Porcine Sample Type Characteristics Associated with Sequencing and Isolation of Influenza A Virus
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Overview of Study Design
2.2. Eligibility Criteria
2.3. Research Outcome
2.4. Veterinary Diagnostic Laboratory Data
2.5. Statistical Analyses
3. Results
3.1. HA Sanger Sequencing Results
3.2. NA Sanger Sequencing Results
3.3. Virus Isolation Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Reeth, K.; Vincent, A.L. Influenza viruses. Dis. Swine 2019, 36, 576–593. [Google Scholar]
- Maclachlan, N.J.; Edward, J.D. (Eds.) Chapter 21—Orthomyxoviridae. In Fenner’s Veterinary Virology, 5th ed.; Academic Press: Boston, MA, USA, 2017; pp. 389–410. [Google Scholar]
- Ferhadian, D.; Contrant, M.; Printz-Schweigert, A.; Smyth, R.P.; Paillart, J.-C.; Marquet, R. Structural and functional motifs in influenza virus RNAs. Front. Microbiol. 2018, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Goneau, L.; Mehta, K.; Wong, J.; L’Huillier, A.; Gubbay, J. Zoonotic influenza and human health—Part 1: Virology and epidemiology of zoonotic influenzas. Curr. Infect. Dis. Rep. 2018, 20, 37. [Google Scholar] [CrossRef] [PubMed]
- Pappaioanou, M.; Gramer, M. Lessons from pandemic H1N1 2009 to improve prevention, detection, and response to influenza pandemics from a One Health perspective. ILAR J. 2010, 51, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Burrough, E.R.; Magstadt, D.R.; Petersen, B.; Timmermans, S.J.; Gauger, P.C.; Zhang, J.; Siepker, C.; Mainenti, M.; Li, G.; Thompson, A.C. Highly pathogenic avian influenza A (H5N1) clade 2.3. 4.4 b virus infection in domestic dairy cattle and cats, United States, 2024. Emerg. Infect. Dis. 2024, 30, 1335. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.L.; Arruda, B.; Palmer, M.V.; Boggiatto, P.; Sarlo Davila, K.; Buckley, A.; Ciacci Zanella, G.; Snyder, C.A.; Anderson, T.K.; Hutter, C.; et al. Experimental reproduction of viral replication and disease in dairy calves and lactating cows inoculated with highly pathogenic avian influenza H5N1 clade 2.3.4.4b. bioRxiv 2024. [Google Scholar] [CrossRef]
- Arruda, B.; Baker, A.L.V.; Buckley, A.; Anderson, T.K.; Torchetti, M.; Bergeson, N.H.; Killian, M.L.; Lantz, K. Divergent pathogenesis and transmission of highly pathogenic avian influenza a (H5N1) in swine. Emerg. Infect. Dis. 2024, 30, 738. [Google Scholar] [CrossRef] [PubMed]
- USDA, APHIS. Federal and State Veterinary Agencies Share Update on HPAI Detections in Oregon Backyard Farm, Including First H5N1 Detections in Swine. Available online: https://www.aphis.usda.gov/news/agency-announcements/federal-state-veterinary-agencies-share-update-hpai-detections-oregon (accessed on 6 November 2024).
- Heil, G.L.; McCarthy, T.; Yoon, K.J.; Liu, S.; Saad, M.D.; Smith, C.B.; Houck, J.A.; Dawson, E.D.; Rowlen, K.L.; Gray, G.C. MChip, a low density microarray, differentiates among seasonal human H1N1, North American swine H1N1, and the 2009 pandemic H1N1. Influenza Other Respir. Viruses 2010, 4, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Latorre-Margalef, N.; Avril, A.; Tolf, C.; Olsen, B.; Waldenström, J. How does sampling methodology influence molecular detection and isolation success in influenza A virus field studies? Appl. Environ. Microbiol. 2016, 82, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gauger, P.C. Isolation of swine influenza virus in cell cultures and embryonated chicken eggs. In Animal Influenza Virus; Springer: New York, NY, USA, 2014; pp. 265–276. [Google Scholar]
- Janke, B. Influenza A virus infections in swine: Pathogenesis and diagnosis. Vet. Pathol. 2014, 51, 410–426. [Google Scholar] [CrossRef] [PubMed]
- Decorte, I.; Steensels, M.; Lambrecht, B.; Cay, A.B.; De Regge, N. Detection and isolation of swine influenza A virus in spiked oral fluid and samples from individually housed, experimentally infected pigs: Potential role of porcine oral fluid in active influenza A virus surveillance in swine. PLoS ONE 2015, 10, e0139586. [Google Scholar] [CrossRef] [PubMed]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Killian, M.L. Avian influenza virus sample types, collection, and handling. In Animal Influenza Virus; Springer: New York, NY, USA, 2014; pp. 83–91. [Google Scholar]
- Sharma, A.; Zeller, M.A.; Souza, C.K.; Anderson, T.K.; Vincent, A.L.; Harmon, K.; Li, G.; Zhang, J.; Gauger, P.C. Characterization of a 2016-2017 human seasonal H3 influenza a virus spillover now endemic to US swine. Msphere 2022, 7, e00809–e00821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gauger, P.C. Isolation of swine influenza A virus in cell cultures and embryonated chicken eggs. In Animal Influenza Virus: Methods and Protocols; Springer: New York, NY, USA, 2020; pp. 281–294. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing: R Foundation for Statistical Computing; R Package Version 3.6.1; R Core Team: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 8 July 2025).
- Lenth, R.; Singmann, H.; Love, J.; Buerkner, P.; Herve, M. Emmeans: Estimated Marginal Means, Aka Least-Squares Means. 2018. Available online: https://github.com/rvlenth/emmeans (accessed on 8 July 2025).
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D.; van Den Brand, T. Ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. 2007. Available online: https://search.r-project.org/CRAN/refmans/ggplot2/html/ggplot2-package.html (accessed on 8 July 2025).
- Wickham, H. Dplyr: A Grammar of Data Manipulation. 2015. Available online: https://dplyr.tidyverse.org (accessed on 8 July 2025).
- Robin, X.; Turck, N.; Hainard, H.; Tiberti, N.; Lisacek, F.; Sanchez, J.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Lele, S.R. A new method for estimation of resource selection probability function. J. Wildl. Manag. 2009, 73, 122–127. [Google Scholar] [CrossRef]
- Signorell, A.; Aho, K.; Alfons, A.; Anderegg, N.; Aragon, T.; Arppe, A.; Baddeley, A.; Barton, K.; Bolker, B.; Borchers, H.W. DescTools: Tools for Descriptive Statistics. 2025. Available online: https://cran.r-project.org/package=DescTools (accessed on 8 July 2025).
- Laboratory, ISU VDL. Applying World-Class Technology to Real-World Problems. Available online: https://vetmed.iastate.edu/vdl/#:~:text=Applying%20world%2Dclass%20technology%20to,than%201.7%20million%20tests%20annually. (accessed on 8 July 2025).
- Arruda, P.H.; Gauger, P. Optimizing sample selection, collection, and submission to optimize diagnostic value. Dis. Swine 2019, 7, 98–111. [Google Scholar]
- Bjustrom-Kraft, J.; Christopher-Hennings, J.; Daly, R.; Main, R.; Torrison, J.; Thurn, M.; Zimmerman, J. The use of oral fluid diagnostics in swine medicine. J. Swine Health Prod. 2018, 26, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Decorte, I.; Van der Stede, Y.; Nauwynck, H.; De Regge, N.; Cay, A.B. Effect of saliva stabilisers on detection of porcine reproductive and respiratory syndrome virus in oral fluid by quantitative reverse transcriptase real-time PCR. Vet. J. 2013, 197, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Hazra, A. Using the confidence interval confidently. J. Thorac. Dis. 2017, 9, 4125. [Google Scholar] [CrossRef] [PubMed]
- Moraes, D.C.; Cezar, G.A.; Magalhães, E.S.; Nicolino, R.R.; Rupasinghe, K.; Chandra, S.; Silva, G.S.; Almeida, M.N.; Crim, B.; Burrough, E.R. Macroepidemiological trends of Influenza A virus detection through reverse transcription real-time polymerase chain reaction (RT-rtPCR) in porcine samples in the United States over the last 20 years. Front. Vet. Sci. 2025, 12, 1572237. [Google Scholar] [CrossRef] [PubMed]
- Henao-Diaz, A.; Giménez-Lirola, L.; Baum, D.H.; Zimmerman, J. Guidelines for oral fluid-based surveillance of viral pathogens in swine. Porc. Health Manag. 2020, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Vereecke, N.; Woźniak, A.; Pauwels, M.; Coppens, S.; Nauwynck, H.; Cybulski, P.; Theuns, S.; Stadejek, T. Successful Whole Genome Nanopore Sequencing of Swine Influenza A Virus (SwIAV) Directly from Oral Fluids Collected in Polish Pig Herds. Viruses 2023, 15, 435. [Google Scholar] [CrossRef] [PubMed]
- Eichelberger, M.C.; Monto, A.S. Neuraminidase, the forgotten surface antigen, emerges as an influenza vaccine target for broadened protection. J. Infect. Dis. 2019, 219, S75–S80. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Mantilla, J.; Alvarez, J.; Culhane, M.; Nirmala, J.; Cano, J.P.; Torremorell, M. Comparison of individual, group and environmental sampling strategies to conduct influenza surveillance in pigs. BMC Vet. Res. 2019, 15, 61. [Google Scholar] [CrossRef] [PubMed]
- Toohey-Kurth, K.L.; Mulrooney, D.M.; Hinkley, S.; Lea Killian, M.; Pedersen, J.C.; Bounpheng, M.A.; Pogranichniy, R.; Bolin, S.; Maes, R.; Tallmadge, R.L. Best practices for performance of real-time PCR assays in veterinary diagnostic laboratories. J. Vet. Diagn. Investig. 2020, 32, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Hierholzer, J.; Killington, R. Virus isolation and quantitation. In Virology Methods Manual; Elsevier: Amsterdam, The Netherlands, 1996; pp. 25–46. [Google Scholar]
- Christopher-Hennings, J.; Erickson, G.A.; Hesse, R.A.; Nelson, E.A.; Rossow, S.; Scaria, J.; Slavic, D. Diagnostic tests, test performance, and considerations for interpretation. Dis. Swine 2019, 6, 75–97. [Google Scholar]
- Detmer, S.; Gramer, M.; Goyal, S.; Torremorell, M.; Torrison, J. Diagnostics and surveillance for swine influenza. In Swine Influenza; Springer: Berlin/Heidelberg, Germany, 2013; pp. 85–112. [Google Scholar]
- Detmer, S.E.; Patnayak, D.P.; Jiang, Y.; Gramer, M.R.; Goyal, S.M. Detection of Influenza A virus in porcine oral fluid samples. J. Vet. Diagn. Investig. 2011, 23, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Goel, V.; Ding, J.; Hatuwal, B.; Giri, E.; Deliberto, T.J.; Lowe, J.; Webby, R.; Emch, M.; Wan, X.-F. Ecological drivers of evolution of swine influenza in the United States: A review. Emerg. Microbes Infect. 2025, 14, 2455598. [Google Scholar] [CrossRef] [PubMed]
- Spackman, E.; Pedersen, J.C.; McKinley, E.T.; Gelb, J. Optimal specimen collection and transport methods for the detection of avian influenza virus and Newcastle disease virus. BMC Vet. Res. 2013, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Asanzhanova, N.; Ryskeldinova, S.Z.; Chervyakova, O.; Khairullin, B.; Kasenov, M.; Tabynov, K. Comparison of different methods of purification and concentration in production of influenza vaccine. Bull. Exp. Biol. Med. 2017, 164, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Pawar, S.D.; Keng, S.S.; Tare, D.S.; Thormothe, A.L.; Sapkal, G.N.; Anukumar, B.; Lole, K.S.; Mullick, J.; Mourya, D.T. A virus precipitation method for concentration & detection of avian influenza viruses from environmental water resources & its possible application in outbreak investigations. Indian J. Med. Res. 2019, 150, 612–619. [Google Scholar] [PubMed]
- Dohoo, I.R. Bias—Is it a problem, and what should we do? Prev. Vet. Med. 2014, 113, 331–337. [Google Scholar] [CrossRef] [PubMed]

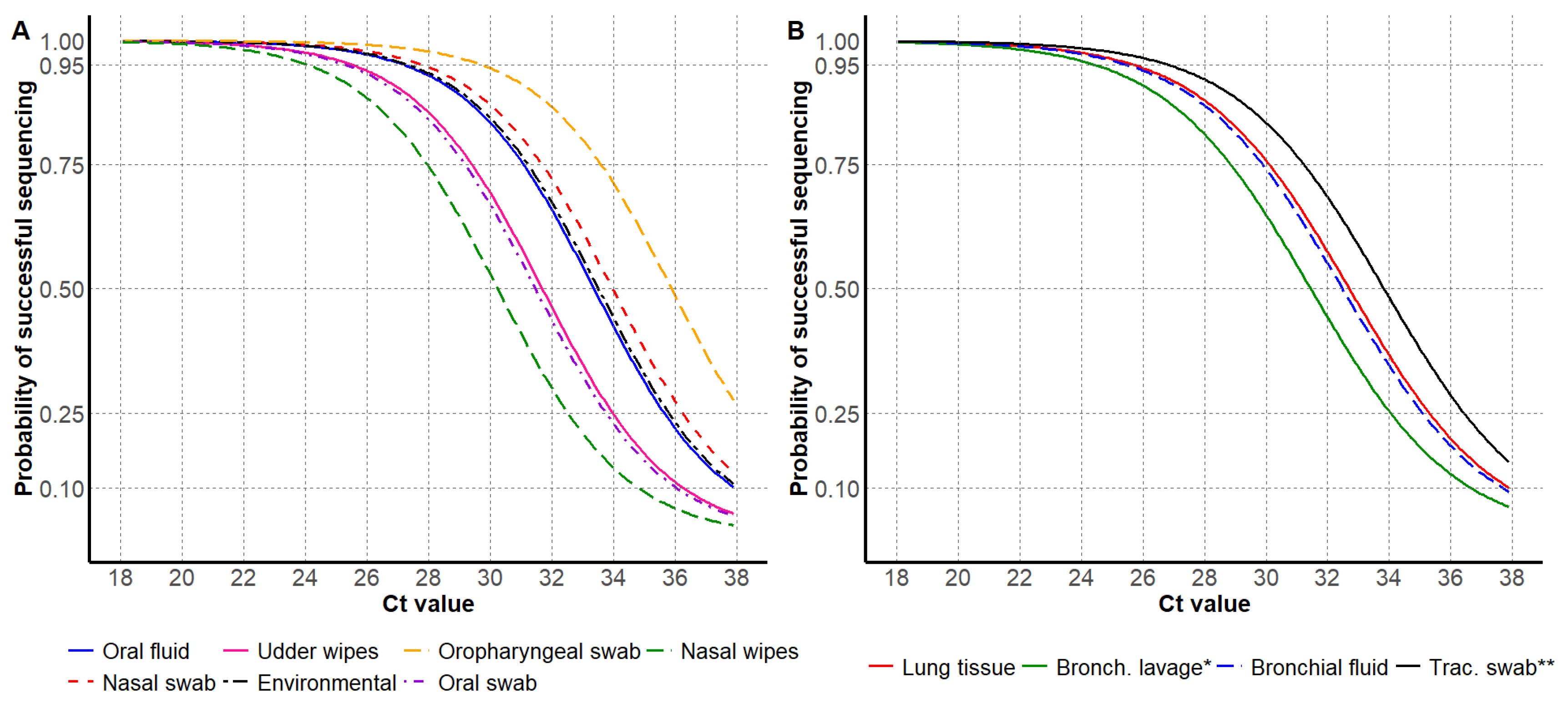
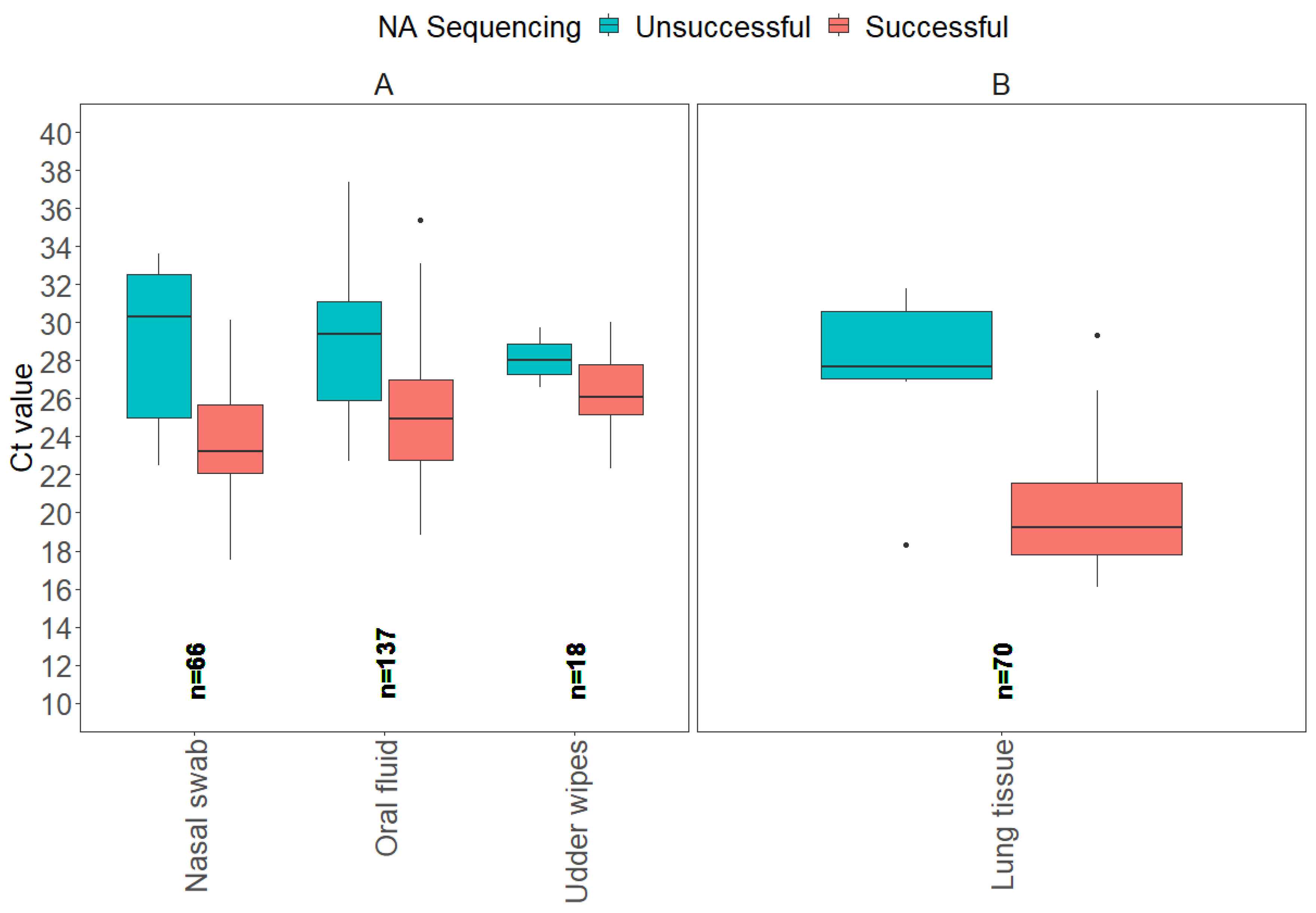
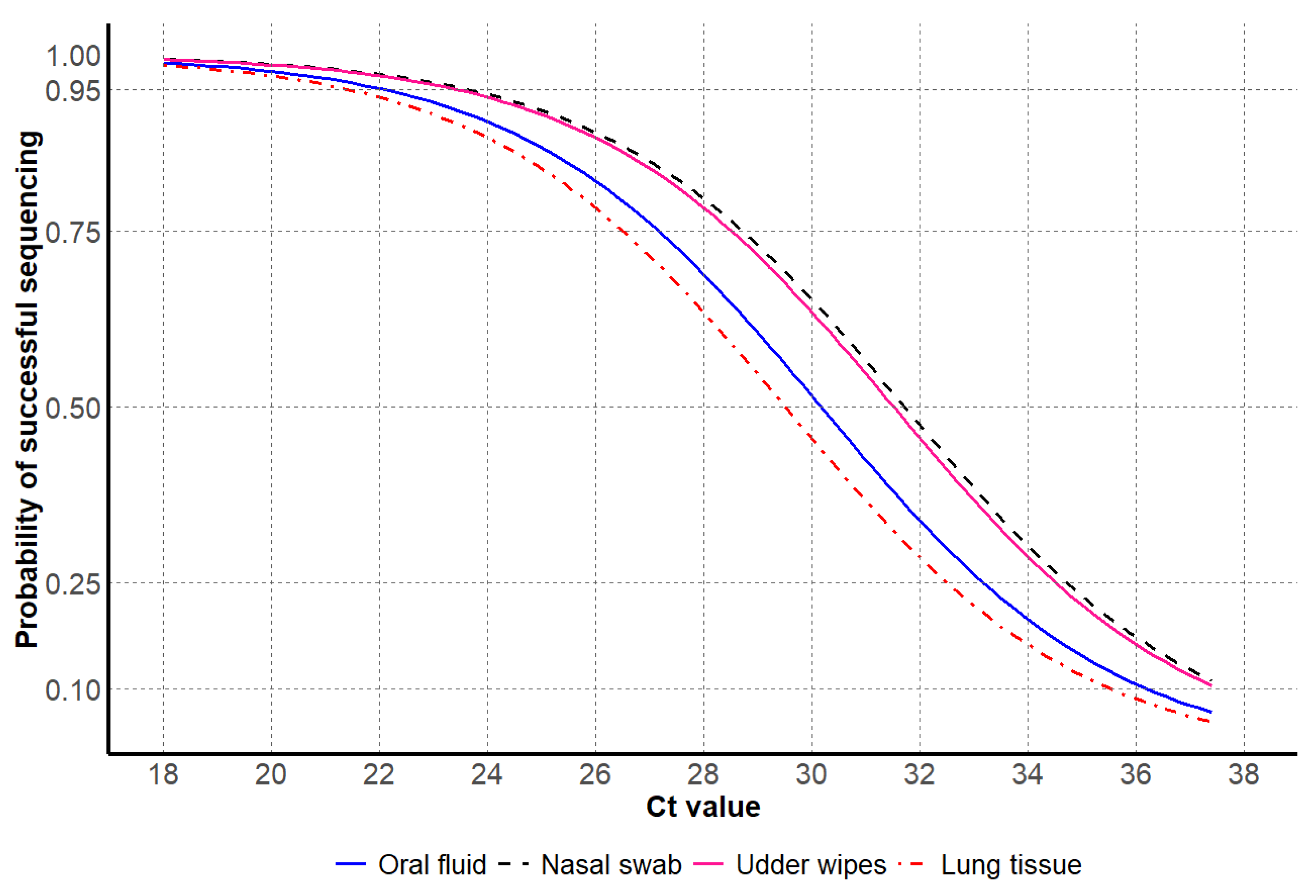
| Sample Type | HA Sequencing Successful | HA Sequencing Unsuccessful |
|---|---|---|
| Antemortem Samples | Ct Value: Average (Min–Max) | Ct Value: Average (Min–Max) |
| Environmental | 27.2 (21.5–32.4) | 34.6 (31.1–37.4) |
| Nasal swab | 24.9 (13.6–37.5) | 32.1 (23.1–37.6) |
| Nasal wipes | 23.7 (20.6–30.4) | 33.4 (29.1–37.8) |
| Oral fluid | 25.9 (15.0–37.8) | 32.4 (21.4–37.9) |
| Oral swab | 27.3 (20.2–34.3) | 27.9 (25.0–29.7) |
| Oropharyngeal swab | 27.2 (20.5–33.8) | 34.7 (32.1–37.3) |
| Udder wipes | 27.2 (22.3–33.1) | 30.6 (26.4–35.9) |
| Postmortem samples | ||
| Bronchial fluid | 22.1 (16.2–33.3) | 31.1 (24.8–36.3) |
| Bronchoalveolar lavage | 21.5 (17.2–28.0) | 34.3 (31.7–35.8) |
| Lung tissue | 21.3 (12.1–37.1) | 30.8 (19.3–37.9) |
| Tracheal swab | 25.4 (21.0–29.6) | 29.6 (25.0–34.6) |
| Sample Type | Ct Value by Probability of Success (C.I. 1) | Total Number | Proportion | ||
|---|---|---|---|---|---|
| Antemortem Samples | 95% | 75% | 50% | ||
| Environmental | 27.4 (25.4, 29.0) | 31.2 (29.4, 32.9) | 33.5 (31.8, 35.3) | 59 | 0.9% |
| Nasal swab | 27.8 (27.1, 28.4) | 31.7 (31.1, 32.4) | 34.0 (33.4, 34.8) | 1046 | 16.3% |
| Nasal wipes | 24.1 (18.8, 29.2) | 28.0 (22.7, 33.1) | 30.3 (25.1, 35.5) | 17 | 0.3% |
| Oral fluid | 27.3 (26.7, 27.5) | 31.1 (30.8, 31.4) | 33.3 (33.1, 33.8) | 3445 | 53.6% |
| Oral swab | 25.4 (21.7, 28.7) | 29.0 (25.7, 32.7) | 31.5 (28.1, 34.9) | 13 | 0.2% |
| Oropharyngeal swab | 29.8 (26.1, 33.3) | 33.6 (30.1, 37.2) | 35.9 (32.4, 37.9) | 30 | 0.5% |
| Udder wipes | 25.5 (24.3, 26.6) | 29.4 (28.3, 30.5) | 31.7 (30.6, 32.9) | 123 | 1.9% |
| Postmortem samples | |||||
| Bronchial fluid | 25.6 (23.6, 28.3) | 29.9 (27.5, 32.3) | 32.5 (29.8, 34.6) | 73 | 1.1% |
| Bronchoalveolar lavage | 24.9 (20.8, 28.9) | 28.8 (24.7, 32.9) | 31.4 (27.1, 35.2) | 29 | 0.5% |
| Lung tissue | 25.7 (25.4, 26.7) | 30.1 (29.4, 30.6) | 32.7 (31.7, 33.0) | 1575 | 24.5% |
| Tracheal swab | 26.9 (22.5, 31.4) | 31.3 (26.5, 35.3) | 33.8 (28.9, 37.7) | 20 | 0.3% |
| Total | 6430 | 100% | |||
| Sample Type | NA Sequencing Successful | NA Sequencing Unsuccessful |
|---|---|---|
| Antemortem Samples | Ct Value: Average (Min–Max) | Ct Value: Average (Min–Max) |
| Nasal swab | 23.9 (17.5–30.1) | 28.8 (22.5–33.6) |
| Oral fluid | 25.1 (18.8–35.4) | 29.0 (22.7–37.4) |
| Udder wipes | 26.5 (22.3–30.0) | 28.1 (26.6–29.7) |
| Postmortem samples | ||
| Lung tissue | 20.1 (16.1–29.3) | 27.3 (18.3–31.8) |
| Sample Type | Ct Value by Probability of Success (C.I. 1) | Total Number | Proportion | ||
|---|---|---|---|---|---|
| Antemortem Samples | 95% | 75% | 50% | ||
| Nasal swab | 23.6 (20.6, 26.1) | 28.8 (26.3, 31.4) | 31.8 (29.2, 35.1) | 66 | 22.8% |
| Oral fluid | 22.1 (19.3, 23.8) | 27.1 (25.8, 28.4) | 30.1 (28.9, 32.0) | 137 | 47.2% |
| Udder wipes | 23.4 (19.3, 27.1) | 28.6 (25.0, 32.4) | 31.6 (28.1, 36.0) | 18 | 5.9% |
| Postmortem samples | |||||
| Lung tissue | 21.5 (18.1, 24.1) | 26.4 (23.8, 29.5) | 29.4 (26.8, 33.1) | 70 | 24.1% |
| Total | 291 | 100% | |||
| Sample Type | Virus Isolation Successful | Virus Isolation Unsuccessful |
|---|---|---|
| Antemortem Samples | Ct Value: Average (Min–Max) | Ct Value: Average (Min–Max) |
| Nasal swab | 24.3 (14.9–37.0) | 30.1 (18.9–37.9) |
| Oral fluid | 22.6 (15.5–34.1) | 26.9 (18.2–37.7) |
| Postmortem samples | ||
| Bronchoalveolar lavage | 21.1 (17.4–26.3) | 24.1 (21.1–28.1) |
| Lung tissue | 19.7 (11.5–33.4) | 22.5 (12.5–37.1) |
| Tracheal swab | 25.5 (22.5–28.6) | 28.6 (26.8–32.2) |
| Sample Type | Ct Value by Probability of Success (C.I.1) | Total Number | Proportion | ||
|---|---|---|---|---|---|
| Antemortem Samples | 95% | 75% | 50% | ||
| Nasal swab | 21.1 (20.1, 21.9) | 27.0 (26.3, 27.8) | 30.6 (29.9, 31.4) | 647 | 8.8% |
| Oral fluid | 13.9 (12.9, 14.9) | 20.0 (19.3, 20.5) | 23.5 (22.9, 24.0) | 760 | 10.3% |
| Postmortem samples | |||||
| Bronchoalveolar lavage | 15.5 (13.0, 17.8) | 21.4 (19.0, 23.7) | 24.8 (22.6, 27.3) | 39 | 0.5% |
| Lung tissue | 18.7 (18.4, 19.2) | 24.8 (24.4, 25.1) | 28.3 (27.8, 29.0) | 5892 | 80.2% |
| Tracheal swab | 15.5 (11.5, 21.4) | 21.4 (15.5, 27.4) | 25.1 (19.1, 31.0) | 6 | 0.1% |
| Total | 7344 | 100% | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moraes, D.C.A.; Osemeke, O.H.; Zeller, M.A.; Baker, A.L.; Silva, G.S.; Trevisan, G.; Linhares, D.C.L.; Gauger, P.C. Porcine Sample Type Characteristics Associated with Sequencing and Isolation of Influenza A Virus. Vet. Sci. 2025, 12, 683. https://doi.org/10.3390/vetsci12070683
Moraes DCA, Osemeke OH, Zeller MA, Baker AL, Silva GS, Trevisan G, Linhares DCL, Gauger PC. Porcine Sample Type Characteristics Associated with Sequencing and Isolation of Influenza A Virus. Veterinary Sciences. 2025; 12(7):683. https://doi.org/10.3390/vetsci12070683
Chicago/Turabian StyleMoraes, Daniel C. A., Onyekachukwu H. Osemeke, Michael A. Zeller, Amy L. Baker, Gustavo S. Silva, Giovani Trevisan, Daniel C. L. Linhares, and Phillip C. Gauger. 2025. "Porcine Sample Type Characteristics Associated with Sequencing and Isolation of Influenza A Virus" Veterinary Sciences 12, no. 7: 683. https://doi.org/10.3390/vetsci12070683
APA StyleMoraes, D. C. A., Osemeke, O. H., Zeller, M. A., Baker, A. L., Silva, G. S., Trevisan, G., Linhares, D. C. L., & Gauger, P. C. (2025). Porcine Sample Type Characteristics Associated with Sequencing and Isolation of Influenza A Virus. Veterinary Sciences, 12(7), 683. https://doi.org/10.3390/vetsci12070683






