Metagenomic Analysis of the Fecal Virome in Wild Mammals Hospitalized in Pisa, Italy
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Metagenomics Analysis
2.3. Bioinformatics Analysis
- Mammalia virus database:
- ○
- Virus: viruses (taxid 10239)
- ○
- Host: Mammalia (taxid 40674), excluding Homo sapiens (taxid 9606)
- Aves virus database:
- ○
- Virus: viruses (taxid 10239)
- ○
- Host: Aves (taxid 8782)
2.4. Bioinformatic and Statistical Parameters
- Length
- Number of identical sites
- Pairwise identity
- Reference sequence coverage
- E-value (vs. reference)
- Consensus length > 150 bp
- Identical sites > 75%
- Pairwise identity > 80%
- E-value ≤ 10−100
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| VTH | Veterinary Teaching Hospital |
| SISPA | sequence-independent single primer amplification |
Appendix A
| Pool | Sampling Date | Municipality | Sex | Admission | |
|---|---|---|---|---|---|
| Fox | Pool 1 | 9 September 2020 | Pisa | M | Intraspecific aggression |
| 28 October 2020 | Vecchiano | M | Debilitation | ||
| 23 July 2021 | Pontedera | M | Road accident | ||
| 30 July 2021 | Pisa | M | Road accident | ||
| Pool 2 | 2 March 2021 | Ponsacco | F | Road accident | |
| 1 March 2021 | Pisa | F | Road accident | ||
| 25 August 2021 | Capannoli | M | Road accident | ||
| 6 July 2021 | Casciana Terme Lari | M | Debilitation | ||
| Pool 3 | 23 October 2020 | San Giuliano Terme | M | Road accident | |
| 19 March 2021 | Vecchiano | M | Road accident | ||
| 17 September 2021 | Vecchiano | M | Road accident | ||
| 25 October 2021 | Castelfranco di sotto | F | Road accident | ||
| Badger | Pool 4 | 10 November 2020 | San Giuliano Terme | M | Road accident |
| 10 February 2021 | Bientina | F | Unknown | ||
| 26 July 2021 | Pisa | M | Road accident | ||
| 9 September 2021 | Calci | M | Road accident | ||
| Pool 5 | 22 March 2021 | Fauglia | M | Road accident | |
| 16 July 2021 | Pisa | F | Intraspecific aggression | ||
| Marten | 15 June 2021 | Santa Luce | M | Debilitation | |
| Porcupines | Pool 6 | 12 October 2020 | Volterra | F | Intraspecific aggression |
| 23 March 2021 | Bientina | M | Road accident | ||
| 16 July 2021 | Pontedera | F | Road accident | ||
| 27 September 2021 | San Miniato | M | Unknown |
References
- Cunningham, A.A. A Walk on the Wild Side—Emerging Wildlife Diseases. BMJ 2005, 331, 1214–1215. [Google Scholar] [CrossRef] [PubMed]
- Kock, R. Drivers of Disease Emergence and Spread: Is Wildlife to Blame? Onderstepoort J. Vet. Res. 2014, 81, a739. [Google Scholar] [CrossRef]
- Dobson, A.; Foufopoulos, J. Emerging Infectious Pathogens of Wildlife. Phil. Trans. R. Soc. Lond. B 2001, 356, 1001–1012. [Google Scholar] [CrossRef]
- Gortázar, C.; Acevedo, P.; Ruiz-Fons, F.; Vicente, J. Disease Risks and Overabundance of Game Species. Eur. J. Wildl. Res. 2006, 52, 81–87. [Google Scholar] [CrossRef]
- Shaheen, M.N.F. The Concept of One Health Applied to the Problem of Zoonotic Diseases. Rev. Med. Virol. 2022, 32, e2326. [Google Scholar] [CrossRef] [PubMed]
- Overviews of Pathogen Emergence: Which Pathogens Emerge, When and Why? In Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2007; pp. 85–111. ISBN 978-3-540-70961-9.
- Wobeser, G.A. Disease in Wild Animals; Springer: Berlin/Heidelberg, Germany, 2007; ISBN 978-3-540-48974-0. [Google Scholar]
- Arumugam, R.; Uli, J.E.; Annavi, G. A Review of the Application of Next Generation Sequencing (NGS) in Wild Terrestrial Vertebrate Research. Annu. Res. Rev. Biol. 2019, 31, 1–9. [Google Scholar] [CrossRef]
- Bodewes, R.; Ruiz-Gonzalez, A.; Schapendonk, C.M.; Van Den Brand, J.M.; Osterhaus, A.D.; Smits, S.L. Viral Metagenomic Analysis of Feces of Wild Small Carnivores. Virol. J. 2014, 11, 89. [Google Scholar] [CrossRef]
- Pacini, M.I.; Mazzei, M.; Sgorbini, M.; D’Alfonso, R.; Papini, R.A. A One-Year Retrospective Analysis of Viral and Parasitological Agents in Wildlife Animals Admitted to a First Aid Hospital. Animals 2023, 13, 931. [Google Scholar] [CrossRef]
- Sarchese, V.; Fruci, P.; Palombieri, A.; Di Profio, F.; Robetto, S.; Ercolini, C.; Orusa, R.; Marsilio, F.; Martella, V.; Di Martino, B. Molecular Identification and Characterization of a Genotype 3 Hepatitis E Virus (HEV) Strain Detected in a Wolf Faecal Sample, Italy. Animals 2021, 11, 3465. [Google Scholar] [CrossRef]
- Djikeng, A.; Halpin, R.; Kuzmickas, R.; DePasse, J.; Feldblyum, J.; Sengamalay, N.; Afonso, C.; Zhang, X.; Anderson, N.G.; Ghedin, E.; et al. Viral Genome Sequencing by Random Priming Methods. BMC Genom. 2008, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Reyes, G.R.; Kim, J.P. Sequence-Independent, Single-Primer Amplification (SISPA) of Complex DNA Populations. Mol. Cell. Probes 1991, 5, 473–481. [Google Scholar] [CrossRef]
- Chrzastek, K.; Lee, D.; Smith, D.; Sharma, P.; Suarez, D.L.; Pantin-Jackwood, M.; Kapczynski, D.R. Use of Sequence-Independent, Single-Primer-Amplification (SISPA) for Rapid Detection, Identification, and Characterization of Avian RNA Viruses. Virology 2017, 509, 159–166. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Rosario, K.; Breitbart, M. Exploring the Viral World through Metagenomics. Curr. Opin. Virol. 2011, 1, 289–297. [Google Scholar] [CrossRef]
- Li, L.; Deng, X.; Delwart, E. Metagenomic Analysis of Viruses in Stool Samples from Children with Acute Flaccid Paralysis. J. Virol. 2015, 89, 4658–4666. [Google Scholar]
- Zhao, G.; Wu, G.; Lim, E.S.; Droit, L.; Krishnamurthy, S.; Barouch, D.H.; Virgin, H.W.; Wang, D. VirusSeeker, a Computational Pipeline for Virus Discovery and Virome Composition Analysis. Virology 2013, 447, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Paez-Espino, D.; Eloe-Fadrosh, E.A.; Pavlopoulos, G.A.; Thomas, A.D.; Huntemann, M.; Mikhailova, N.; Rubin, E.; Ivanova, N.N.; Kyrpides, N.C. Uncovering Earth’s Virome. Nature 2016, 536, 425–430. [Google Scholar] [CrossRef]
- Wommack, K.E.; Bhavsar, J.; Ravel, J. Metagenomics: Read Length Matters. Appl. Environ. Microbiol. 2012, 74, 1453–1463. [Google Scholar] [CrossRef]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining Viral Signal from Microbial Genomic Data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Brum, J.R.; Ignacio-Espinoza, J.C.; Roux, S.; Doulcier, G.; Acinas, S.G.; Alberti, A.; Chaffron, S.; Cruaud, C.; de Vargas, C.; Gasol, J.M.; et al. Patterns and Ecological Drivers of Ocean Viral Communities. Science 2015, 348, 1261498. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, A.B.R.; Ounit, R.; Afshinnekoo, E.; Prill, R.J.; Hénaff, E.; Alexander, N.; Minot, S.; Mason, C.E. Comprehensive benchmarking and ensemble approaches for metagenomic classifiers. Genome Biol. 2017, 18, 182. [Google Scholar] [CrossRef]
- Wiethoelter, A.K.; Beltrán-Alcrudo, D.; Kock, R.; Mor, S.M. Global Trends in Infectious Diseases at the Wildlife–Livestock Interface. Proc. Natl. Acad. Sci. USA 2015, 112, 9662–9667. [Google Scholar] [CrossRef]
- Martin, C.; Pastoret, P.-P.; Brochier, B.; Humblet, M.-F.; Saegerman, C. A Survey of the Transmission of Infectious Diseases/Infections between Wild and Domestic Ungulates in Europe. Vet. Res. 2011, 42, 70. [Google Scholar] [CrossRef]
- Walker, D.; Abbondati, E.; Cox, A.L.; Mitchell, G.B.B.; Pizzi, R.; Sharp, C.P.; Philbey, A.W. Infectious Canine Hepatitis in Red Foxes (Vulpes vulpes) in Wildlife Rescue Centres in the UK. Vet. Rec. 2016, 178, 421. [Google Scholar] [CrossRef]
- Pyke, G.H.; Szabo, J.K. Conservation and the 4 Rs, Which Are Rescue, Rehabilitation, Release, and Research. Conserv. Biol. 2018, 32, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Gourlay, P.; Decors, A.; Moinet, M.; Lambert, O.; Lawson, B.; Beaudeau, F.; Assié, S. The Potential Capacity of French Wildlife Rescue Centres for Wild Bird Disease Surveillance. Eur. J. Wildl. Res. 2014, 60, 865–873. [Google Scholar] [CrossRef]
- Pacini, M.I.; Bonelli, F.; Briganti, A.; Citi, S.; Perrucci, S.; Papini, R.A.; Sgorbini, M. Wildlife Ungulate Rescue and Emergency Services in the Pisa Area (Tuscany, Italy): Evaluation of a 9-Years Period (2010–2018). Front. Vet. Sci. 2020, 7, 626. [Google Scholar] [CrossRef]
- Delogu, M.; Cotti, C.; Lelli, D.; Sozzi, E.; Trogu, T.; Lavazza, A.; Garuti, G.; Castrucci, M.R.; Vaccari, G.; De Marco, M.A.; et al. Eco-Virological Preliminary Study of Potentially Emerging Pathogens in Hedgehogs (Erinaceus Europaeus) Recovered at a Wildlife Treatment and Rehabilitation Center in Northern Italy. Animals 2020, 10, 407. [Google Scholar] [CrossRef] [PubMed]
- Romero, F.; Espinoza, A.; Sallaberry-Pincheira, N.; Napolitano, C. A Five-Year Retrospective Study on Patterns of Casuistry and Insights on the Current Status of Wildlife Rescue and Rehabilitation Centers in Chile. Rev. Chil. Hist. Nat. 2019, 92, 6. [Google Scholar] [CrossRef]
- Rutjes, S.A.; Lodder-Verschoor, F.; Lodder, W.J.; Van Der Giessen, J.; Reesink, H.; Bouwknegt, M.; De Roda Husman, A.M. Seroprevalence and Molecular Detection of Hepatitis E Virus in Wild Boar and Red Deer in The Netherlands. J. Virol. Methods 2010, 168, 197–206. [Google Scholar] [CrossRef]
- Bergner, L.M.; Orton, R.J.; Da Silva Filipe, A.; Shaw, A.E.; Becker, D.J.; Tello, C.; Biek, R.; Streicker, D.G. Using Noninvasive Metagenomics to Characterize Viral Communities from Wildlife. Mol. Ecol. Resour. 2019, 19, 128–143. [Google Scholar] [CrossRef]
- Van Den Brand, J.M.A.; Van Leeuwen, M.; Schapendonk, C.M.; Simon, J.H.; Haagmans, B.L.; Osterhaus, A.D.M.E.; Smits, S.L. Metagenomic Analysis of the Viral Flora of Pine Marten and European Badger Feces. J. Virol. 2012, 86, 2360–2365. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.G.; Kapusinszky, B.; Wang, C.; Rose, R.K.; Lipton, H.L.; Delwart, E.L. The Fecal Viral Flora of Wild Rodents. PLoS Pathog. 2011, 7, e1002218. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Martínez, L.A.; Loza-Rubio, E.; Mosqueda, J.; González-Garay, M.L.; García-Espinosa, G. Fecal Virome Composition of Migratory Wild Duck Species. PLoS ONE 2018, 13, e0206970. [Google Scholar] [CrossRef] [PubMed]
- Ao, Y.; Xu, J.; Duan, Z. A Novel Cardiovirus Species Identified in Feces of Wild Himalayan Marmots. Infect. Genet. Evol. 2022, 103, 105347. [Google Scholar] [CrossRef]
- Huaman, J.L.; Pacioni, C.; Sarker, S.; Doyle, M.; Forsyth, D.M.; Pople, A.; Carvalho, T.G.; Helbig, K.J. Novel Picornavirus Detected in Wild Deer: Identification, Genomic Characterisation, and Prevalence in Australia. Viruses 2021, 13, 2412. [Google Scholar] [CrossRef]
- Vibin, J.; Chamings, A.; Collier, F.; Klaassen, M.; Nelson, T.M.; Alexandersen, S. Metagenomics Detection and Characterisation of Viruses in Faecal Samples from Australian Wild Birds. Sci. Rep. 2018, 8, 8686. [Google Scholar] [CrossRef]
- Reuter, G.; Boros, Á.; Földvári, G.; Szekeres, S.; Mátics, R.; Kapusinszky, B.; Delwart, E.; Pankovics, P. Dicipivirus (Family Picornaviridae) in Wild Northern White-Breasted Hedgehog (Erinaceus roumanicus). Arch. Virol. 2018, 163, 175–181. [Google Scholar] [CrossRef]
- Yang, S.; Liu, D.; Wang, Y.; Qu, F.; He, Y.; Sun, Z.; Shen, Q.; Li, W.; Fu, X.; Deng, X.; et al. Bufavirus Protoparvovirus in Feces of Wild Rats in China. Virus Genes. 2016, 52, 130–133. [Google Scholar] [CrossRef]
- Bodewes, R.; Van Der Giessen, J.; Haagmans, B.L.; Osterhaus, A.D.M.E.; Smits, S.L. Identification of Multiple Novel Viruses, Including a Parvovirus and a Hepevirus, in Feces of Red Foxes. J. Virol. 2013, 87, 7758–7764. [Google Scholar] [CrossRef]
- Bradley, C.A.; Altizer, S. Urbanization and the Ecology of Wildlife Diseases. Trends Ecol. Evol. 2007, 22, 95–102. [Google Scholar] [CrossRef]
- Bateman, P.W.; Fleming, P.A. Big City Life: Carnivores in Urban Environments. J. Zool. 2012, 287, 1–23. [Google Scholar] [CrossRef]
- Garcês, A.; Pires, I. Secrets of the Astute Red Fox (Vulpes Vulpes, Linnaeus, 1758): An Inside-Ecosystem Secret Agent Serving One Health. Environments 2021, 8, 103. [Google Scholar] [CrossRef]
- Corsolini, S.; Burrini, L.; Focardi, S.; Lovari, S. How Can We Use the Red Fox as a Bioindicator of Organochlorines? Arch. Environ. Contam. Toxicol. 2000, 39, 547–556. [Google Scholar] [CrossRef]
- Kalisińska, E.; Palczewska-Komsa, M. Teeth of the Red Fox Vulpes Vulpes (L., 1758) as a Bioindicator in Studies on Fluoride Pollution. Acta Theriol. 2011, 56, 343–351. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mo, S.S.; Urdahl, A.M.; Madslien, K.; Sunde, M.; Nesse, L.L.; Slettemeås, J.S.; Norström, M. What Does the Fox Say? Monitoring Antimicrobial Resistance in the Environment Using Wild Red Foxes as an Indicator. PLoS ONE 2018, 13, e0198019. [Google Scholar] [CrossRef]
- Campbell, S.J.; Ashley, W.; Gil-Fernandez, M.; Newsome, T.M.; Di Giallonardo, F.; Ortiz-Baez, A.S.; Mahar, J.E.; Towerton, A.L.; Gillings, M.; Holmes, E.C.; et al. Red Fox Viromes in Urban and Rural Landscapes. Virus Evol. 2020, 6, veaa065. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.I.; Blanton, J.D.; Gilbert, A.; Castrodale, L.; Hueffer, K.; Slate, D.; Rupprecht, C.E. A Conceptual Model for the Impact of Climate Change on Fox Rabies in Alaska, 1980–2010. Zoonoses Public Health 2014, 61, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Donato, C.; Vijaykrishna, D. The Broad Host Range and Genetic Diversity of Mammalian and Avian Astroviruses. Viruses 2017, 9, 102. [Google Scholar] [CrossRef]
- Koci, M.D.; Schultz-Cherry, S. Avian Astroviruses. Avian Pathol. 2002, 31, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Sun, M.; Liao, M. A Review of Emerging Goose Astrovirus Causing Gout. BioMed Res. Int. 2022, 2022, 1635373. [Google Scholar] [CrossRef]
- Kofstad, T.; Jonassen, C.M. Screening of Feral and Wood Pigeons for Viruses Harbouring a Conserved Mobile Viral Element: Characterization of Novel Astroviruses and Picornaviruses. PLoS ONE 2011, 6, e25964. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, Y.; Li, H.; Chen, Z.; Zhou, J.; Liu, G.; Wang, Y. Genomic Characterization and Phylogenetic Analysis of a New Canine Picornavirus Variant in the Mainland of China. Virus Res. 2021, 296, 198351. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Choi, G.K.Y.; Yip, C.C.Y.; Huang, Y.; Tsoi, H.-W.; Yuen, K.-Y. Complete Genome Sequence of a Novel Picornavirus, Canine Picornavirus, Discovered in Dogs. J. Virol. 2012, 86, 3402–3403. [Google Scholar] [CrossRef]
- Yang, S.; He, Y.; Chen, X.; Kalim, U.; Wang, Y.; Yang, S.; Qi, H.; Cheng, H.; Lu, X.; Wang, X.; et al. Viral Metagenomics Reveals Diverse Viruses in the Feces Samples of Raccoon Dogs. Front. Vet. Sci. 2021, 8, 693564. [Google Scholar] [CrossRef]
- Raue, R.; Schmidt, V.; Freick, M.; Reinhardt, B.; Johne, R.; Kamphausen, L.; Kaleta, E.F.; Müller, H.; Krautwald-Junghanns, M.-E.; Diger Raue, R.; et al. A Disease Complex Associated with Pigeon Circovirus Infection, Young Pigeon Disease Syndrome. Avian Pathol. 2007, 34, 418–425. [Google Scholar] [CrossRef]
- Silva, B.B.I.; Urzo, M.L.R.; Encabo, J.R.; Simbulan, A.M.; Lunaria, A.J.D.; Sedano, S.A.; Hsu, K.-C.; Chen, C.-C.; Tyan, Y.-C.; Chuang, K.-P. Pigeon Circovirus over Three Decades of Research: Bibliometrics, Scoping Review, and Perspectives. Viruses 2022, 14, 1498. [Google Scholar] [CrossRef] [PubMed]
- Franciosini, M.P.; Fringuelli, E.; Tarhuni, O.; Guelfi, G.; Todd, D.; Proietti, P.C.; Falocci, N.; Asdrubali, G. Development of a Polymerase Chain Reaction-Based In Vivo Method in the Diagnosis of Subclinical Pigeon Circovirus Infection. Avian Dis. 2005, 49, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Todd, D.; Fringuelli, E.; Scott, A.N.J.; Borghmans, B.J.; Duchatel, J.P.; Shivaprasad, H.L.; Raidal, S.R.; Abadie, J.X.; Franciosini, M.P.; Smyth, J.A. Sequence Comparison of Pigeon Circoviruses. Res. Vet. Sci. 2008, 84, 311–319. [Google Scholar] [CrossRef]
- Hayward, J.A.; Tachedjian, G. Retroviruses of Bats: A Threat Waiting in the Wings? mBio 2021, 12, e0194121. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Tachedjian, M.; Wang, L.; Tachedjian, G.; Wang, L.-F.; Zhang, S. Discovery of Retroviral Homologs in Bats: Implications for the Origin of Mammalian Gammaretroviruses. J. Virol. 2012, 86, 4288–4293. [Google Scholar] [CrossRef]
- Cui, J.; Tachedjian, G.; Wang, L.-F. Bats and Rodents Shape Mammalian Retroviral Phylogeny. Sci. Rep. 2015, 5, 16561. [Google Scholar] [CrossRef]
- Tomonaga, K.; Coffin, J.M. Structure and Distribution of Endogenous Nonecotropic Murine Leukemia Viruses in Wild Mice. J. Virol. 1998, 72, 8289–8300. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, O.E.; Belák, S.; Granberg, F. The Effect of Preprocessing by Sequence-Independent, Single-Primer Amplification (SISPA) on Metagenomic Detection of Viruses. Biosecurity Bioterrorism Biodefense Strategy Pract. Sci. 2013, 11, S227–S234. [Google Scholar] [CrossRef] [PubMed]

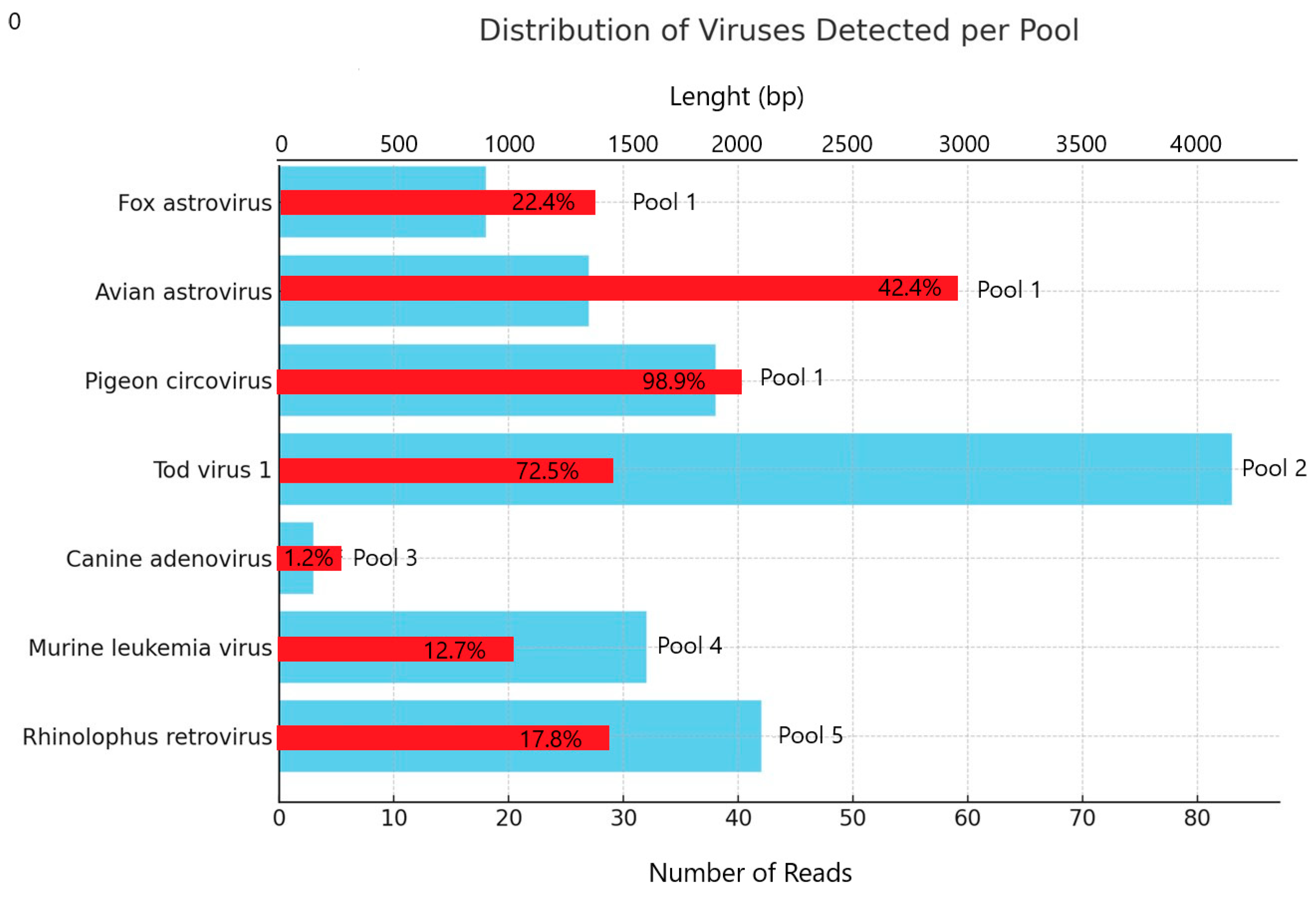
| Pool Number | N° of Sequences | Classified Sequences * (%) | Unclassified Sequences * (%) | Bacteria Sequences * (%) | Viral Sequences * (%) |
|---|---|---|---|---|---|
| 1 | 2.54 × 107 | 43 | 57 | 83 | 0.6 |
| 2 | 2.58 × 107 | 29 | 71 | 86 | 1.7 |
| 3 | 2.03 × 107 | 46 | 54 | 95 | 0.6 |
| 4 | 5.35 × 107 | 89 | 11 | 97 | 1.3 |
| 5 | 3.91 × 107 | 59 | 41 | 89 | 1.0 |
| 6 | 1.90 × 107 | 48 | 52 | 80 | 0.4 |
| Host | Virus | Ref-Seq | n° Reads | Length (bp) | Accession Number Contig > 150 bp | Ref-Seq Coverage | Identical Sites | Pairwise Identity | E Value |
|---|---|---|---|---|---|---|---|---|---|
| Pool 1 Vulpes vulpes | Fox astrovirus | KC692365.1 | 18 | 1443 | PV999255 PV999256 | 1443/6456 22.4% | 100% | 100% | 0.0 |
| Avian astrovirus | MF768270 | 27 | 2917 | PV999257 PV999258 PV999259 | 2917/6872 42.4% | 90.7% | 96.1% | 0.0 | |
| Pigeon circovirus | MW656109 | 38 | 2015 | PX067712 | 2015/2038 98.9% | 92.4% | 96.4% | 0.0 | |
| Pool 2 Vulpes vulpes | Tod virus (Canine picodicistrovirus) | MT833880 | 83 | 1492 | PV999260 PV999261 PV999262 | 1492/2058 72.5% | 88.6% | 97% | 0.0 |
| Pool 3 Vulpes vulpes | Canine adenovirus | Y07760.1 | 3 | 361 | PX067713 | 361/30,536 1.2% | 100% | 100% | 0.0 |
| Pool 4 Meles meles | No significant viral sequences detected | ||||||||
| Pool 5 Meles meles | Murine leukemiavirus | KY574516 | 32 | 1039 | PV999263 PV999264 | 1039/8191 12.7% | 90.4% | 93.1% | 0.0 |
| Rhinolophus ferrumequinum retrovirus | JQ303225 | 42 | 1493 | PV999265 PV999266 PV999267 PV999268 | 1493/8389 17.8% | 98.4% | 99.7% | 0.0 | |
| Pool 6 Hystrix cristata | No significant viral sequences detected | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pacini, M.I.; Forzan, M.; Sgorbini, M.; Cingottini, D.; Mazzei, M. Metagenomic Analysis of the Fecal Virome in Wild Mammals Hospitalized in Pisa, Italy. Vet. Sci. 2025, 12, 820. https://doi.org/10.3390/vetsci12090820
Pacini MI, Forzan M, Sgorbini M, Cingottini D, Mazzei M. Metagenomic Analysis of the Fecal Virome in Wild Mammals Hospitalized in Pisa, Italy. Veterinary Sciences. 2025; 12(9):820. https://doi.org/10.3390/vetsci12090820
Chicago/Turabian StylePacini, Maria Irene, Mario Forzan, Micaela Sgorbini, Dania Cingottini, and Maurizio Mazzei. 2025. "Metagenomic Analysis of the Fecal Virome in Wild Mammals Hospitalized in Pisa, Italy" Veterinary Sciences 12, no. 9: 820. https://doi.org/10.3390/vetsci12090820
APA StylePacini, M. I., Forzan, M., Sgorbini, M., Cingottini, D., & Mazzei, M. (2025). Metagenomic Analysis of the Fecal Virome in Wild Mammals Hospitalized in Pisa, Italy. Veterinary Sciences, 12(9), 820. https://doi.org/10.3390/vetsci12090820





