
| Copper (μg/dL) | 149.00 | 100% | 80% | 5 | 83.33% | 100% | 90% | −41.16% |
| Zinc (μg/dL) | 138.60 | 100% | 85% | 6.67 | 86.96% | 100% | 92.50% | −31.15% |
| TAC (Mm/L) | 1.30 | 100% | 95% | 20 | 95.24% | 100% | 97.50% | −63.58% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darwish, A.A.; Alqahtani, H.A.; Tahoun, A.; Ateya, A.; Helmy, N.A.; Hafez, A.A.; Alharbi, H.M.; Alwutayd, K.M.; Babaker, M.A.; AL-Farga, A.; et al. Sheep Pox Susceptibility: Role of Genetic Variants, Gene Expression, and Immune-Oxidative Markers. Vet. Sci. 2025, 12, 867. https://doi.org/10.3390/vetsci12090867
Darwish AA, Alqahtani HA, Tahoun A, Ateya A, Helmy NA, Hafez AA, Alharbi HM, Alwutayd KM, Babaker MA, AL-Farga A, et al. Sheep Pox Susceptibility: Role of Genetic Variants, Gene Expression, and Immune-Oxidative Markers. Veterinary Sciences. 2025; 12(9):867. https://doi.org/10.3390/vetsci12090867
Chicago/Turabian StyleDarwish, Asmaa A., Huda A. Alqahtani, Amin Tahoun, Ahmed Ateya, Noha A. Helmy, Amani A. Hafez, Hanan M. Alharbi, Khairiah M. Alwutayd, Manal A. Babaker, Ammar AL-Farga, and et al. 2025. "Sheep Pox Susceptibility: Role of Genetic Variants, Gene Expression, and Immune-Oxidative Markers" Veterinary Sciences 12, no. 9: 867. https://doi.org/10.3390/vetsci12090867
APA StyleDarwish, A. A., Alqahtani, H. A., Tahoun, A., Ateya, A., Helmy, N. A., Hafez, A. A., Alharbi, H. M., Alwutayd, K. M., Babaker, M. A., AL-Farga, A., Al-Shahari, E. A., Salih, Z. A., Al-Duais, M. A., & El-Sayed, A. (2025). Sheep Pox Susceptibility: Role of Genetic Variants, Gene Expression, and Immune-Oxidative Markers. Veterinary Sciences, 12(9), 867. https://doi.org/10.3390/vetsci12090867







