Multidimensional Phenotypic and Microbiome Studies Uncover an Association Between Reduced Feed Efficiency in Sheep During Mycoplasmal Pneumonia and Microbial Crosstalk Within the Rumen-Lung Axis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal and Management
2.2. Blood Sample Collection and Hematological Measurements
2.3. Sample Collection
2.4. Histological Analysis
2.5. Animal Grouping
2.6. Sequencing and Analysis of 16S rRNA Gene
2.7. Statistical Analysis
3. Results
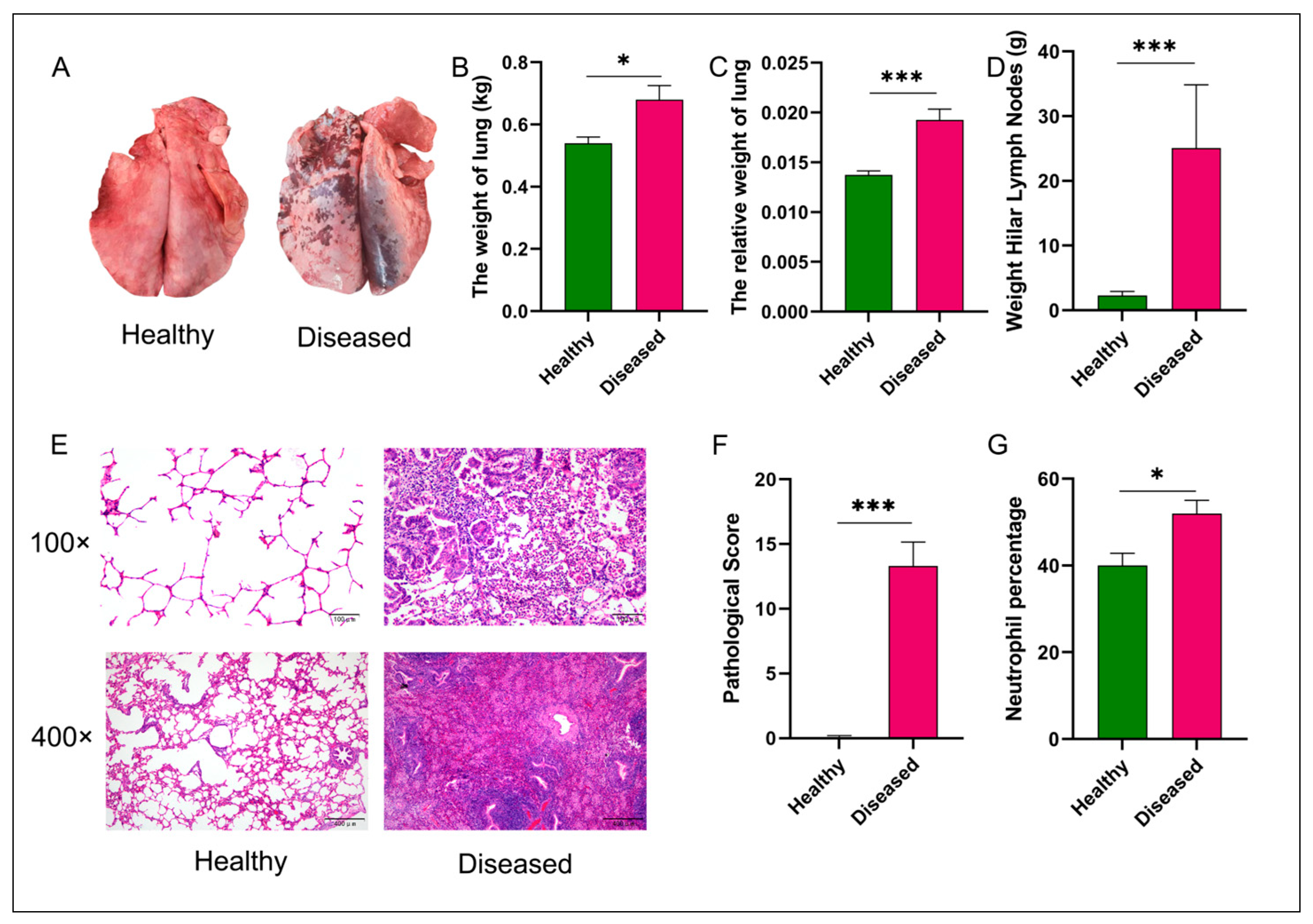
3.1. Statistical Comparison of Gross Lung Lesions, Pulmonary Lymph Node Weights, and Histopathological Scores Between Diseased and Healthy Groups
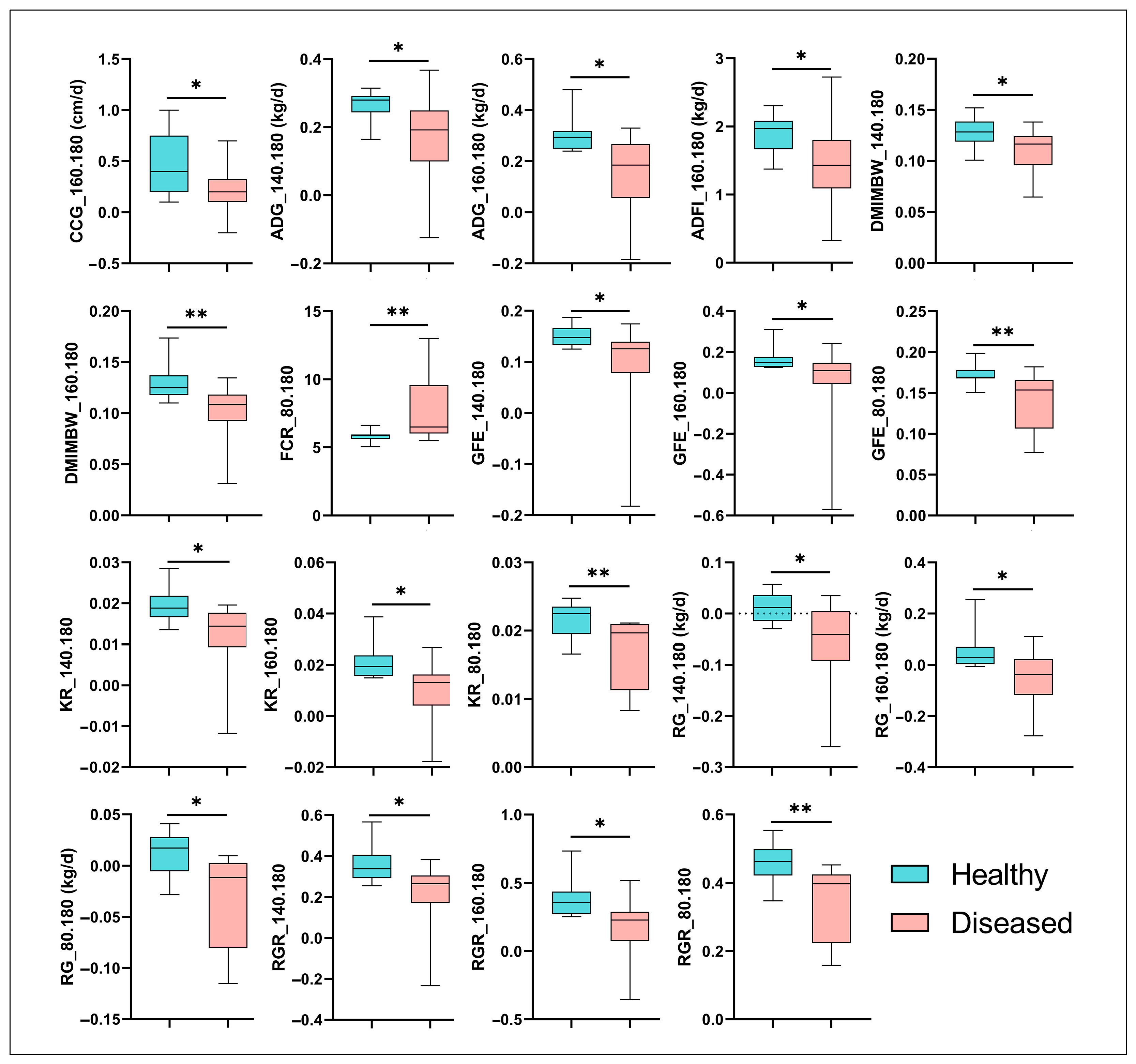
3.2. Growth Performance and Feed Efficiency of Hu Sheep Affected by MPS
3.3. Identification of Mesomycoplasma Ovipneumoniae as a Key Pathogen in MPS via 16S rRNA Gene Sequencing
3.4. Comparative Analysis of Pulmonary and Rumen Microbiota Diversity in Healthy and MPS-Sheep
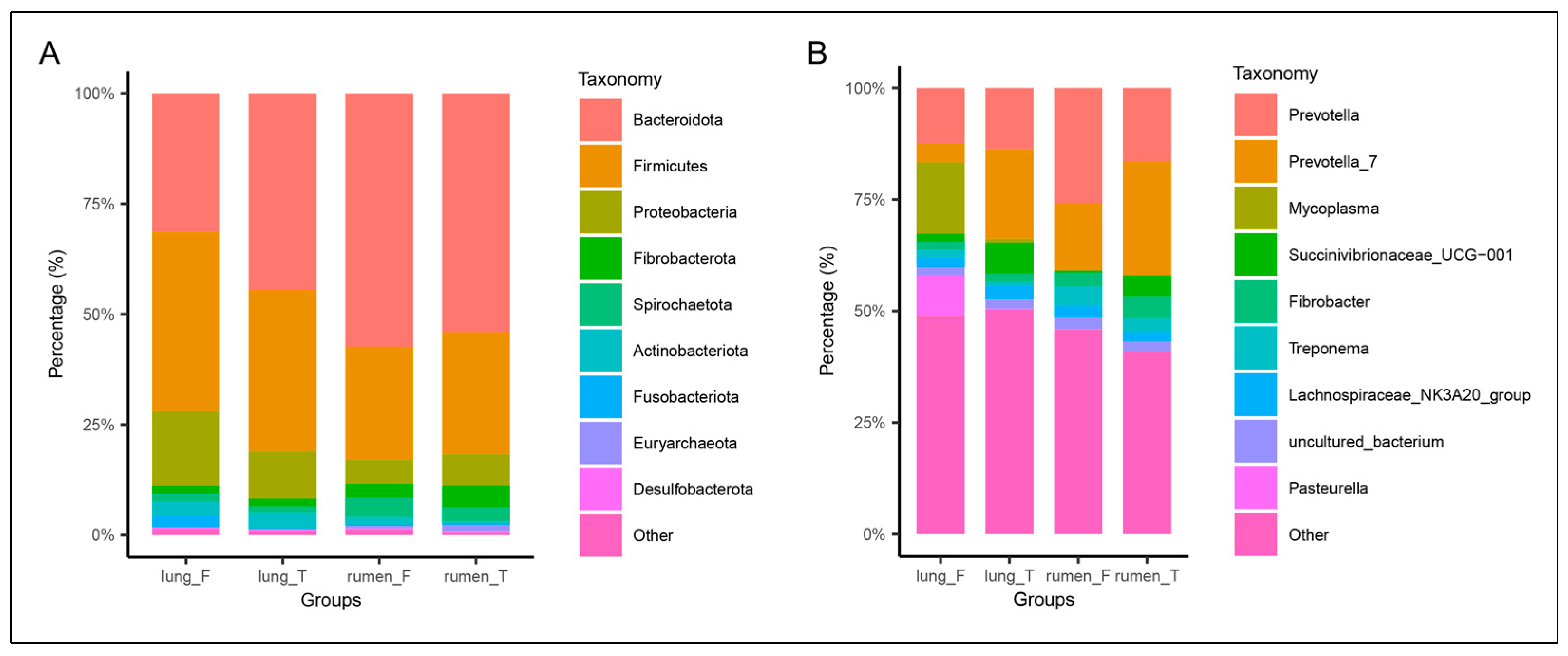
3.5. Taxonomic Composition of Lung and Rumen Microbiota in Healthy vs. Diseased Groups
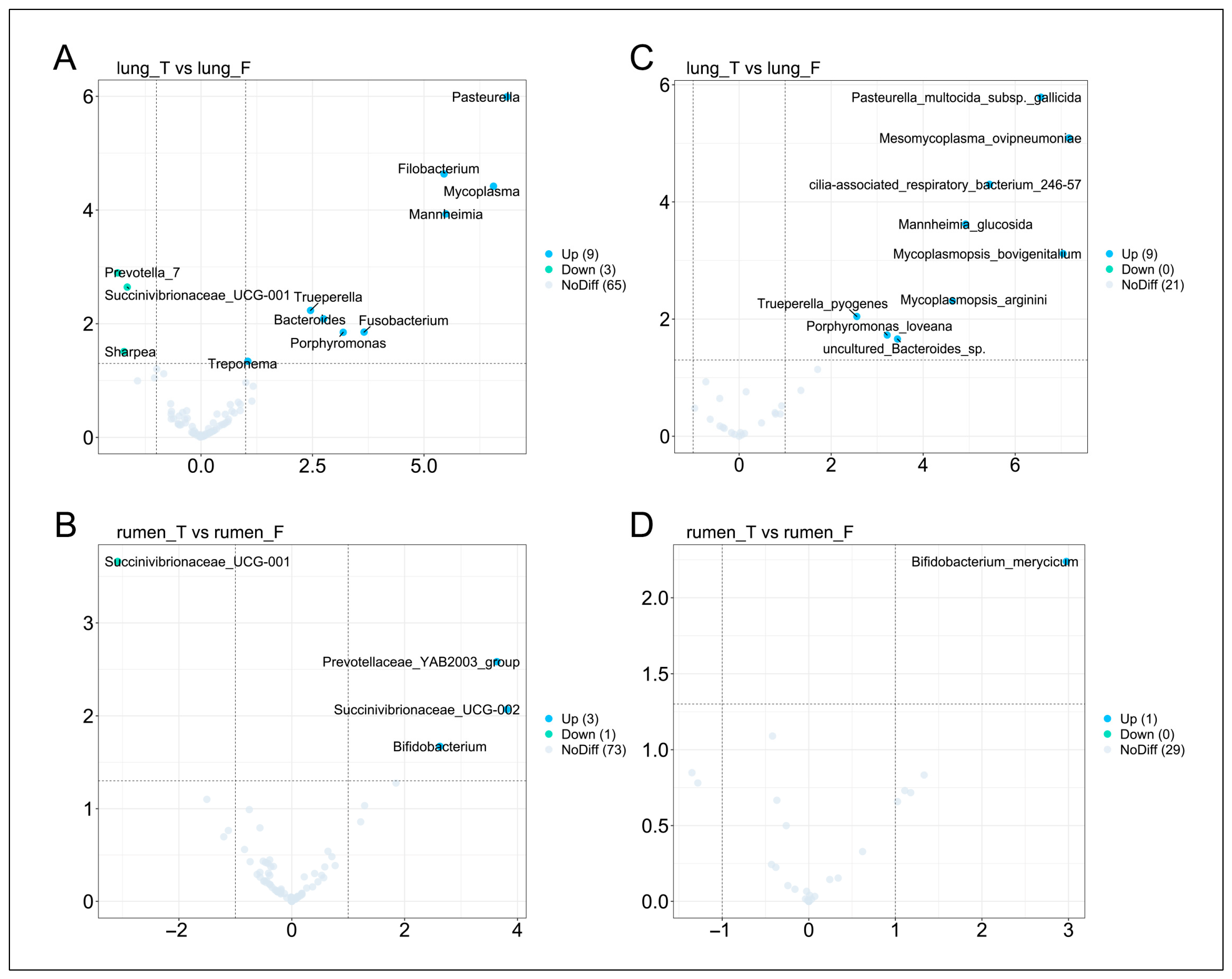
3.6. Differential Microbial Analysis of Lung and Rumen Between Healthy and MPS-Sheep
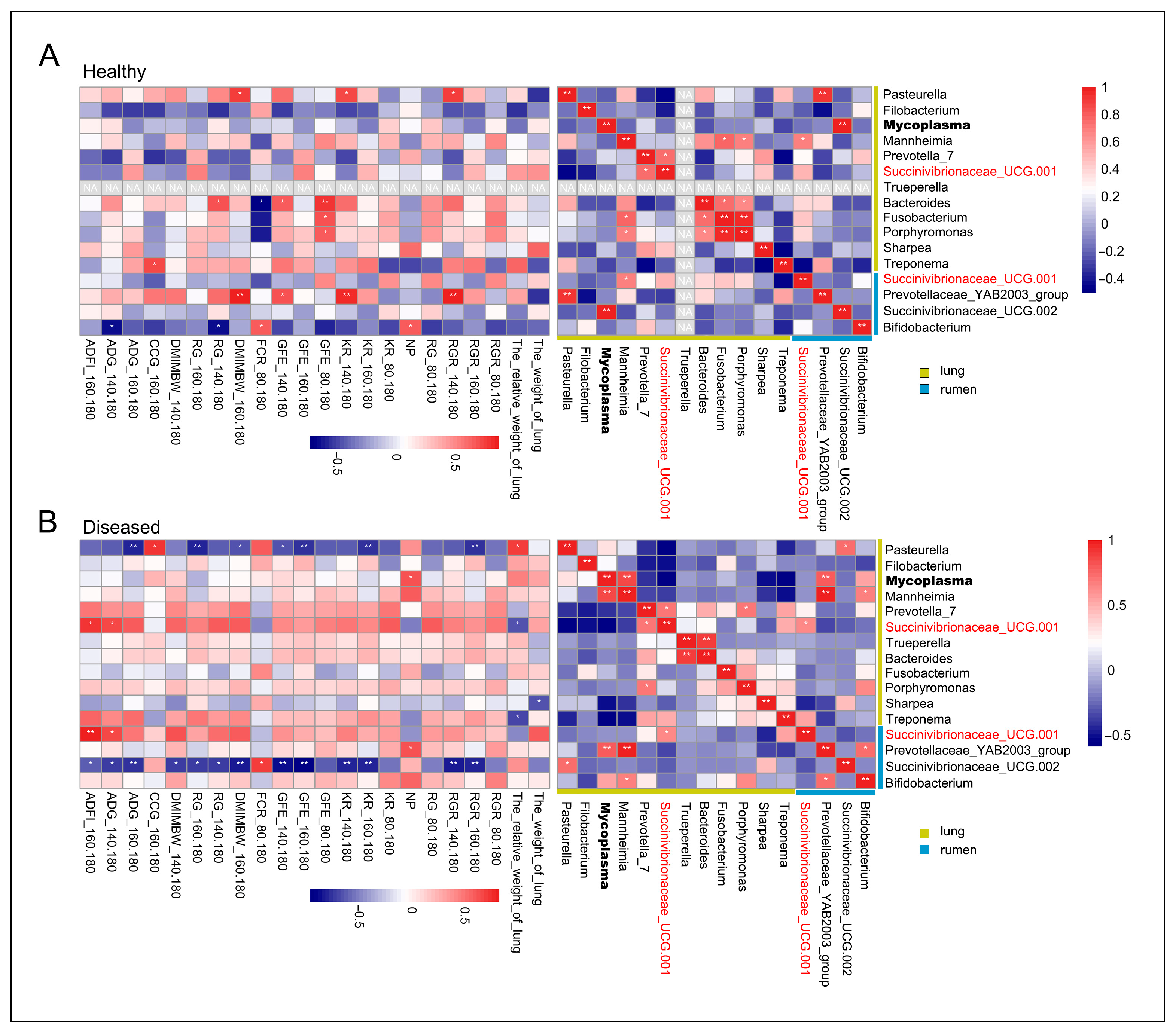
3.7. Correlation Analysis Between Differentially Abundant Lung Microbes, Differentially Abundant Rumen Microbes, and Growth Performance/Feed Efficiency
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MPS | Mycoplasmal pneumonia of sheep |
| BW | Body weight |
| ADG | Average daily gain |
| ADFI | Average daily feed intake |
| MBW | Metabolic body weight |
| FCR | Feed conversion ratio |
| KR | Kleiber ratio |
| GFE | Gross feed efficiency |
| DMIMBW | Dry matter intake per unit metabolic body weight |
| RG | Residual gain |
| RGR | Relative growth rate |
| ASV | Amplicon sequence variant |
| NP | Neutrophil percentage |
| CCG | Chest circumference gain |
References
- Asp Tauni, F. Association of Mycoplasma ovipneumoniae Infection with Respiratory Disease in Swedish Sheep. Master’s Thesis, SLU, Department of Biomedical Sciences and Veterinary Public Health (until 231231), Uppsala, Schweden, 2017. Volume 2017. p. 38. Available online: https://slu.primo.exlibrisgroup.com/permalink/46SLUB_INST/1jctjau/alma9920864777605121 (accessed on 24 July 2025).
- Manlove, K.; Branan, M.; Baker, K.; Bradway, D.; Cassirer, E.F.; Marshall, K.L.; Miller, R.S.; Sweeney, S.; Cross, P.C.; Besser, T.E. Risk factors and productivity losses associated with Mycoplasma ovipneumoniae infection in United States domestic sheep operations. Prev. Vet. Med. 2019, 168, 30–38. [Google Scholar] [CrossRef]
- Jaÿ, M.; Ambroset, C.; Tricot, A.; Colin, A.; Tardy, F. Population structure and antimicrobial susceptibility of Mycoplasma ovipneumoniae isolates in France. Vet. Microbiol. 2020, 248, 108828. [Google Scholar] [CrossRef]
- Dassanayake, R.P.; Shanthalingam, S.; Herndon, C.N.; Subramaniam, R.; Lawrence, P.K.; Bavananthasivam, J.; Cassirer, E.F.; Haldorson, G.J.; Foreyt, W.J.; Rurangirwa, F.R.; et al. Mycoplasma ovipneumoniae can predispose bighorn sheep to fatal Mannheimia haemolytica pneumonia. Vet. Microbiol. 2010, 145, 354–359. [Google Scholar] [CrossRef]
- López-Olvera, J.R.; Besser, T.E.; Levy, J.; Ackerman, M.; Nelson, D.; Manlove, K.; Potter, K.A.; Busboom, J.; Benson, M. A pilot study of the effects of Mycoplasma ovipneumoniae exposure on domestic lamb growth and performance. PLoS ONE 2019, 14, e0207420. [Google Scholar] [CrossRef]
- Maksimović, Z.; Rifatbegović, M.; Loria, G.R.; Nicholas, R.A.J. Mycoplasma ovipneumoniae: A Most Variable Pathogen. Pathogens 2022, 11, 1477. [Google Scholar] [CrossRef]
- Allen, I.C.; Handeland, K.; Tengs, T.; Kokotovic, B.; Vikøren, T.; Ayling, R.D.; Bergsjø, B.; Sigurðardóttir, Ó.G.; Bretten, T. Mycoplasma ovipneumoniae—A Primary Cause of Severe Pneumonia Epizootics in the Norwegian Muskox (Ovibos moschatus) Population. PLoS ONE 2014, 9, e106116. [Google Scholar] [CrossRef]
- Niang, M.; Rosenbusch, R.F.; Lopez-Virella, J.; Kaeberle, M.L. Expression of functions by normal sheep alveolar macrophages and their alteration by interaction with Mycoplasma ovipneumoniae. Vet. Microbiol. 1977, 58, 31–43. [Google Scholar] [CrossRef]
- Shahzad, W.; Ajuwape, A.T.P.; Rosenbusch, R.F. Global suppression of mitogen-activated ovine peripheral blood mononuclear cells by surface protein activity from Mycoplasma ovipneumoniae. Vet. Immunol. Immunopathol. 2010, 136, 116–121. [Google Scholar] [CrossRef]
- Miles, R.J.; Taylor, R.; Varsani, H. Oxygen uptake and H2O2 production by fermentative Mycoplasma spp. J. Med. Microbiol. 1999, 34, 219–223. [Google Scholar] [CrossRef]
- Yan, C.; Dong, T.; Shan, Y.; Zhao, B.; Yang, H.; Cai, Y.; Li, S.; Liu, Q.; Chu, Y.; Hao, H.; et al. Mycoplasma ovipnuemoniae impairs the immune response of sheep and suppresses neutrophil function by inhibiting S100A9. Vet. Microbiol. 2025, 303, 110446. [Google Scholar] [CrossRef]
- Xue, D.; Ma, Y.; Li, M.; Li, Y.; Luo, H.; Liu, X.; Wang, Y. Mycoplasma ovipneumoniae induces inflammatory response in sheep airway epithelial cells via a MyD88-dependent TLR signaling pathway. Vet. Immunol. Immunopathol. 2015, 163, 57–66. [Google Scholar] [CrossRef]
- Jiang, Z.; Song, F.; Li, Y.; Xue, D.; Zhao, N.; Zhang, J.; Deng, G.; Li, M.; Liu, X.; Wang, Y.; et al. Capsular Polysaccharide of Mycoplasma ovipneumoniae Induces Sheep Airway Epithelial Cell Apoptosis via ROS-Dependent JNK/P38 MAPK Pathways. Oxidative Med. Cell. Longev. 2017, 2017, 6175841. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, Z.; Xue, D.; Deng, G.; Li, M.; Liu, X.; Wang, Y. Mycoplasma ovipneumoniae induces sheep airway epithelial cell apoptosis through an ERK signalling-mediated mitochondria pathway. BMC Microbiol. 2016, 16, 222. [Google Scholar] [CrossRef]
- El Tekle, G.; Garrett, W.S. Bacteria in cancer initiation, promotion and progression. Nat. Rev. Cancer 2023, 23, 600–618. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, Y.; Yan, Z.; Liu, H.; Chen, B.; Liang, Z.; Wang, F.; Miller, B.E.; Tal-Singer, R.; Yi, X.; et al. Multi-omic meta-analysis identifies functional signatures of airway microbiome in chronic obstructive pulmonary disease. ISME J. 2020, 14, 2748–2765. [Google Scholar] [CrossRef]
- Wang, Z.; Locantore, N.; Haldar, K.; Ramsheh, M.Y.; Beech, A.S.; Ma, W.; Brown, J.R.; Tal-Singer, R.; Barer, M.R.; Bafadhel, M.; et al. Inflammatory Endotype—Associated Airway Microbiome in Chronic Obstructive Pulmonary Disease Clinical Stability and Exacerbations: A Multicohort Longitudinal Analysis. Am. J. Respir. Crit. Care Med. 2021, 203, 1488–1502. [Google Scholar] [CrossRef]
- Vázquez-Pérez, J.A.; Carrillo, C.O.; Iñiguez-García, M.A.; Romero-Espinoza, I.; Márquez-García, J.E.; Falcón, L.I.; Torres, M.; Herrera, M.T. Alveolar microbiota profile in patients with human pulmonary tuberculosis and interstitial pneumonia. Microb. Pathog. 2020, 139, 103851. [Google Scholar] [CrossRef]
- Johnston, D.; Earley, B.; Cormican, P.; Murray, G.; Kenny, D.A.; Waters, S.M.; McGee, M.; Kelly, A.K.; McCabe, M.S. Illumina MiSeq 16S amplicon sequence analysis of bovine respiratory disease associated bacteria in lung and mediastinal lymph node tissue. BMC Vet. Res. 2017, 13, 118. [Google Scholar] [CrossRef]
- Miao, Y.; Zhao, X.; Lei, J.; Ding, J.; Feng, H.; Wu, K.; Liu, J.; Wang, C.; Ye, D.; Wang, X.; et al. Characterization of Lung Microbiomes in Pneumonic Hu Sheep Using Culture Technique and 16S rRNA Gene Sequencing. Animals 2023, 13, 2763. [Google Scholar] [CrossRef]
- Depner, M.; Taft, D.H.; Kirjavainen, P.V.; Kalanetra, K.M.; Karvonen, A.M.; Peschel, S.; Schmausser-Hechfellner, E.; Roduit, C.; Frei, R.; Lauener, R.; et al. Maturation of the gut microbiome during the first year of life contributes to the protective farm effect on childhood asthma. Nat. Med. 2020, 26, 1766–1775. [Google Scholar] [CrossRef]
- Liu, N.-N.; Ma, Q.; Ge, Y.; Yi, C.-X.; Wei, L.-Q.; Tan, J.-C.; Chu, Q.; Li, J.-Q.; Zhang, P.; Wang, H. Microbiome dysbiosis in lung cancer: From composition to therapy. npj Precis. Oncol. 2020, 4, 33. [Google Scholar] [CrossRef]
- Özçam, M.; Lynch, S.V. The gut–airway microbiome axis in health and respiratory diseases. Nat. Rev. Microbiol. 2024, 22, 492–506. [Google Scholar] [CrossRef]
- Tang, J.; Xu, L.; Zeng, Y.; Gong, F. Effect of gut microbiota on LPS-induced acute lung injury by regulating the TLR4/NF-kB signaling pathway. Int. Immunopharmacol. 2021, 91, 107272. [Google Scholar] [CrossRef]
- Li, R.; Li, J.; Zhou, X. Lung microbiome: New insights into the pathogenesis of respiratory diseases. Signal Transduct. Target. Ther. 2024, 9, 19. [Google Scholar] [CrossRef]
- Jia, M.; Zhu, S.; Xue, M.-Y.; Chen, H.; Xu, J.; Song, M.; Tang, Y.; Liu, X.; Tao, Y.; Zhang, T.; et al. Single-cell transcriptomics across 2534 microbial species reveals functional heterogeneity in the rumen microbiome. Nat. Microbiol. 2024, 9, 1884–1898. [Google Scholar] [CrossRef]
- Wang, D.; Chen, L.; Tang, G.; Yu, J.; Chen, J.; Li, Z.; Cao, Y.; Lei, X.; Deng, L.; Wu, S.; et al. Multi-omics revealed the long-term effect of ruminal keystone bacteria and the microbial metabolome on lactation performance in adult dairy goats. Microbiome 2023, 11, 215. [Google Scholar] [CrossRef]
- Hu, X.; Li, S.; Fu, Y.; Zhang, N. Targeting gut microbiota as a possible therapy for mastitis. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1409–1423. [Google Scholar] [CrossRef]
- Kong, F.; Wang, S.; Zhang, Y.; Li, C.; Dai, D.; Guo, C.; Wang, Y.; Cao, Z.; Yang, H.; Bi, Y.; et al. Rumen microbiome associates with postpartum ketosis development in dairy cows: A prospective nested case–control study. Microbiome 2025, 13, 69. [Google Scholar] [CrossRef]
- Wang, Y.; Nan, X.; Zhao, Y.; Jiang, L.; Wang, H.; Zhang, F.; Hua, D.; Liu, J.; Yao, J.; Yang, L.; et al. Dietary Supplementation of Inulin Ameliorates Subclinical Mastitis via Regulation of Rumen Microbial Community and Metabolites in Dairy Cows. Microbiol Spectr. 2021, 9, e0010521. [Google Scholar] [CrossRef]
- Gaowa, N.; Panke-Buisse, K.; Wang, S.; Wang, H.; Cao, Z.; Wang, Y.; Yao, K.; Li, S. Brisket Disease Is Associated with Lower Volatile Fatty Acid Production and Altered Rumen Microbiome in Holstein Heifers. Animals 2020, 10, 1712. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Liu, Y.X.; Chen, L.; Ma, T.; Li, X.; Zheng, M.; Zhou, X.; Chen, L.; Qian, X.; Xi, J.; Lu, H.; et al. EasyAmplicon: An easy-to-use, open-source, reproducible, and community-based pipeline for amplicon data analysis in microbiome research. iMeta 2023, 2, e83. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Martin, A.M.; Hogg, J.T.; Manlove, K.R.; LaSharr, T.N.; Shannon, J.M.; McWhirter, D.E.; Miyasaki, H.; Monteith, K.L.; Cross, P.C. Disease and secondary sexual traits: Effects of pneumonia on horn size of bighorn sheep. J. Wildl. Manag. 2022, 86, e22154. [Google Scholar] [CrossRef]
- Pointon, A.M.; Byrt, D.; Heap, P. Effect of enzootic pneumonia of pigs on growth performance. Aust. Vet. J. 1985, 62, 13–18. [Google Scholar] [CrossRef]
- Barbara, E.; Straw, S.J.S.; Amy, E. Effect of pneumonia on growth rate and feed efficiency of minimal disease pigs exposed to Actinobacillus pleuropneumoniae and Mycoplasma hyopneumoniae. Prev. Vet. Med. 1990, 9, 287–294. [Google Scholar] [CrossRef]
- Buczinski, S.; Achard, D.; Timsit, E. Effects of calfhood respiratory disease on health and performance of dairy cattle: A systematic review and meta-analysis. J. Dairy Sci. 2021, 104, 8214–8227. [Google Scholar] [CrossRef]
- Li, N.; Yang, Z.; Liao, B.; Pan, T.; Pu, J.; Hao, B.; Fu, Z.; Cao, W.; Zhou, Y.; He, F.; et al. Chronic exposure to ambient particulate matter induces gut microbial dysbiosis in a rat COPD model. Respir. Res. 2020, 21, 271. [Google Scholar] [CrossRef]
- Wang, W.; Wei, Z.; Li, Z.; Ren, J.; Song, Y.; Xu, J.; Liu, A.; Li, X.; Li, M.; Fan, H.; et al. Integrating genome- and transcriptome-wide association studies to uncover the host–microbiome interactions in bovine rumen methanogenesis. iMeta 2024, 3, e234. [Google Scholar] [CrossRef]
- Zhuang, Y.; Gao, D.; Jiang, W.; Xu, Y.; Liu, G.; Hou, G.; Chen, T.; Li, S.; Zhang, S.; Liu, S.; et al. Core microbe Bifidobacterium in the hindgut of calves improves the growth phenotype of young hosts by regulating microbial functions and host metabolism. Microbiome 2025, 13, 13. [Google Scholar] [CrossRef]
- Di, Y.P.; Yun, Y.; Srinivas, G.; Kuenzel, S.; Linnenbrink, M.; Alnahas, S.; Bruce, K.D.; Steinhoff, U.; Baines, J.F.; Schaible, U.E. Environmentally Determined Differences in the Murine Lung Microbiota and Their Relation to Alveolar Architecture. PLoS ONE 2014, 9, e113466. [Google Scholar] [CrossRef]
- Huffnagle, G.B.; Dickson, R.P.; Lukacs, N.W. The respiratory tract microbiome and lung inflammation: A two-way street. Mucosal Immunol. 2017, 10, 299–306. [Google Scholar] [CrossRef]
- Yoon, H.-Y.; Moon, S.-J.; Song, J.W. Lung Tissue Microbiome Is Associated with Clinical Outcomes of Idiopathic Pulmonary Fibrosis. Front. Med. 2021, 8, 744523. [Google Scholar] [CrossRef]
- Sze, M.A.; Dimitriu, P.A.; Hayashi, S.; Elliott, W.M.; McDonough, J.E.; Gosselink, J.V.; Cooper, J.; Sin, D.D.; Mohn, W.W.; Hogg, J.C. The Lung Tissue Microbiome in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2012, 185, 1073–1080. [Google Scholar] [CrossRef]
- Das, S.; Bernasconi, E.; Koutsokera, A.; Wurlod, D.-A.; Tripathi, V.; Bonilla-Rosso, G.; Aubert, J.-D.; Derkenne, M.-F.; Mercier, L.; Pattaroni, C.; et al. A prevalent and culturable microbiota links ecological balance to clinical stability of the human lung after transplantation. Nat. Commun. 2021, 12, 2126. [Google Scholar] [CrossRef]
- Manfredo Vieira, S.; Hiltensperger, M.; Kumar, V.; Zegarra-Ruiz, D.; Dehner, C.; Khan, N.; Costa, F.R.C.; Tiniakou, E.; Greiling, T.; Ruff, W.; et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018, 359, 1156–1161. [Google Scholar] [CrossRef]
- Jan, L.; Boute, P.; Mouawad, F. Pasteurella multocida acute epiglottitis. Eur. Ann. Otorhinolaryngol. Head Neck Dis. 2021, 138, 100–102. [Google Scholar] [CrossRef]
- Glendinning, L.; Collie, D.; Wright, S.; Rutherford, K.M.D.; McLachlan, G. Comparing microbiotas in the upper aerodigestive and lower respiratory tracts of lambs. Microbiome 2017, 5, 145. [Google Scholar] [CrossRef]
- Malmberg, J.L.; Allen, S.E.; Jennings-Gaines, J.E.; Johnson, M.; Luukkonen, K.L.; Robbins, K.M.; Cornish, T.E.; Smiley, R.A.; Wagler, B.L.; Gregory, Z.; et al. Pathology of chronic Mycoplasma ovipneumoniae carriers in a declining bighorn sheep (Ovis canadensis) population. J. Wildl. Dis. 2024, 60, 448–460, 413. [Google Scholar]
- Kew, M.C. Clinical, pathologic, and etiologic heterogeneity in hepatocellular carcinoma: Evidence from southern Africa. Hepatology 1981, 1, 366–369. [Google Scholar] [CrossRef]
- Alley, M.R.; Ionas, G.; Clarke, J.K. Chronic non-progressive pneumonia of sheep in New Zealand—A review of the role of Mycoplasma ovipneumoniae. N. Z. Vet. J. 1999, 47, 155–160. [Google Scholar] [CrossRef]
- Rifatbegović, M.; Maksimović, Z.; Hulaj, B. Mycoplasma ovipneumoniae associated with severe respiratory disease in goats. Vet. Rec. 2011, 168, 565. [Google Scholar] [CrossRef]
- Nicholas, R.A.J.; Ayling, R.D.; Loria, G.R. Ovine mycoplasmal infections. Small Rumin. Res. 2008, 76, 92–98. [Google Scholar] [CrossRef]
- Legesse, A.; Abayneh, T.; Mamo, G.; Gelaye, E.; Tesfaw, L.; Yami, M.; Belay, A. Molecular characterization of Mannheimia haemolytica isolates associated with pneumonic cases of sheep in selected areas of Central Ethiopia. BMC Microbiol. 2018, 18, 205. [Google Scholar] [CrossRef]
- Gharib mombeni, E.; Gharibi, D.; Ghorbanpoor, M.; Jabbari, A.R.; Cid, D. Molecular characterization of Mannheimia haemolytica associated with ovine and caprine pneumonic lung lesions. Microb. Pathog. 2021, 153, 104791. [Google Scholar] [CrossRef]
- Rainbolt, S.; Pillai, D.K.; Lubbers, B.V.; Moore, M.; Davis, R.; Amrine, D.; Mosier, D. Comparison of Mannheimia haemolytica isolates from an outbreak of bovine respiratory disease. Vet. Microbiol. 2016, 182, 82–86. [Google Scholar] [CrossRef]
- Hidalgo-Cantabrana, C.; Delgado, S.; Ruiz, L.; Ruas-Madiedo, P.; Sánchez, B.; Margolles, A.; Britton, R.A.; Cani, P.D. Bifidobacteria and Their Health-Promoting Effects. Microbiol. Spectr. 2017, 5, 10. [Google Scholar] [CrossRef]
- Huang, S.; Ji, S.; Suen, G.; Wang, F.; Li, S. The Rumen Bacterial Community in Dairy Cows Is Correlated to Production Traits During Freshening Period. Front. Microbiol. 2021, 12, 630605. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, C.; Niu, X.; Zhang, Z.; Li, F.; Li, F. An intensive milk replacer feeding program benefits immune response and intestinal microbiota of lambs during weaning. BMC Vet. Res. 2018, 14, 366. [Google Scholar] [CrossRef]
- Wang, X.; Martin, G.B.; Wen, Q.; Liu, S.; Li, Y.; Shi, B.; Guo, X.; Zhao, Y.; Guo, Y.; Yan, S. Palm oil protects α-linolenic acid from rumen biohydrogenation and muscle oxidation in cashmere goat kids. J. Anim. Sci. Biotechnol. 2020, 11, 100. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, L.; Zhang, Y.; Zhang, X.; Li, F.; Huang, K.; Zhang, D.; Ma, Z.; Yan, C.; Zhang, Q.; Pu, M.; et al. Multidimensional Phenotypic and Microbiome Studies Uncover an Association Between Reduced Feed Efficiency in Sheep During Mycoplasmal Pneumonia and Microbial Crosstalk Within the Rumen-Lung Axis. Vet. Sci. 2025, 12, 741. https://doi.org/10.3390/vetsci12080741
Feng L, Zhang Y, Zhang X, Li F, Huang K, Zhang D, Ma Z, Yan C, Zhang Q, Pu M, et al. Multidimensional Phenotypic and Microbiome Studies Uncover an Association Between Reduced Feed Efficiency in Sheep During Mycoplasmal Pneumonia and Microbial Crosstalk Within the Rumen-Lung Axis. Veterinary Sciences. 2025; 12(8):741. https://doi.org/10.3390/vetsci12080741
Chicago/Turabian StyleFeng, Lianjun, Yukun Zhang, Xiaoxue Zhang, Fadi Li, Kai Huang, Deyin Zhang, Zongwu Ma, Chengqi Yan, Qi Zhang, Mengru Pu, and et al. 2025. "Multidimensional Phenotypic and Microbiome Studies Uncover an Association Between Reduced Feed Efficiency in Sheep During Mycoplasmal Pneumonia and Microbial Crosstalk Within the Rumen-Lung Axis" Veterinary Sciences 12, no. 8: 741. https://doi.org/10.3390/vetsci12080741
APA StyleFeng, L., Zhang, Y., Zhang, X., Li, F., Huang, K., Zhang, D., Ma, Z., Yan, C., Zhang, Q., Pu, M., Xiao, Z., Gao, L., Lin, C., Wu, W., Wang, W., & Tian, H. (2025). Multidimensional Phenotypic and Microbiome Studies Uncover an Association Between Reduced Feed Efficiency in Sheep During Mycoplasmal Pneumonia and Microbial Crosstalk Within the Rumen-Lung Axis. Veterinary Sciences, 12(8), 741. https://doi.org/10.3390/vetsci12080741







