Escherichia coli Strains Originating from Raw Sheep Milk, with Special Reference to Their Genomic Characterization, Such as Virulence Factors (VFs) and Antimicrobial Resistance (AMR) Genes, Using Whole-Genome Sequencing (WGS)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbial Dataset
2.2. Whole-Genome Sequencing and Assembly
2.3. Bioinformatic Analysis
2.4. Antimicrobial Susceptibility Testing
3. Results and Discussion
3.1. Genome Assembly and Annotation
3.2. Phylogenetic Analysis and Genotyping
3.3. Pangenome Analysis
3.4. Antimicrobial Resistance
3.5. Virulence Factors and Mobile Genetic Elements
- i.
- All strains were negative for stx (shigatoxin—Shiga Toxin-producing E. coli (STEC) strains or EHEC), LAA PAI (Locus of Adhesion and Autoaggregation Pathogenicity Island), primarily found in LEE-negative (Locus of Enterocyte Effacement) STEC strains, estA (the gene encodes a heat-stable enterotoxin in ETEC), eltAB (the genes encode the A and B subunits of a heat-labile enterotoxin in ETEC), and pEAF/bfp [EPEC adherence factor plasmid, bundle-forming pilus operon, and plasmid-encoded regulator (perABC) gene cluster, which constitutes the adherence factor in typical EPEC].
- ii.
- All strains were eae-positive (the adhesion factor of EPEC and EHEC). The intimin-encoding gene eae is crucial in the production of the attaching and effacing (A/E) lesions (LEE PAI). The esp genes, such as espL, espR, espX, and espY, which encode proteins that are secreted by the Type III secretion system and are involved in various steps of the infection process, including attaching to and damaging host cells, were also identified. These genes are crucial for the virulence of EPEC and EHEC.
- iii.
- The LEE PAI was partially identified in all strains; it is found in EPEC and EHEC. The PAI plays an important role in the attachment and effacing (A/E) lesion formation on intestinal epithelial cells. Partially identified means that the PAI was not completely detected (only 15 to 39% of the PAI was identified). The examined genomes were not complete but in contigs/scaffolds, meaning that several genes could be fragmented and, therefore, difficult to be detected and recorded by the program, as the identification value was far below the software’s threshold.
- iv.
- The strains S3, S11, S22, S37, S45, and S50 were positive to the presence of the ETT2 PAI and its regulator (etrA) found in EHEC and STEC, but also in atypical EPEC, EAEC, and ExPEC strains. The PAI encodes a type III secretion system (T3SS) known as ETT2 T3SS, which is involved in the production of several effectors and regulatory proteins but also extends its function by affecting the expression of other virulence genes outside the PAI [69,70,71].
- v.
- The ETT2-negative strains (S4, S24, S30, S33, and S35) possessed alternative mechanisms of adherence and aggregation such as the fim gene cluster (fimABCDEFGHI), papCD genes, and focC gene (except for S4).
- vi.
- All strains were astA-positive (the gene encodes a heat-stable enterotoxin in EHEC, EAEC, and atypical EPEC).
- vii.
- All strains shared ExPEC-like genetic determinants such as ybtP (iron), irp1 and irp2 (invasins), hlyE (toxin), fimF, fimG, fimH, yagVWXYZ/ecpABCDE, ykgK/ecpR, papCD and focC (adhesins), and ompA, ompC, ompD, ompF, ompG, and ompT (serum resistance proteins).
- viii.
- The strains S24, S30, S33, and S35 were aatA-positive (the gene encodes a dispersing protein).
- ix.
- The strain S3 was PAI IV- and HPI-positive. The first PAI contains various VFs related to inflammation, adhesion, colonization, and protein secretion (type I secretion system—T1SS). This PAI is characterized by its proximity to tRNA-encoding genes and the presence of integrase which facilitates its movement within the prokaryotic genome and/or between other microbes through horizontal gene transfer [72]. The second PAI is involved in iron uptake through the production of a siderophore (yersiniabactin), enhanced autophagy, and other virulence mechanisms (flagellum-mediated motility) [73].
- x.
- The invasin ibeB, detected in all strains, plays a crucial role in bacterial invasion. It is frequently associated with other VFs such as ibeA (the gene was not detected) and ompA, causing tissue penetration, including of host cells coating the blood–brain barrier, indicating the high pathogenicity potential of the strains [10].
| Strain ID | Virulence Islands 1 | Iron | Protease | Adhesins | Invasins | Toxins |
|---|---|---|---|---|---|---|
| S3 | PAI IV (68.42%), HPI (66.67%), EET2 (70.27%) plus etrA, and LEE (21.95%) plus eae | fecA-E 2, ybtP | ompACDFGT | fimA-I, fdeC, yagV-Z/ecpA-E, ykgK/ecpR, nlpADEI, yehA-D, lpfA, papCD, and espLRXY | csgA-G, aslA, fyuA, gadBCEWX, ibeB, irp1, irp2, and hha | hlyE and astA |
| S4 | LEE (21.95%) plus eae | fecA-E, ybtP | ompACDFGT | fimA-I, fdeC, yagV-Z/ecpA-E, ykgK/ecpR, nlpADEI, yehA-D, lpfA, papCD, and espLRXY | csgA-G, aslA, fyuA, gadBCEWX, ibeB, irp1, irp2, and hha | hlyE and astA |
| S11 | EET2 (97.30%) plus etrA and LEE (39.02%) plus eae | - | ompACDFG | fimA-I, fdeC, yagV-Z/ecpA-E, ykgK/ecpR, nlpADEI, yehA-D, lpfA, papCD, focC, and espLRXY | csgA-G, aslA, chuU-W, gadBCEWX, ibeB, hha, and tra | hlyE and astA |
| S22 | EET2 (70.27%) plus etrA and LEE (14.63%) plus eae | - | ompACDFG | aaiADF, fimA-I, fdeC, faeC-G, yagV-Z/ecpA-E, ykgK/ecpR, nlpADEI, yehA-D, papCD, focC, upaG/ehaG, and espLRXY | csgA-G, gadBCEWX, ibeB, hha, and tra | hlyE and astA |
| S24 | LEE (17.07%) plus eae | fecA-E | ompACDFGT | aatA, fimA-I, nlpADEI, yehA-D, lpfA, papCD, focC, and espLRXY | csgA-G, gadBCEWX, ibeB, hha, and tra | hlyE and astA |
| S30 | LEE (17.07%) plus eae | fecA-E | ompACDFGT | aatA, fimA-I, nlpADEI, yehA-D, lpfA, papCD, focC, and espLRXY | csgA-G, gadBCEWX, ibeB, hha, and tra | hlyE and astA |
| S33 | LEE (17.07%) plus eae | fecA-E | ompACDFGT | aatA, fimA-I, nlpADEI, yehA-D, lpfA, papCD, focC, and espLRXY | csgA-G, gadBCEWX, ibeB, hha, and tra | hlyE and astA |
| S35 | LEE (17.07%) plus eae | fecA-E | ompACDFGT | aatA, fimA-I, nlpADEI, yehA-D, lpfA, papCD, focC, and espLRXY | csgA-G, gadBCEWX, ibeB, hha, and tra | hlyE and astA |
| S37 | EET2 (70.27%) plus etrA and LEE (17.07%) plus eae | fecA-E | ompACDFG | fimA-I, fdeC, yagV-Z/ecpA-E, ykgK/ecpR, nlpADEI, yehA-D, lpfA, papCD, upaG/ehaG, and espLRXY | csgA-G, aslA, gadBCEWX, ibeB, hha, and tra | hlyE and astA |
| S45 | EET2 (70.27%) plus etrA and LEE (17.07%) plus eae | fecA-E | ompACDFG | fimA-I, fdeC, yagV-Z/ecpA-E, ykgK/ecpR, nlpADEI, yehA-D, lpfA, papCD, upaG/ehaG, and espLRXY | csgA-G, aslA, gadBCEWX, ibeB, and hha | hlyE and astA |
| S50 | EET2 (70.27%) plus etrA and LEE (17.07%) plus eae | fecA-E | ompACDFG | fimA-I, fdeC, yagV-Z/ecpA-E, ykgK/ecpR, nlpADEI, yehA-D, lpfA, papCD, upaG/ehaG, and espLRXY | csgA-G, aslA, gadBCEWX, ibeB, and hha | hlyE and astA |
- pEAF/bfp—negative: The EPEC adherence factor (EAF) plasmid and bfpA gene are both absent in aEPEC, which encode the bundle-forming pili, a protein involved in localized adherence to host cells.
- eae—positive: The aEPEC, similar to tEPEC, harbors the eae gene, which is a key gene for the formation of A/E lesions, a crucial virulence factor of EPEC.
- stx—negative: The presence of stx gene is the trademark of STEC. Although some EPEC may possess the shiga toxin gene, the aEPEC do not produce shiga toxins.
- Genetic similarity to STEC: The aEPEC are genetically closer to STEC than tEPEC, showing similarities in serotypes and other epidemiological aspects. The five serotypes identified in this work (O179:H40, O169:H46, O18ac:H7, ONT:H26, and O107:H27), all were STEC serotypes.
- Diversity in genetic background: The aEPEC displays genetic diversity with some strains exhibiting closer relationships to other E. coli pathotypes such as ETEC or ExPEC (Figure 1b).
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| VFs | Virulence Factors |
| AMR | Antimicrobial Resistance |
| CNS | Coagulase-Negative Staphylococci |
| SCM | Subclinical Mastitis |
| CM | Clinical Mastitis |
| STs | Sequence Types |
| MEcS | Mastitis Escherichia coli Strains |
| MGEs | Mobile Genetic Elements |
| IPEC | Intestinal Pathogenic Escherichia coli |
| ExPEC | Extraintestinal Pathogenic Escherichia coli |
| EAEC | Enteroaggregative Escherichia coli |
| EPEC | Enteropathogenic Escherichia coli |
| EIEC | Enteroinvasive Escherichia coli |
| EHEC | Enterohemorrhagic Escherichia coli |
| ETEC | Enterotoxigenic Escherichia coli |
| STEC | Shiga Toxin-producing Escherichia coli strains |
| BHI | Brain Heart Infusion |
| SCC | Somatic Cell Count |
| EUCAST | European Committee on Antimicrobial Susceptibility Testing |
| CLSI | Clinical and Laboratory Standards Institute |
| AST | Antimicrobial Susceptibility Testing |
| ANI | Average Nucleotide Identity |
| MLST | Multi-Locus Sequence Typing |
| cgMLST | core-genome Multi-Locus Sequence Typing |
| CGE | Center for Genomic Epidemiology |
| iTOL | Interactive Tree of Life |
| ARGANNOT | Antibiotic Resistance Gene Annotation |
| CARD | Comprehensive Antibiotic Resistance Database |
| EcOH | Escherichia coli O-groups and H-types |
| NCBI | National Center for Biotechnology Information |
| TYGS | Type (Strain) Genome Server |
| GTDB-Tk | Genome Taxonomy Database Toolkit |
| MEGARes | Microbial Ecology Group Antimicrobial Resistances |
| IPGA | Integrated Prokaryotes Genome and pan-genome Analysis service |
| ARGs | Antimicrobial Resistance Genes |
| WGS | Whole-Genome Sequencing |
| CDS | Coding DNA Sequence |
| rRNA | Ribosomal RNA |
| tRNA | Transfer RNA |
| tmRNA | Transfer-Messenger RNA |
| dDDH | digital DNA–DNA Hybridization |
| BRIG | BLAST Ring Image Generator |
| BLAST | Basic Local Alignment Search Tool |
| COGs | Clusters of Orthologous Groups/Genes |
| SNPs | Single Nucleotide Polymorphisms |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| ESBL | Extended-Spectrum Beta-Lactamase |
| MFS | Major Facilitator Superfamily |
| LEE | Locus of Enterocyte Effacement |
| LAA | Locus of Adhesion and Autoaggregation |
| PAI | Pathogenicity Island |
References
- dos Reis, C.B.M.; Barreiro, J.R.; Moreno, J.F.G.; Porcionato, M.A.F.; Santos, M.V. Evaluation of somatic cell count thresholds to detect subclinical mastitis in Gyr cows. J. Dairy Sci. 2011, 94, 4406–4412. [Google Scholar] [CrossRef] [PubMed]
- Dairy-Cattle. Escherichia coli—A Practical Summary for Controlling Mastitis. 2019. Available online: https://dairy-cattle.extension.org/escherichia-coli-a-practical-summary-for-controlling-mastitis/ (accessed on 3 April 2025).
- Olson, M.A.; Cullimore, C.; Hutchison, W.D.; Grimsrud, A.; Nobrega, D.; De Buck, J.; Barkema, H.W.; Wilson, E.; Pickett, B.E.; Erickson, D.L. Genes associated with fitness and disease severity in the pan-genome of mastitis-associated Escherichia coli. Front. Microbiol. 2024, 15, 1452007. [Google Scholar] [CrossRef] [PubMed]
- Blum, S.E.; Leitner, G. Genotyping and virulence factors assessment of bovine mastitis Escherichia coli. Vet. Microbiol. 2013, 163, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Keane, O.M. Genetic diversity, the virulence gene profile and antimicrobial resistance of clinical mastitis-associated Escherichia coli. Res. Microbiol. 2016, 167, 678–684. [Google Scholar] [CrossRef]
- Kempf, F.; Slugocki, C.; Blum, S.E.; Leitner, G.; Germon, P. Genomic Comparative Study of Bovine Mastitis Escherichia coli. PLoS ONE 2016, 11, e0147954. [Google Scholar] [CrossRef] [PubMed]
- Nüesch-Inderbinen, M.; Käppeli, N.; Morach, M.; Eicher, C.; Corti, S.; Stephan, R. Molecular types, virulence profiles and antimicrobial resistance of Escherichia coli causing bovine mastitis. Vet. Rec. Open. 2019, 17, e000369. [Google Scholar] [CrossRef]
- Leimbach, A.; Poehlein, A.; Vollmers, J.; Gorlich, D.; Daniel, R.; Dobrindt, U. No evidence for a bovine mastitis Escherichia coli pathotype. BMC Genom. 2017, 18, 359. [Google Scholar] [CrossRef]
- Balbuena-Alonso, M.G.; Cortés-Cortés, G.; Kim, J.W.; Lozano-Zarain, P.; Camps, M.; Rocha-Gracia, R.d.C. Genomic analysis of plasmid content in food isolates of E. coli strongly supports its role as a reservoir for the horizontal transfer of virulence and antibiotic resistance genes. Plasmid 2022, 123–124, 102650. [Google Scholar] [CrossRef]
- Orsi, H.; Guimarães, F.F.; Leite, D.S.; Guerra, S.T.; Joaquim, S.F.; Pantoja, J.C.F.; Hernandes, R.T.; Lucheis, S.B.; Ribeiro, M.G.; Langoni, H.; et al. Characterization of mammary pathogenic Escherichia coli reveals the diversity of Escherichia coli isolates associated with bovine clinical mastitis in Brazil. J. Dairy Sci. 2023, 106, 1403–1413. [Google Scholar] [CrossRef]
- Mora, A.; Herrrera, A.; López, C.; Dahbi, G.; Mamani, R.; Pita, J.M.; Alonso, M.P.; Llovo, J.; Bernárdez, M.I.; Blanco, J.E.; et al. Characteristics of the Shiga-toxin-producing enteroaggregative Escherichia coli O104:H4 German outbreak strain and of STEC strains isolated in Spain. Int. Microbiol. 2011, 14, 121–141. [Google Scholar] [CrossRef]
- Lindstedt, B.-A.; Finton, M.D.; Porcellato, D.; Brandal, L.T. High frequency of hybrid Escherichia coli strains with combined Intestinal Pathogenic Escherichia coli (IPEC) and Extraintestinal Pathogenic Escherichia coli (ExPEC) virulence factors isolated from human faecal samples. BMC Infect. Dis. 2018, 18, 544. [Google Scholar] [CrossRef]
- Mangroliya, D.; Adhyaru, H.; Kabariya, J.; Ramani, V. Genomic insights into plasmid mediated AMR genes, virulence factors and mobile genetic elements in raw milk Escherichia coli from Gujarat, India. Sci. Rep. 2025, 15, 6320. [Google Scholar] [CrossRef] [PubMed]
- Sarba, E.J.; Wirtu, W.; Gebremedhin, E.Z.; Borena, B.M.; Marami, L.M. Occurrence and antimicrobial susceptibility patterns of Escherichia coli and Escherichia coli O157 isolated from cow milk and milk products, Ethiopia. Sci. Rep. 2023, 13, 16018. [Google Scholar] [CrossRef] [PubMed]
- Bello, A.; Ning, S.; Zhang, Q.; Hu, S. Genomic analysis of multidrug-resistant Escherichia coli isolated from dairy cows in Shihezi city, Xinjiang, China. Front. Microbiol. 2025, 16, 1527546. [Google Scholar] [CrossRef] [PubMed]
- Syrokou, M.K.; Paramithiotis, S.; Skandamis, P.N.; Drosinos, E.H.; Bosnea, L.; Mataragas, M. High-quality draft genome sequence data of six Lactiplantibacillus plantarum subsp. argentoratensis strains isolated from various Greek wheat sourdoughs. Data Br. 2021, 37, 107172. [Google Scholar] [CrossRef]
- Apostolakos, I.; Skarlatoudi, T.; Vatavali, K.; Giannouli, A.; Bosnea, L.; Mataragas, M. Genomic and phenotypic characterization of mastitis-causing staphylococci and probiotic lactic acid bacteria isolated from raw sheep’s milk. Int. J. Mol. Sci. 2023, 24, 13883. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Lee, I.; Ouk Kim, Y.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Cosentino, S.; Voldby Larsen, M.; Møller Aarestrup, F.; Lund, O. PathogenFinder—Distinguishing friend from foe using bacterial Whole Genome Sequence data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef]
- Seemann, T. Abricate, Github. 2020. Available online: https://github.com/tseemann/abricate (accessed on 12 April 2024).
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Bharat, A.; Petkau, A.; Avery, B.P.; Chen, J.C.; Folster, J.P.; Carson, C.A.; Kearney, A.; Nadon, C.; Mabon, P.; Thiessen, J.; et al. Correlation between phenotypic and in silico detection of antimicrobial resistance in Salmonella enterica in Canada using staramr. Microorganisms 2022, 10, 292. [Google Scholar] [CrossRef]
- Sherry, N.L.; Horan, K.A.; Ballard, S.A.; da Silva, A.G.; Gorrie, C.L.; Schultz, M.B.; Stevens, K.; Valcanis, M.; Sait, M.L.; Stinear, T.P.; et al. An ISO-certified genomics workflow for identification and surveillance of antimicrobial resistance. Nat. Commun. 2023, 14, 60. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef]
- Ingle, D.J.; Valcanis, M.; Kuzevski, A.; Tauschek, M.; Inouye, M.; Stinear, T.; Levine, M.M.; Robins-Browne, R.M.; Holt, K.E. In silico serotyping of E. coli from short read data identifies limited novel O-loci but extensive diversity of O:H serotype combinations within and between pathogenic lineages. Microb. Genom. 2016, 2, e000064. [Google Scholar] [CrossRef]
- Public Health Agency of Canada—National Microbiology Laboratory. Escherichia coli Virulence Factors, Github 2017. Available online: https://github.com/phac-nml/ecoli_vf (accessed on 12 April 2024).
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Bonin, N.; Doster, E.; Worley, H.; Pinnell, L.J.; Bravo, J.E.; Ferm, P.; Marini, S.; Prosperi, M.; Noyes, N.; Morley, P.S.; et al. MEGARes and AMR++, v3.0: An updated comprehensive database of antimicrobial resistance determinants and an improved software pipeline for classification using high-throughput sequencing. Nucleic Acids Res. 2023, 51, D744–D752. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garciá-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In silico detection and typing of plasmids using Plasmidfinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Roer, L.; Tchesnokova, V.; Allesoe, R.; Muradova, M.; Chattopadhyay, S.; Ahrenfeldt, J.; Thomsen, M.C.F.; Lund, O.; Hansen, F.; Hammerum, A.M.; et al. Development of a web tool for Escherichia coli subtyping based on fimH alleles. J. Clin. Microbiol. 2017, 55, 2538–2543. [Google Scholar] [CrossRef]
- Roer, L.; Johannesen, T.B.; Hansen, F.; Stegger, M.; Tchesnokova, V.; Sokurenko, E.; Garibay, N.; Allesøe, R.; Thomsen, M.C.F.; Lund, O.; et al. CHTyper, a web tool for subtyping of extraintestinal pathogenic Escherichia coli based on the fumC and fimH alleles. J. Clin. Microbiol. 2018, 56, e00063-18. [Google Scholar] [CrossRef]
- Joensen, K.G.; Tetzschner, A.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and easy in silico serotyping of Escherichia coli using whole genome sequencing (WGS) data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; the Agama Study Group; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pangenome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Gardner, S.N.; Slezak, T.; Hall, B.G. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 2015, 31, 2877–2878. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Beghain, J.; Bridier-Nahmias, A.; Le Nagard, H.; Denamur, E.; Clermont, O. Clermontyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 2018, 4, e000192. [Google Scholar] [CrossRef]
- Clermont, O.; Dixit, O.V.A.; Vangchhia, B.; Condamine, B.; Dion, S.; Bridier-Nahmias, A.; Denamur, E.; Gordon, D. Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ. Microbiol. 2019, 21, 3107–3117. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, Y.; Fan, G.; Sun, D.; Zhang, X.; Yu, Z.; Wang, J.; Wu, L.; Shi, W.; Ma, J. IPGA: A handy integrated prokaryotes genome and pan-genome analysis web service. iMeta 2022, 1, e55. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Mendes, I.; Griffiths, E.; Manuele, A.; Fornika, D.; Tausch, S.H.; Le-Viet, T.; Phelan, J.; Meehan, C.J.; Raphenya, A.R.; Alcock, B.; et al. hAMRonization: Enhancing antimicrobial resistance prediction using the PHA4GE AMR detection specification and tooling. bioRxiv 2024. [Google Scholar] [CrossRef]
- Steinig, E.; Wirth, W. Brick, Github 2024. Available online: https://github.com/esteinig/brick (accessed on 5 May 2025).
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Camargo, A.P.; Roux, S.; Schulz, F.; Babinski, M.; Xu, Y.; Hu, B.; Chain, P.S.G.; Nayfach, S.; Kyrpides, N.C. Identification of mobile genetic elements with geNomad. Nat. Biotechnol. 2024, 42, 1303–1312. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 1 March 2025).
- Sitter, T.L.; Vaughan, A.L.; Schoof, M.; Jackson, S.A.; Glare, T.R.; Cox, M.P.; Fineran, P.C.; Gardner, P.P.; Hurst, M.R.H. Evolution of virulence in a novel family of transmissible mega-plasmids. Environ. Microbiol. 2021, 23, 5289–5304. [Google Scholar] [CrossRef]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef]
- Chen, T.; Liu, Y.-X.; Chen, T.; Yang, M.; Fan, S.; Shi, M.; Wei, B.; Lv, H.; Cao, W.; Wang, C.; et al. ImageGP 2 for enhanced data visualization and reproducible analysis in biomedical research. iMeta 2024, 3, e239. [Google Scholar] [CrossRef]
- Higgins, J.; Hohn, C.; Hornor, S.; Frana, M.; Denver, M.; Joerger, R. Genotyping of Escherichia coli from environmental and animal samples. J. Microbiol. Methods 2007, 70, 227–235. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, Z.; Huang, C.; Gao, X.; Wang, Z.; Liu, Y.; Tian, C.; Hong, W.; Niu, S.; Liu, M. The phylogenetic group, antimicrobial susceptibility, and virulence genes of Escherichia coli from clinical bovine mastitis. J. Dairy Sci. 2018, 101, 572–580. [Google Scholar] [CrossRef]
- Wu, G.; Ehricht, R.; Mafura, M.; Stokes, M.; Smith, N.; Pritchard, G.C.; Woodward, M.J. Escherichia coli isolates from extraintestinal organs of livestock animals harbour diverse virulence genes and belong to multiple genetic lineages. Vet. Microbiol. 2012, 160, 197–206. [Google Scholar] [CrossRef]
- Koita, K.; Rao, C.V. Identification and analysis of the putative pentose sugar efflux transporters in Escherichia coli. PLoS ONE 2012, 7, e43700. [Google Scholar] [CrossRef]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, X.; Xu, X.; Zhao, Y.; Yang, D.; Han, X.; Tian, M.; Ding, C.; Peng, D.; Yu, S. Escherichia coli type III secretion system 2 (ETT2) is widely distributed in avian pathogenic Escherichia coli isolates from Eastern China. Epidemiol. Infect. 2016, 144, 2824–2830. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhu, H.; Hu, J.; Zhang, B.; Guo, W.; Wang, Z.; Wang, D.; Qi, J.; Tian, M.; Bao, Y.; et al. Genetic distribution, characterization, and function of Escherichia coli type III secretion system 2 (ETT2). iScience 2024, 27, 109763. [Google Scholar] [CrossRef] [PubMed]
- Shulman, A.; Yair, Y.; Biran, D.; Sura, T.; Otto, A.; Gophna, U.; Becher, D.; Hecker, M.; Ron, E.Z. The Escherichia coli type III secretion system 2 has a global effect on cell surface. mBio 2018, 9, e01070-18. [Google Scholar] [CrossRef]
- Gal-Mor, O.; Finlay, B.B. Pathogenicity islands: A molecular toolbox for bacterial virulence. Cell Microbiol. 2006, 8, 1707–1719. [Google Scholar] [CrossRef]
- Zhao, W.; Gao, B.; Liu, C.; Zhang, B.; Shan, C.; Deng, J.; Wan, Q.; Wang, X.; Zhao, R.; Gao, L.; et al. High pathogenicity island is associated with enhanced autophagy in pathogenic Escherichia coli HPI—Infected macrophages. Res. Vet. Sci. 2021, 135, 113–120. [Google Scholar] [CrossRef]
- Guerra, S.T.; Dalanezi, F.M.; de Paula, C.L.; Hernandes, R.T.; Pantoja, J.C.F.; Listoni, F.J.P.; Langoni, H.; Ribeiro, M.G. Putative virulence factors of extra-intestinal Escherichia coli isolated from bovine mastitis with different clinical scores. Lett. Appl. Microbiol. 2019, 68, 403–408. [Google Scholar] [CrossRef]
- Kathayat, D.; Lokesh, D.; Ranjit, S.; Rajashekara, G. Avian pathogenic Escherichia coli (APEC): An overview of virulence and pathogenesis factors, zoonotic potential, and control strategies. Pathogens 2021, 10, 467. [Google Scholar] [CrossRef]
- Lee, W.; Ha, J.; Choi, J.; Jung, Y.; Kim, E.; An, E.S.; Kim, S.H.; Shin, H.; Ryu, S.; Kim, S.H.; et al. Genetic and virulence characteristics of hybrid Shiga-toxin-producing and atypical enteropathogenic Escherichia coli strains isolated in South Korea. Front. Microbiol. 2024, 15, 1398262. [Google Scholar] [CrossRef]
- Watson, V.E.; Hazen, T.H.; Rasko, D.A.; Jacob, M.E.; Elfenbein, J.R.; Stauffer, S.H.; Gookin, J.L. Comparative genomics of atypical enteropathogenic Escherichia coli from kittens and children identified bacterial factors associated with virulence in kittens. Infect. Immun. 2021, 89, e00619–e00620. [Google Scholar] [CrossRef] [PubMed]
- Dulguer, M.V.; Fabbricotti, S.H.; Bando, S.Y.; Moreira-Filho, C.A.; Fagundes-Neto, U.; Scaletsky, I.C.A. Atypical enteropathogenic Escherichia coli strains: Phenotypic and genetic profiling reveals a strong association between enteroaggregative E. coli heat-stable enterotoxin and diarrhea. J. Infect. Dis. 2003, 188, 1685–1694. [Google Scholar] [CrossRef]
- Gelalcha, B.D.; Mohammed, R.I.; Gelgie, A.E.; Dego, O.K. 2023. Molecular epidemiology and pathogenomics of extended-spectrum beta-lactamase producing- Escherichia coli and—Klebsiella pneumoniae isolates from bulk tank milk in Tennessee, USA. Front. Microbiol. 2023, 14, 1283165. [Google Scholar] [CrossRef] [PubMed]
- Afema, J.A.; Ahmed, S.; Besser, T.E.; Jones, L.P.; Sischo, W.M.; Davis, M.A. Molecular epidemiology of dairy cattle-associated Escherichia coli carrying blaCTX-M genes in Washington state. Appl. Environ. Microbiol. 2018, 84, e02430. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 2227–2238. [Google Scholar] [CrossRef]
- Lamberti, M.F.T.; Terán, L.C.; Lopez, F.E.; Pescaretti, M.M.; Delgado, M.A. Genomic and proteomic characterization of two strains of Shigella flexneri 2 isolated from infants’ stool samples in Argentina. BMC Genom. 2022, 23, 495. [Google Scholar] [CrossRef]
- Bamidele, O.; Jiang, Z.-D.; Dupont, D. Occurrence of putative virulence-related genes, aatA, aggR and aaiC, of enteroaggregative Escherichia coli (EAEC) among adults with travelers’ diarrhea acquired in Guatemala and Mexico. Microb. Pathog. 2019, 128, 97–99. [Google Scholar] [CrossRef]
- Tanih, N.F.; Bolick, D.T.; Samie, A.; Nyathi, E.; Dillingham, R.; Pinkerton, R.C.; Guerrant, R.L.; Bessong, P.O. Prevalence of virulence genes in enteroaggregative Escherichia coli isolates from young children from rural South Africa. Am. J. Trop. Med. Hyg. 2019, 101, 1027–1033. [Google Scholar] [CrossRef]
- Zhang, R.; Gu, D.-X.; Huang, Y.-L.; Chan, E.W.-C.; Chen, G.-X.; Chen, S. Comparative genetic characterization of enteroaggregative Escherichia coli strains recovered from clinical and non-clinical settings. Sci. Rep. 2016, 6, 24321. [Google Scholar] [CrossRef]
- de Sousa, C.P.; Dubreuil, J.D. Distribution and expression of the astA gene (EAST1 toxin) in Escherichia coli and Salmonella. Int. J. Med. Microbiol. 2001, 291, 15–20. [Google Scholar] [CrossRef]
- Tanabe, R.H.S.; Dias, R.C.B.; Orsi, H.; de Lira, D.R.P.; Vieira, M.A.; dos Santos, L.F.; Ferreira, A.M.; Rall, V.L.M.; Mondelli, A.L.; Gomes, T.A.T.; et al. Characterization of uropathogenic Escherichia coli reveals hybrid isolates of uropathogenic and diarrheagenic (UPEC/DEC) E. coli. Microorganisms 2022, 10, 645. [Google Scholar] [CrossRef] [PubMed]
- Condoleo, R.; Giangolini, G.; Chiaverini, A.; Patriarca, D.; Scaramozzino, P.; Mezher, Z. Occurrence of Listeria monocytogenes and Escherichia coli in Raw Sheep’s Milk from Farm Bulk Tanks in Central Italy. J Food Prot. 2020, 83, 1929–1933. [Google Scholar] [CrossRef] [PubMed]
- Roșu, R.-D.; Morar, A.; Ban-Cucerzan, A.; Imre, M.; Sallam, K.I.; Maha, A.-A.A.; Abd-Elghany, S.M.; Popa, S.A.; Pătrînjan, R.-T.; Morar, D.; et al. The Microbiological Quality of Raw Ovine Milk in the Banat Region of Romania with a Focus on Escherichia coli and Its Pathogenic Potential and Antimicrobial Resistance. Vet. Sci. 2024, 11, 562. [Google Scholar] [CrossRef] [PubMed]
- van den Brom, R.; de Jong, A.; van Engelen, E.; Heuvelink, A.; Vellema, P. Zoonotic risks of pathogens from sheep and their milk borne transmission. Small Rumin. Res. 2020, 189, 106123. [Google Scholar] [CrossRef]
- Caro, I.; Fernández-Barata, V.M.; Alonso-Llamazares, A.; García-Armesto, M.R. Detection, occurrence, and characterization of Escherichia coli O157:H7 from raw ewe’s milk in Spain. J. Food Prot. 2006, 69, 920–924. [Google Scholar] [CrossRef]

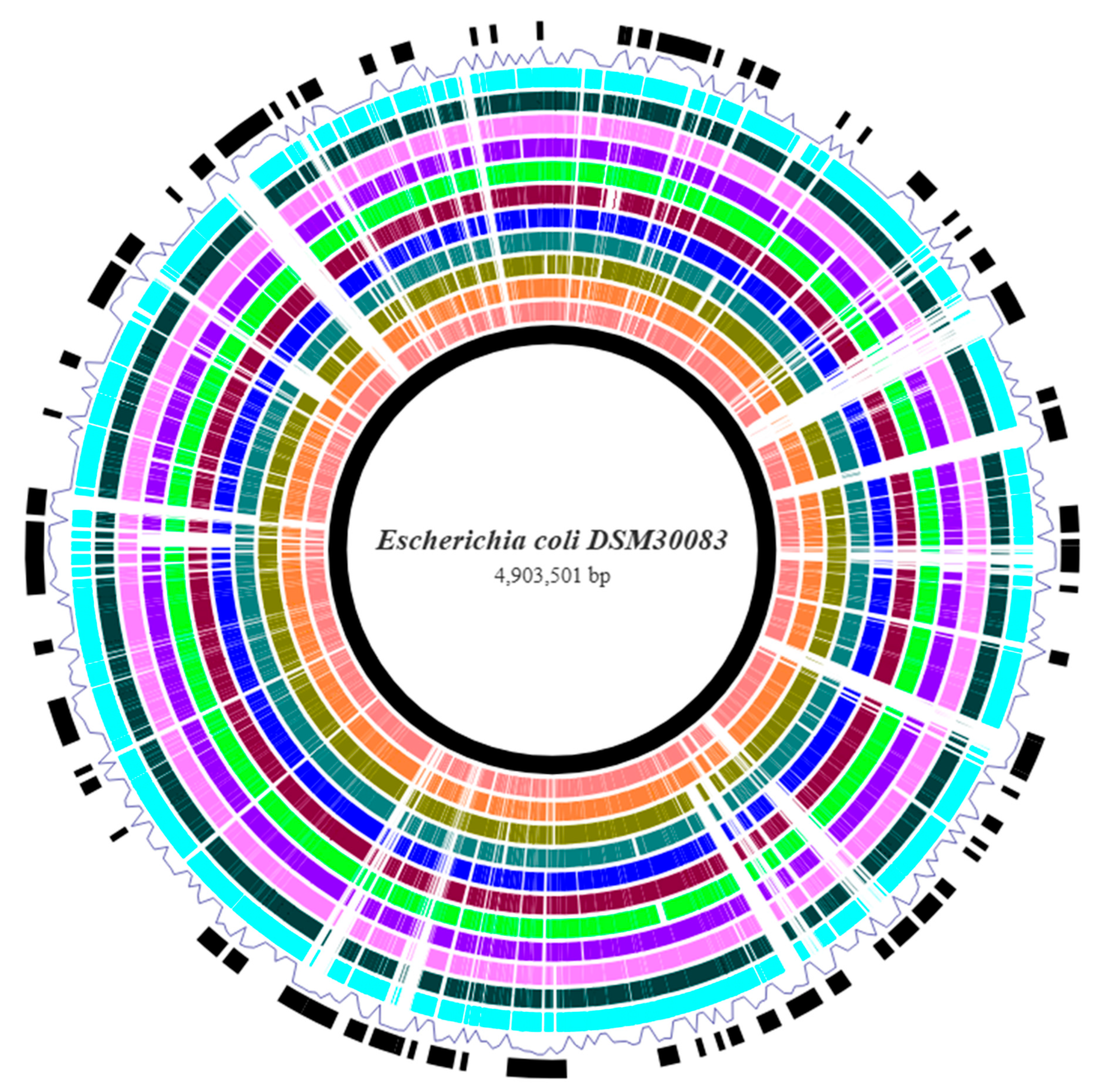
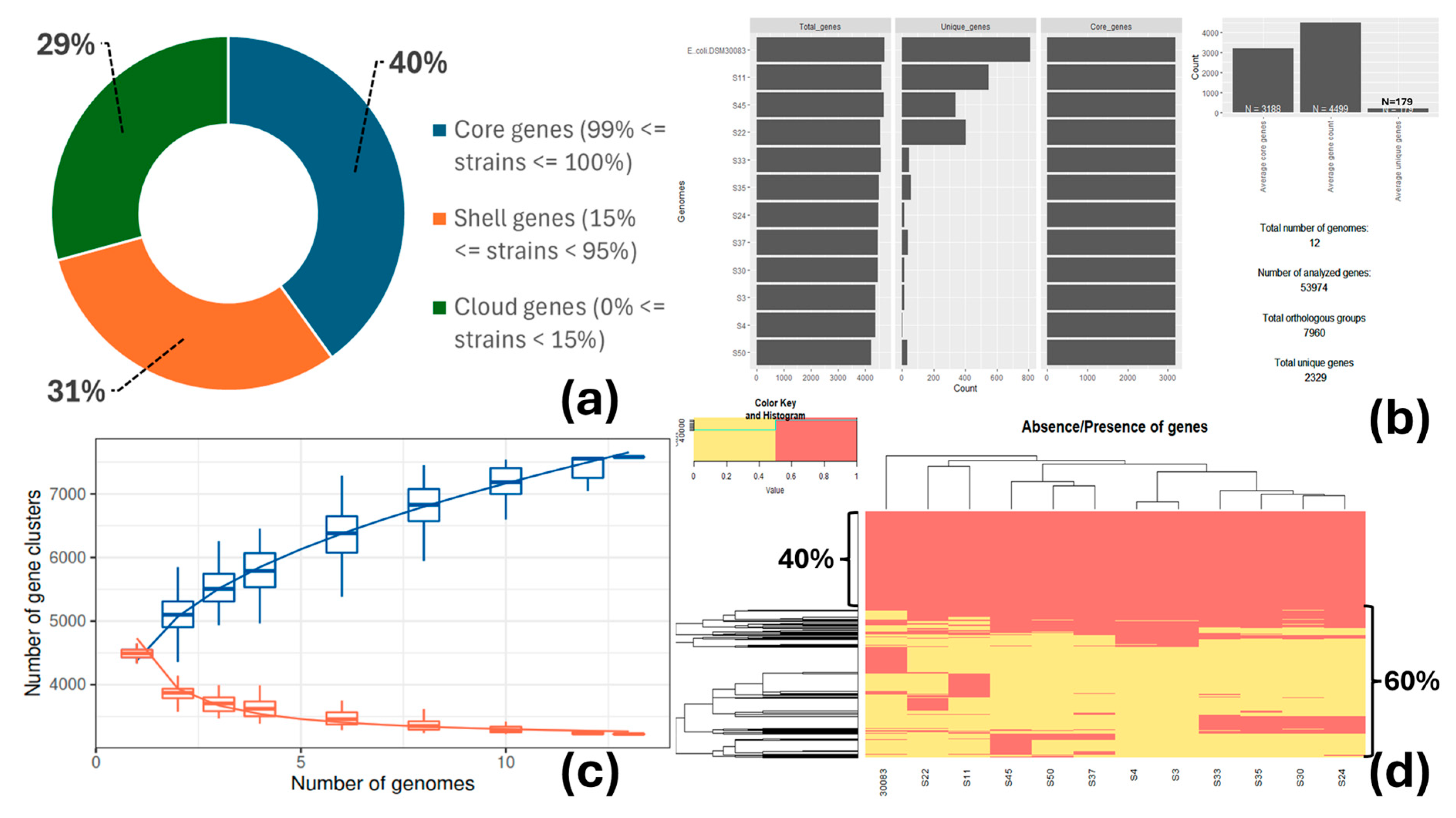
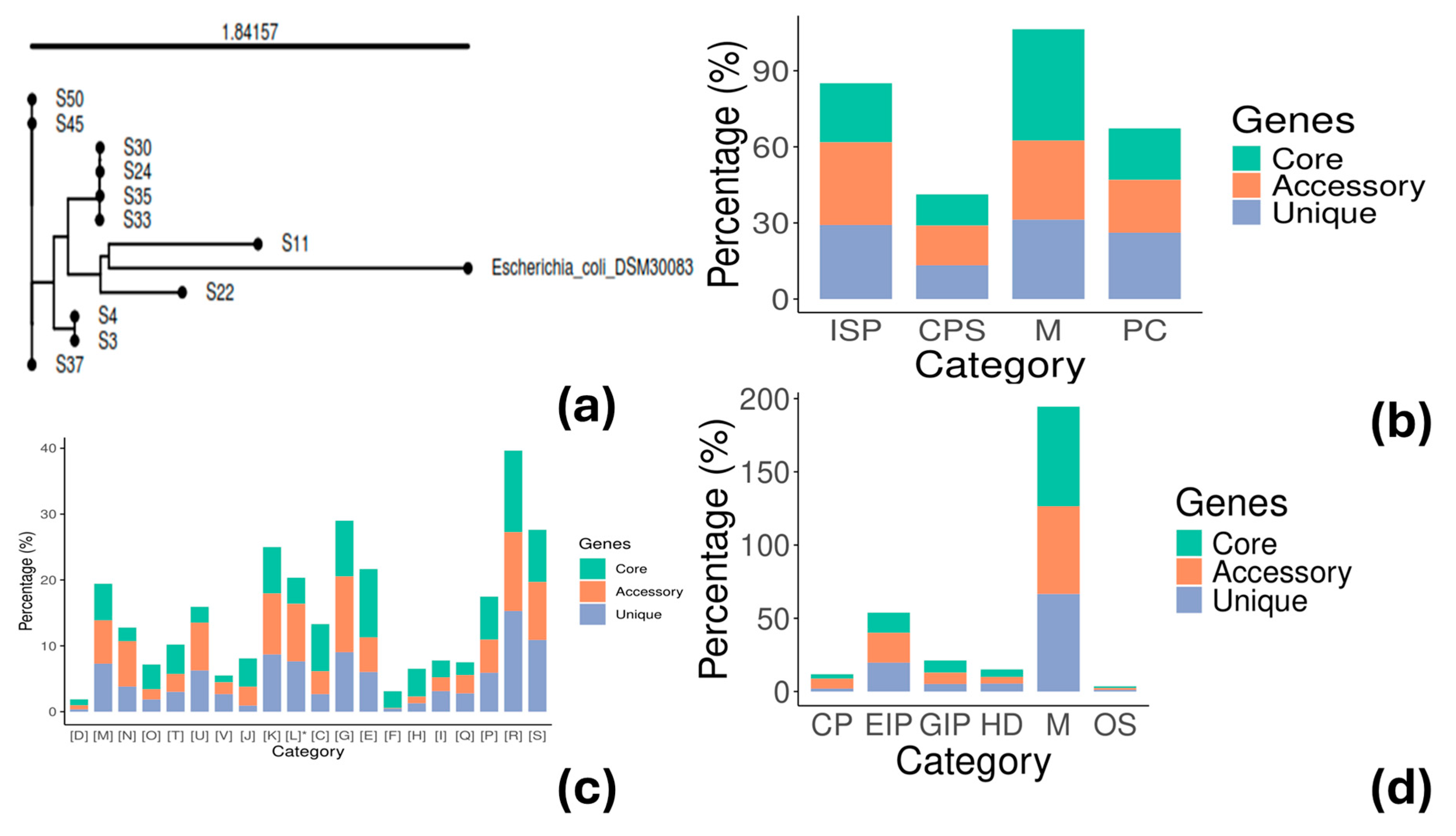
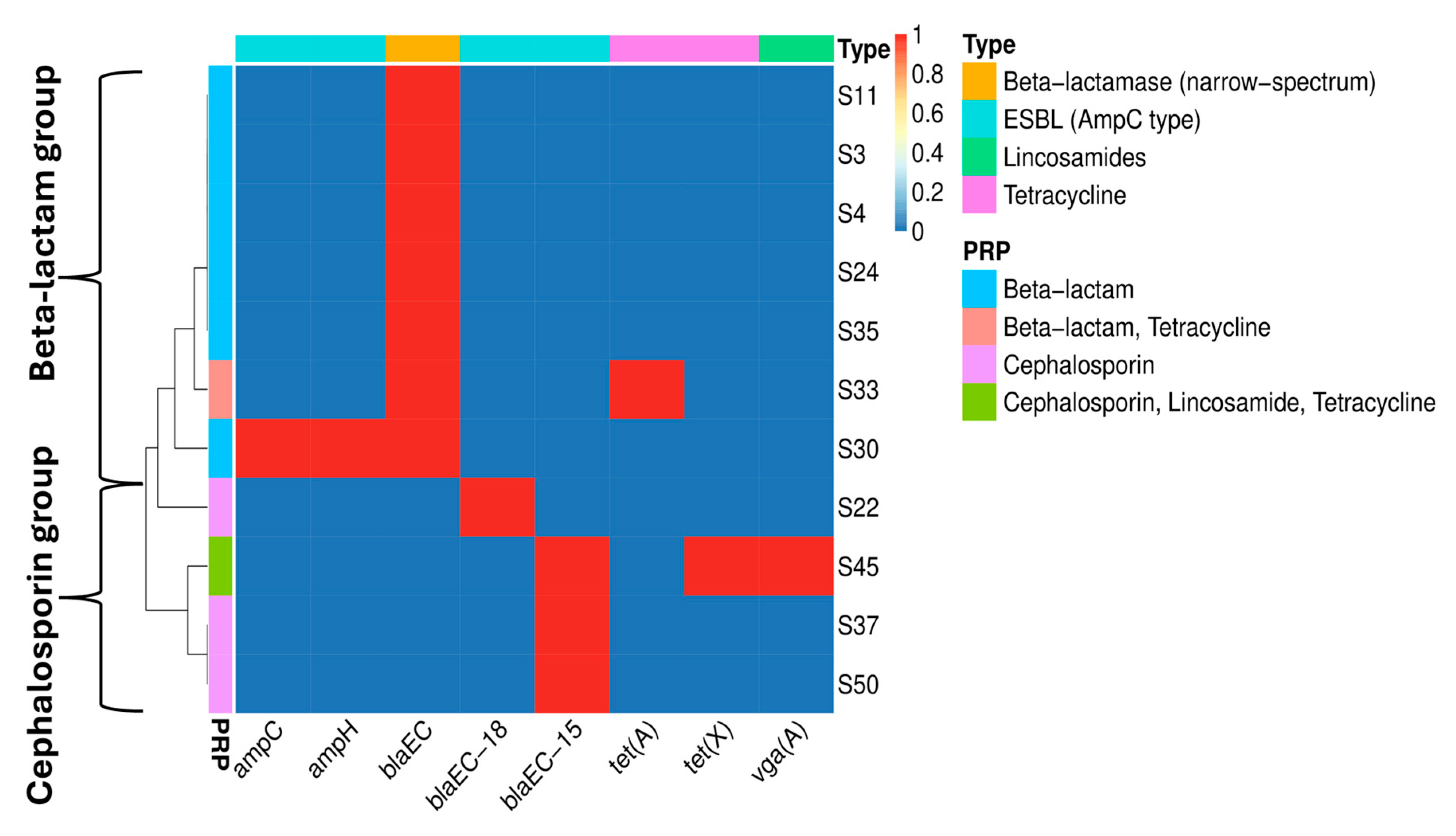
| Strain ID 1 | Phylogroup | O:H Serotype | MLST (Achtman) | cgMLST | FimType | CHType | Human Pathogen |
|---|---|---|---|---|---|---|---|
| S3 | A | O179:H40 | ST10 | 23273 | fimH137 | fumC11 | Yes (0.940) 2 |
| S4 | A | O179:H40 | ST10 | 23273 | fimH137 | fumC11 | Yes (0.941) |
| S11 | E | O169:H46 | ST1131 | 34239 | fimH31 | fumC23 | Yes (0.938) |
| S22 | B1 | O18ac:H7 | ST351 | 148610 | fimH31 | fumC95 | Yes (0.942) |
| S24 | A | ONT:H26 | ST4977 | 56618 | fimH27 | fumC11 | Yes (0.847) |
| S30 | A | ONT:H26 | ST4977 | 56618 | fimH27 | fumC11 | Yes (0.933) |
| S33 | A | ONT:H26 | ST4977 | 56618 | fimH27 | fumC11 | Yes (0.932) |
| S35 | A | ONT:H26 | ST4977 | 56618 | fimH27 | fumC11 | Yes (0.842) |
| S37 | A | O107:H27 | ST10 | 23653 | fimH54 | fumC11 | Yes (0.934) |
| S45 | A | O107:H27 | ST10 | 23653 | fimH54 | fumC11 | Yes (0.932) |
| S50 | A | O107:H27 | ST10 | 23653 | fimH54 | fumC11 | Yes (0.874) |
| Antibiotic/Drug | Abbreviation | S30 | S45 | Antibiotic Class |
|---|---|---|---|---|
| Ampicillin | AMP | I | S | Penicillin (Beta-lactam) |
| Ceftiofur | XNL | R | S | Cephalosporin (Beta-lactam) |
| Cephalothin | CEP | R | S | Cephalosporin (Beta-lactam) |
| Erythromycin | ERY | S | S | Macrolides |
| Oxacillin + 2% NaCl | OXA+ | R | S | Penicillin (Beta-lactam) |
| Penicillin | PEN | I | S | Penicillin (Beta-lactam) |
| Penicillin/Novobiocin | P/N | S | S | Beta-lactam/Aminocoumarin |
| Pirlamycin | PIRL | I | S | Lincosamide |
| Sulphadimethoxine | SDM | R | R | Sulfonamide |
| Tetracycline | TET | S | R | Tetracyclines |
| Strain ID | Plasmids | Phages |
|---|---|---|
| S3 | Col156 | - |
| S4 | - | - |
| S11 | ColpVC and IncFII(pCoo) | Lambdavirus and peduovirus |
| S22 | IncFII(pCoo), IncFIA, and IncFIB(AP001918) | Lambdavirus and peduovirus |
| S24 | ColpVC, Col(MG828), Col156, Col8282, IncFII(29)_pUTI89, IncI1(Alpha), and Col(KPHS6) | - |
| S30 | ColpVC, Col(MG828), Col156, Col8282, IncFII(29)_pUTI89, IncI1(Alpha), and Col(KPHS6) | - |
| S33 | ColpVC, Col(MG828), Col156, Col8282, IncFII(29)_pUTI89, IncI1(Alpha), Col(KPHS6), Col440I, and ColRNAI | Lambdavirus |
| S35 | ColpVC, Col(MG828), Col156, Col8282, IncFII(29)_pUTI89, IncI1(Alpha), Col(KPHS6), and rep33_rep(pSMA198) | - |
| S37 | IncY | - |
| S45 | rep19b_repA(SAP105A) and rep5b_rep(pUR2355) | - |
| S50 | ColpVC | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skarlatoudi, T.; Anagnostou, G.-M.; Theodorakis, V.; Bosnea, L.; Mataragas, M. Escherichia coli Strains Originating from Raw Sheep Milk, with Special Reference to Their Genomic Characterization, Such as Virulence Factors (VFs) and Antimicrobial Resistance (AMR) Genes, Using Whole-Genome Sequencing (WGS). Vet. Sci. 2025, 12, 744. https://doi.org/10.3390/vetsci12080744
Skarlatoudi T, Anagnostou G-M, Theodorakis V, Bosnea L, Mataragas M. Escherichia coli Strains Originating from Raw Sheep Milk, with Special Reference to Their Genomic Characterization, Such as Virulence Factors (VFs) and Antimicrobial Resistance (AMR) Genes, Using Whole-Genome Sequencing (WGS). Veterinary Sciences. 2025; 12(8):744. https://doi.org/10.3390/vetsci12080744
Chicago/Turabian StyleSkarlatoudi, Theodora, Glykeria-Myrto Anagnostou, Vasileios Theodorakis, Loulouda Bosnea, and Marios Mataragas. 2025. "Escherichia coli Strains Originating from Raw Sheep Milk, with Special Reference to Their Genomic Characterization, Such as Virulence Factors (VFs) and Antimicrobial Resistance (AMR) Genes, Using Whole-Genome Sequencing (WGS)" Veterinary Sciences 12, no. 8: 744. https://doi.org/10.3390/vetsci12080744
APA StyleSkarlatoudi, T., Anagnostou, G.-M., Theodorakis, V., Bosnea, L., & Mataragas, M. (2025). Escherichia coli Strains Originating from Raw Sheep Milk, with Special Reference to Their Genomic Characterization, Such as Virulence Factors (VFs) and Antimicrobial Resistance (AMR) Genes, Using Whole-Genome Sequencing (WGS). Veterinary Sciences, 12(8), 744. https://doi.org/10.3390/vetsci12080744







