Comparative Analysis of Bacterial Diversity and Composition in Oral Fluid from Pigs of Different Ages and Water Pipe Wall Biofilms
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Sample Detection
2.3. Construction and Sequencing of Library
2.4. Sequencing Data Quality Control, Assembly, and Analysis
2.5. Sequencing and Data Processing of Oral Microbial Genomic DNA
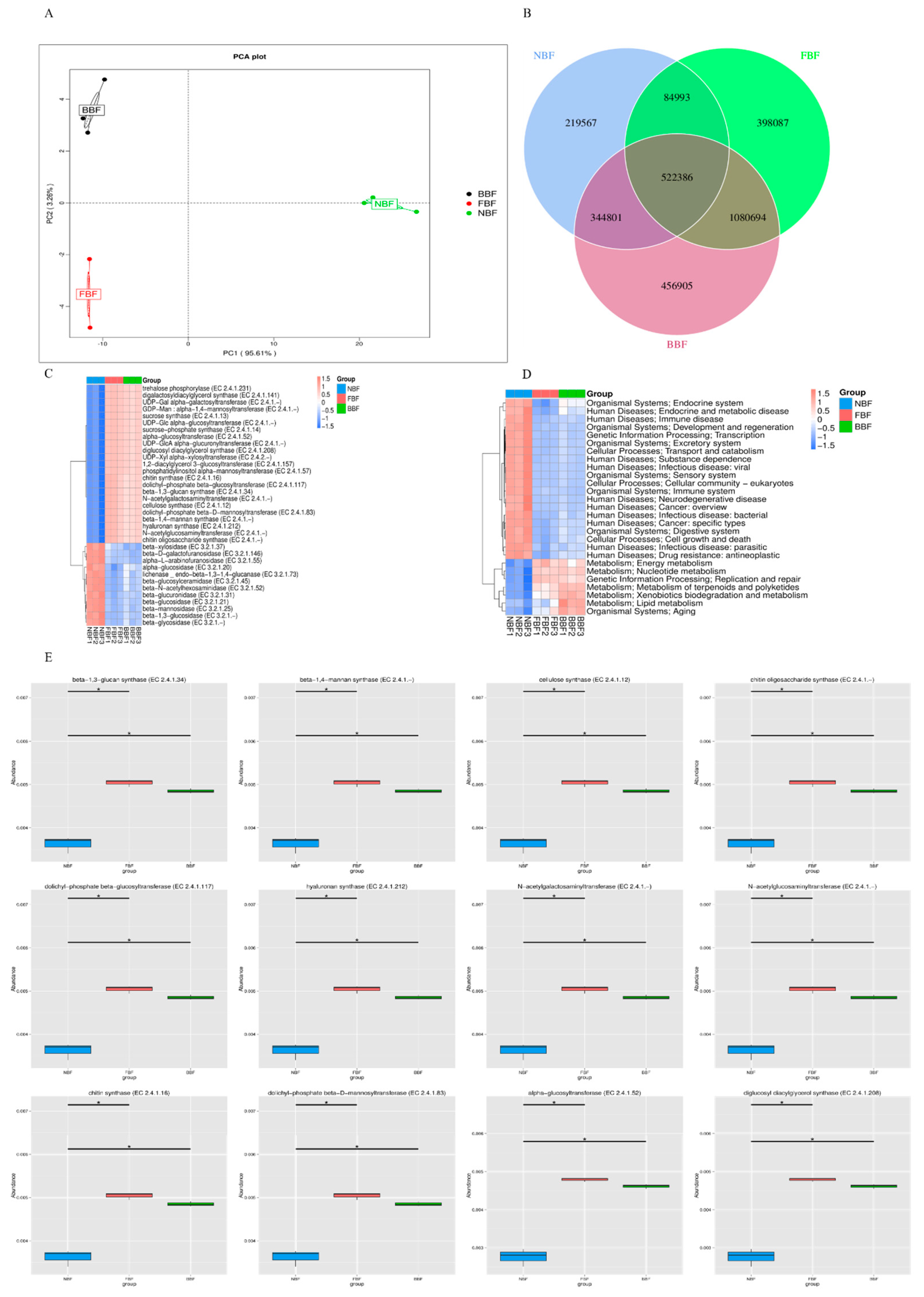
2.6. Oral Microbial Function Prediction Methods
2.7. Statistical Analysis
3. Results
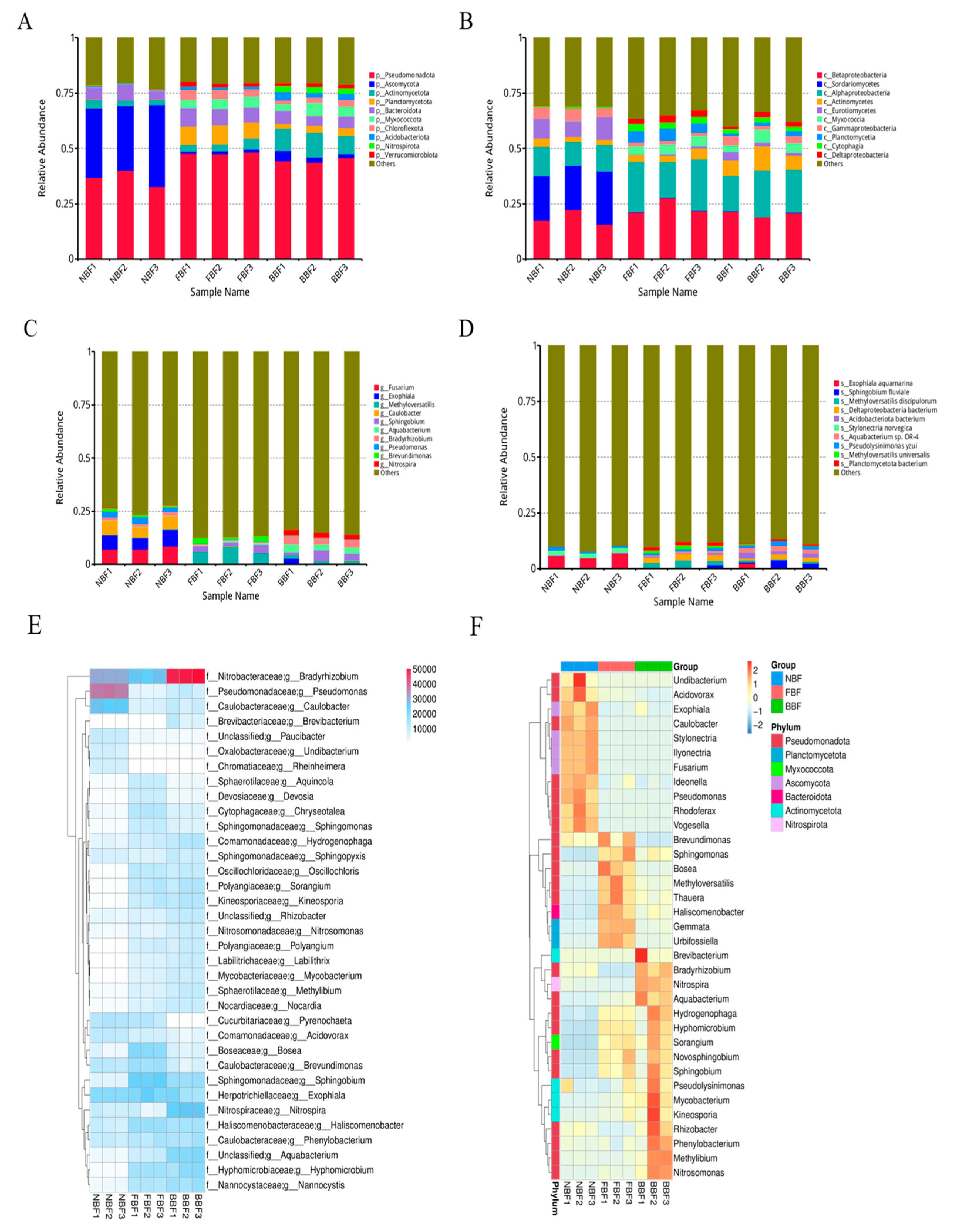
3.1. Pre-Feeding Waterline Bacteria Test
3.2. Quality Control of Data and Metagenome Assembly
3.3. Differences in Gene Distribution Among Samples from Three Pig Houses
3.4. Correlation Analysis Between Samples Based on Gene Number
3.5. Taxonomic Analysis
3.6. Phylogenetic Analysis
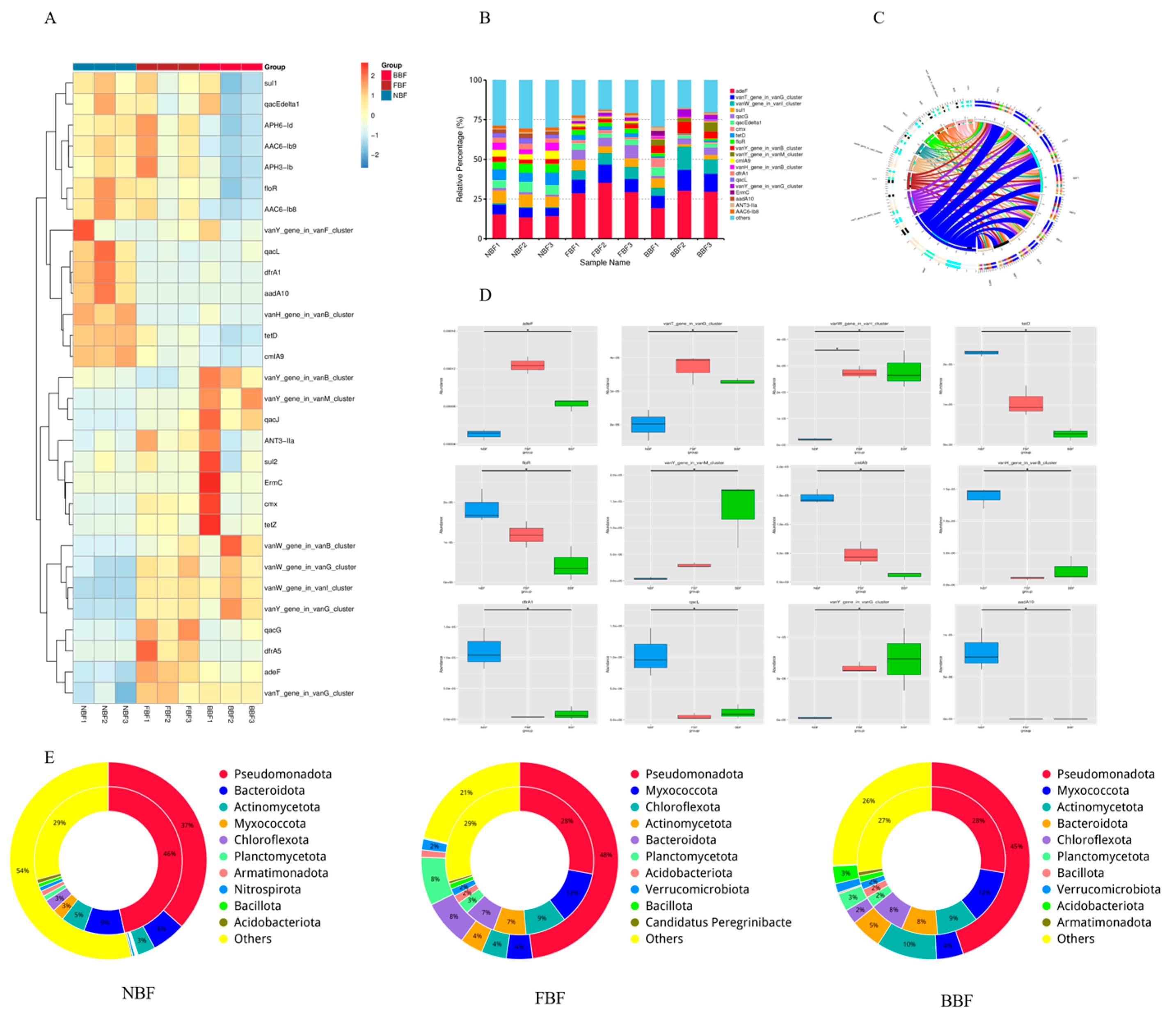
3.7. ARGs Analysis
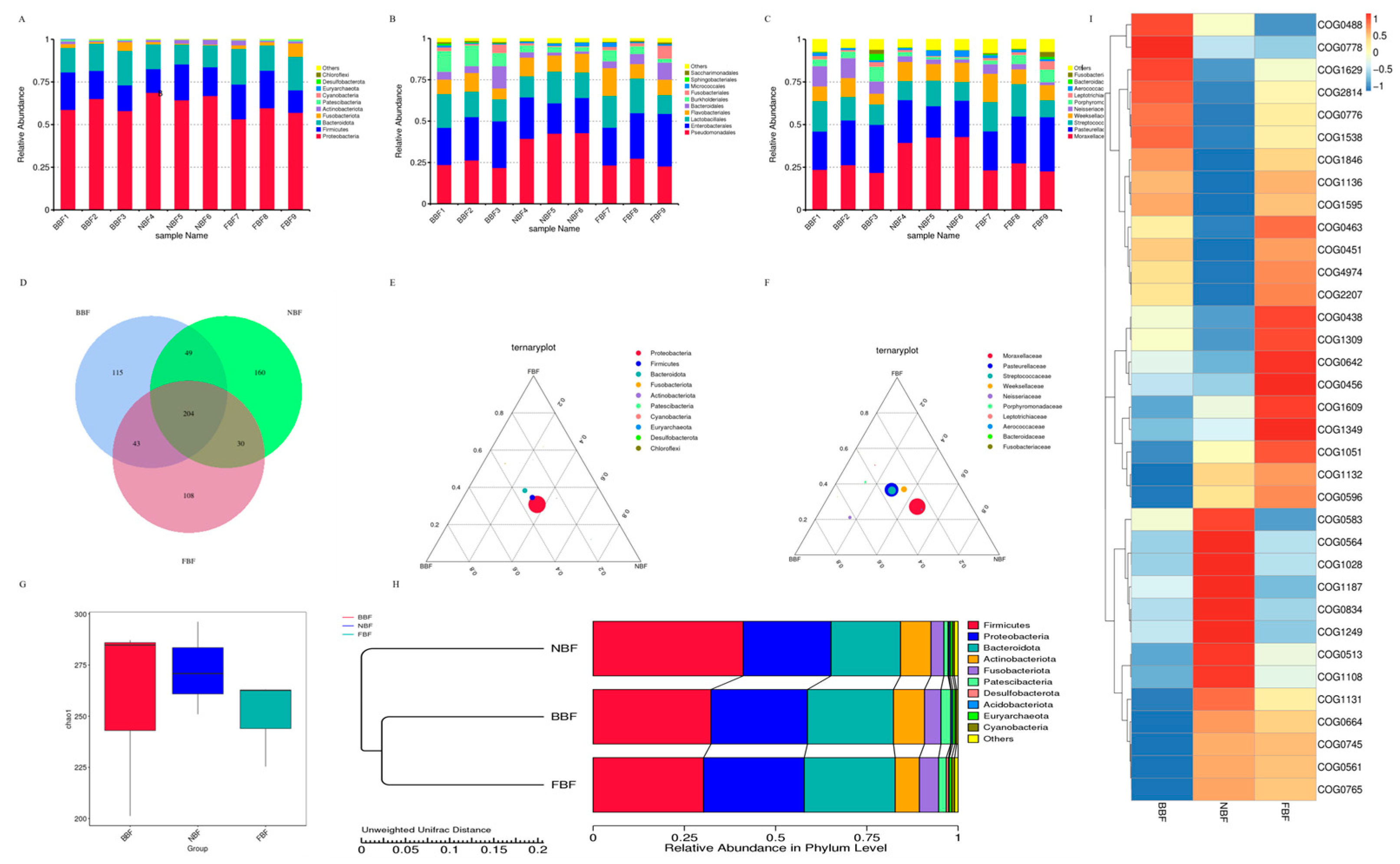
3.8. Amplicon Detection of Saliva Samples from Pigs at Different Stages
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Karlsson, F.H.; Tremaroli, V.; Nookaew, I.; Bergström, G.; Behre, C.J.; Fagerberg, B.; Nielsen, J.; Bäckhed, F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013, 498, 99–103. [Google Scholar] [CrossRef]
- Karlsson, F.H.; Fåk, F.; Nookaew, I.; Tremaroli, V.; Fagerberg, B.; Petranovic, D.; Bäckhed, F.; Nielsen, J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012, 3, 1245. [Google Scholar] [CrossRef]
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013, 2, e01202. [Google Scholar] [CrossRef]
- Zheng, L.; Duarte, M.E.; Sevarolli Loftus, A.; Kim, S.W. Intestinal health of pigs upon weaning: Challenges and nutritional intervention. Front. Vet. Sci. 2021, 8, 628258. [Google Scholar] [CrossRef]
- Hattab, J.; Marruchella, G.; Pallavicini, A.; Gionechetti, F.; Mosca, F.; Trachtman, A.R.; Lanci, L.; Gabrielli, L.; Tiscar, P.G. Insights into the oral bacterial microbiota of sows. Microorganisms 2021, 9, 2314. [Google Scholar] [CrossRef]
- de Vries, H.J.; Beyer, F.; Jarzembowska, M.; Lipińska, J.; van den Brink, P.; Zwijnenburg, A.; Timmers, P.H.A.; Stams, A.J.M.; Plugge, C.M. Isolation and characterization of Sphingomonadaceae from fouled membranes. NPJ Biofilms Microbiomes 2019, 5, 6. [Google Scholar] [CrossRef]
- Xu, M.; Li, H.; Li, S.; Li, C.; Li, J.; Ma, Y. The presence of tetracyclines and sulfonamides in swine feeds and feces: Dependence on the antibiotic type and swine growth stages. Environ. Sci. Pollut. Res. Int. 2020, 27, 43093–43102. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, X.; Yang, X.; Que, J.; Du, Q.; Zhang, Q.; Zou, J. Oral health, caries risk profiles, and oral microbiome of pediatric patients with leukemia submitted to chemotherapy. Biomed. Res. Int. 2021, 2021, 6637503. [Google Scholar] [CrossRef]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef]
- He, K.; Xiong, J.; Yang, W.; Zhao, L.; Wang, T.; Qian, W.; Hu, S.; Wang, Q.; Aleem, M.T.; Miao, W.; et al. Metagenome of gut microbiota provides a novel insight into the pathogenicity of balantioides coli in weaned piglets. Int. J. Mol. Sci. 2023, 24, 10791. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.Q.; Vu, H.P.; Nguyen, L.N.; Wang, Q.; Djordjevic, S.P.; Donner, E.; Yin, H.; Nghiem, L.D. Monitoring antibiotic resistance genes in wastewater treatment: Current strategies and future challenges. Sci. Total Environ. 2021, 783, 146964. [Google Scholar] [CrossRef]
- Zheng, D.; Yin, G.; Liu, M.; Chen, C.; Jiang, Y.; Hou, L.; Zheng, Y. A systematic review of antibiotics and antibiotic resistance genes in estuarine and coastal environments. Sci. Total Environ. 2021, 777, 146009. [Google Scholar] [CrossRef]
- Li, B.; Qiu, Y.; Song, Y.; Lin, H.; Yin, H. Dissecting horizontal and vertical gene transfer of antibiotic resistance plasmid in bacterial community using microfluidics. Environ. Int. 2019, 131, 105007. [Google Scholar] [CrossRef]
- Han, Z.; Zhang, Y.; An, W.; Lu, J.; Hu, J.; Yang, M. Antibiotic resistomes in drinking water sources across a large geographical scale: Multiple drivers and co-occurrence with opportunistic bacterial pathogens. Water Res. 2020, 183, 116088. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, R.; Chen, B.; Zhang, T.; Hu, L.; Zou, S. Characterization of airborne antibiotic resistance genes from typical bioaerosol emission sources in the urban environment using metagenomic approach. Chemosphere 2018, 213, 463–471. [Google Scholar] [CrossRef]
- Liu, K.; Wang, M.; Zhang, Y.; Fang, C.; Zhang, R.; Fang, L.; Sun, J.; Liu, Y.; Liao, X. Distribution of antibiotic resistance genes and their pathogen hosts in duck farm environments in south-east coastal China. Appl. Microbiol. Biotechnol. 2024, 108, 136. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Hess, M.; Sczyrba, A.; Egan, R.; Kim, T.W.; Chokhawala, H.; Schroth, G.; Luo, S.; Clark, D.S.; Chen, F.; Zhang, T.; et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 2011, 331, 463–467. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The carbohydrate-active enzymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Leão, I.; de Carvalho, T.B.; Henriques, V.; Ferreira, C.; Sampaio-Maia, B.; Manaia, C.M. Pseudomonadota in the oral cavity: A glimpse into the environment-human nexus. Appl. Microbiol. Biotechnol. 2023, 107, 517–534. [Google Scholar] [CrossRef]
- Fishbein, S.R.S.; Mahmud, B.; Dantas, G. Antibiotic perturbations to the gut microbiome. Nat. Rev. Microbiol. 2023, 21, 772–788. [Google Scholar] [CrossRef]
- Wang, H.; Masters, S.; Hong, Y.; Stallings, J.; Falkinham, J.O., 3rd; Edwards, M.A.; Pruden, A. Effect of disinfectant, water age, and pipe material on occurrence and persistence of Legionella, mycobacteria, Pseudomonas aeruginosa, and two amoebas. Environ. Sci. Technol. 2012, 46, 11566–11574. [Google Scholar] [CrossRef]
- Cerrato, J.M.; Falkinham, J.O., 3rd; Dietrich, A.M.; Knocke, W.R.; McKinney, C.W.; Pruden, A. Manganese-oxidizing and -reducing microorganisms isolated from biofilms in chlorinated drinking water systems. Water Res. 2010, 44, 3935–3945. [Google Scholar] [CrossRef]
- He, H.; Yang, M.; Li, W.; Lu, Z.; Wang, Y.; Jin, M. Fecal microbial and metabolic characteristics of swine from birth to market. Front. Microbiol. 2023, 14, 1191392. [Google Scholar] [CrossRef]
- Adhikari, B.; Kim, S.W.; Kwon, Y.M. Characterization of Microbiota Associated with Digesta and Mucosa in Different Regions of Gastrointestinal Tract of Nursery Pigs. Int. J. Mol. Sci. 2019, 20, 1630. [Google Scholar] [CrossRef]
- Wardman, J.F.; Bains, R.K.; Rahfeld, P.; Withers, S.G. Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat. Rev. Microbiol. 2022, 20, 542–556. [Google Scholar] [CrossRef]
- Cui, H.; Zhang, C.; Liu, J.; Dong, S.; Zhao, K.; Chen, L.; Chen, Z.; Sun, Y.; Guo, Z. The distribution characteristics of aerosol bacteria in different types of pig houses. Animals 2022, 12, 1540. [Google Scholar] [CrossRef]
- Kimbell, L.K.; LaMartina, E.L.; Kappell, A.D.; Huo, J.; Wang, Y.; Newton, R.J.; McNamara, P.J. Cast iron drinking water pipe biofilms support diverse microbial communities containing antibiotic resistance genes, metal resistance genes, and class 1 integrons. Environ. Sci. Water Res. Technol. 2021, 7, 584–598. [Google Scholar] [CrossRef]
- Coyne, S.; Rosenfeld, N.; Lambert, T.; Courvalin, P.; Périchon, B. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2010, 54, 4389–4393. [Google Scholar] [CrossRef]
- Zhang, X.X.; Zhang, T.; Fang, H.H. Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 2009, 82, 397–414. [Google Scholar] [CrossRef]
- Cameron, E.A.; Sperandio, V.; Dunny, G.M. Enterococcus faecalis Enhances Expression and Activity of the Enterohemorrhagic Escherichia coli Type III Secretion System. mBio 2019, 10, e02547-19. [Google Scholar] [CrossRef]
- Ulloa, E.R.; Singh, K.V.; Geriak, M.; Haddad, F.; Murray, B.E.; Nizet, V.; Sakoulas, G. Cefazolin and Ertapenem Salvage Therapy Rapidly Clears Persistent Methicillin-Susceptible Staphylococcus aureus Bacteremia. Clin. Infect. Dis. 2020, 71, 1413–1418. [Google Scholar] [CrossRef]
- Shin, H.; Kwak, Y.; Jo, S.K.; Kim, S.H.; Huh, J.H. Applicability evaluation of a demand-controlled ventilation system in livestock. Comput. Electron. Agric. 2022, 196, 106907. [Google Scholar] [CrossRef]
- Li, X.; Fields, F.R.; Ho, M.; Marshall-Hudson, A.; Gross, R.; Casser, M.E.; Naito, M. Safety assessment of Streptococcus salivarius DB-B5 as a probiotic candidate for oral health. Food Chem. Toxicol. 2021, 153, 112277. [Google Scholar] [CrossRef]
- Baek, D.H.; Lee, S.H. Anti-Inflammatory Efficacy of Human-Derived Streptococcus salivarius on Periodontopathogen-Induced Inflammation. J. Microbiol. Biotechnol. 2023, 33, 998–1005. [Google Scholar] [CrossRef]
- Shiroma, H.; Darzi, Y.; Terajima, E.; Nakagawa, Z.; Tsuchikura, H.; Tsukuda, N.; Moriya, Y.; Okuda, S.; Goto, S.; Yamada, T. Enteropathway: The metabolic pathway database for the human gut microbiota. Brief. Bioinform. 2024, 25, bbae419. [Google Scholar] [CrossRef]
- Nagakubo, D.; Kaibori, Y. Oral Microbiota: The Influences and Interactions of Saliva, IgA, and Dietary Factors in Health and Disease. Microorganisms 2023, 11, 2307. [Google Scholar] [CrossRef]
- Zhang, S.M.; Hung, J.H.; Yen, T.N.; Huang, S.L. Mutualistic interactions of lactate-producing lactobacilli and lactate-utilizing Veillonella dispar: Lactate and glutamate cross-feeding for the enhanced growth and short-chain fatty acid production. Microb. Biotechnol. 2024, 17, e14484. [Google Scholar] [CrossRef]
- Xin, C.; Khu, S.T.; Wang, T.; Zuo, X.; Zhang, Y. Effect of flow fluctuation on water pollution in drinking water distribution systems. Environ. Res. 2024, 246, 118142. [Google Scholar] [CrossRef]
- Cugini, C.; Shanmugam, M.; Landge, N.; Ramasubbu, N. The Role of Exopolysaccharides in Oral Biofilms. J. Dent. Res. 2019, 98, 739–745. [Google Scholar] [CrossRef]
- Chen, W.; Ma, Q.; Li, Y.; Wei, L.; Zhang, Z.; Khan, A.; Khan, M.Z.; Wang, C. Butyrate Supplementation Improves Intestinal Health and Growth Performance in Livestock: A Review. Biomolecules 2025, 15, 85. [Google Scholar] [CrossRef]
- Wake, N.; Asahi, Y.; Noiri, Y.; Hayashi, M.; Motooka, D.; Nakamura, S.; Gotoh, K.; Miura, J.; Machi, H.; Iida, T.; et al. Temporal dynamics of bacterial microbiota in the human oral cavity determined using an in situ model of dental biofilms. NPJ Biofilms Microbiomes 2016, 2, 16018. [Google Scholar] [CrossRef]


| Sample | Insert Size (bp) | Raw Data | Clean Data | Clean Q20 | Clean Q30 | Clean GC (%) |
|---|---|---|---|---|---|---|
| NBF1 | 350 | 13,363.08 | 13,341.93 | 97.58 | 93.47 | 54.56 |
| NBF2 | 350 | 12,937.60 | 12,916.54 | 97.52 | 93.35 | 54.04 |
| NBF3 | 350 | 12,710.49 | 12,688.42 | 97.57 | 93.51 | 53.66 |
| FBF1 | 350 | 13,019.04 | 13,000.19 | 97.34 | 93.07 | 61.09 |
| FBF2 | 350 | 12,451.71 | 12,428.16 | 97.31 | 92.94 | 61.41 |
| FBF3 | 350 | 12,998.80 | 12,970.01 | 97.52 | 93.48 | 61.84 |
| BBF1 | 350 | 12,957.69 | 12,926.97 | 97.61 | 93.38 | 60.17 |
| BBF2 | 350 | 12,974.73 | 12,939.75 | 97.66 | 93.55 | 62.41 |
| BBF3 | 350 | 13,330.60 | 13,292.03 | 97.68 | 93.64 | 61.51 |
| Sample ID | Total Length (bp) | Number of Scaffolds | Average Length (bp) | N50 Length (bp) | N90 Length (bp) | Max Length (bp) |
|---|---|---|---|---|---|---|
| NBF1 | 348,804,097 | 250,672 | 1391.48 | 1791 | 601 | 259,671 |
| NBF2 | 359,763,692 | 260,253 | 1382.36 | 1752 | 602 | 130,849 |
| NBF3 | 287,773,422 | 217,356 | 1323.97 | 1587 | 596 | 200,836 |
| FBF1 | 677,294,730 | 459,987 | 1472.42 | 2037 | 599 | 639,520 |
| FBF2 | 626,882,810 | 421,083 | 1488.74 | 2061 | 603 | 694,772 |
| FBF3 | 684,005,321 | 494,497 | 1383.23 | 1696 | 596 | 397,351 |
| BBF1 | 662,760,463 | 520,064 | 1274.38 | 1426 | 577 | 578,482 |
| BBF2 | 740,577,707 | 533,990 | 1386.88 | 1744 | 606 | 298,203 |
| BBF3 | 772,220,995 | 607,619 | 1270.90 | 1466 | 589 | 389,864 |
| Sample | NBF1 | NBF2 | NBF3 | FBF1 | FBF2 | FBF3 | BBF1 | BBF2 | BBF3 |
|---|---|---|---|---|---|---|---|---|---|
| Gene_number | 912,055 | 862,381 | 811,137 | 1,684,947 | 1,646,640 | 1,757,003 | 1,816,062 | 1,840,404 | 1,980,463 |
| COG ID | Description |
|---|---|
| COG0488 | ABC cassette proteins with duplicated ATPase domains, Uup/ABCF family |
| COG0778 | Nitroreductase |
| COG1629 | Outer membrane receptor protein, Fe transport |
| COG2814 | Predicted arabinose efflux permease AraJ, MFS family |
| COG0776 | DNA-binding chromatin protein HU or IHF, alpha or beta variants |
| COG1538 | Outer membrane protein TolC |
| COG1846 | DNA-binding transcriptional regulator, MarR family |
| COG1136 | ABC-type lipoprotein targeting system ATPase component LolD |
| COG1595 | DNA-directed RNA polymerase specialized sigma subunit, sigma24 family |
| COG0463 | Glycosyltransferase involved in cell wall bisynthesis |
| COG0451 | Nucleoside-diphosphate-sugar epimerase |
| COG4974 | Site-specific tyrosine recombinase XerD |
| COG2207 | AraC-type DNA-binding domain and AraC-containing proteins |
| COG0438 | Lipopolysaccharide 1,6-galactosyltransferase, GT1 family |
| COG1309 | DNA-binding protein, AcrR family, includes nucleoid occlusion protein SlmA |
| COG0642 | Signal transduction histidine kinase |
| COG0456 | Ribosomal protein S18 acetylase RimI and related acetyltransferases |
| COG1609 | DNA-binding transcriptional regulator, LacI/PurR family G1349—DNA-binding transcriptional regulator of sugar metabolism, DeoR/GlpR family |
| COG1051 | ADP-ribose pyrophosphatase YjhB, NUDIX family |
| COG1132 | ABC-type multidrug and LPS transport system, ATPase and permease component MsbA |
| COG0596 | 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase MenH/undecaprenyl monophosphate-sugar esterase UshA/YqjL |
| COG0583 | DNA-binding transcriptional regulator, LysR family |
| COG0564 | Pseudouridine synthase RluA, 23S rRNA- or tRNA-specific |
| COG1028 | NAD(P)-dependent dehydrogenase, short-chain alcohol dehydrogenase family |
| COG1187 | Pseudouridylate synthase RsuA/RluF, specific for 16S rRNA U516, 23S rRNA U2604/U2605/U2457, and tRNA(Tyr)-35 |
| COG0834 | ABC-type amino acid transport/signal transduction system, periplasmic component/domain |
| COG1249 | Dihydrolipoamide dehydrogenase (E3) component of pyruvate/2-oxoglutarate dehydrogenase complex or glutathione oxidoreductase |
| COG0513 | Superfamily II DNA and RNA helicase |
| COG1108 | ABC-type Mn2+/Zn2+ transport system, permease component |
| COG1131 | Ribosome-associated ATPase or ATPase component of an ABC-type multidrug transport system |
| COG0664 | cAMP-binding domain of CRP or a regulatory subunit of cAMP-dependent protein kinases |
| COG0745 | DNA-binding response regulator, OmpR family, contains REC and winged-helix (wHTH) domain |
| COG0561 | Hydroxymethylpyrimidine pyrophosphatase and other HAD family phosphatases |
| COG0765 | ABC-type amino acid transport system, permease component |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, Q.; Lu, W.; Zhang, T.; Hao, S.; Wang, J.; Xu, X.; Wang, F.; Huang, Z.; Lei, X.; Cao, S.; et al. Comparative Analysis of Bacterial Diversity and Composition in Oral Fluid from Pigs of Different Ages and Water Pipe Wall Biofilms. Vet. Sci. 2025, 12, 1022. https://doi.org/10.3390/vetsci12111022
Ren Q, Lu W, Zhang T, Hao S, Wang J, Xu X, Wang F, Huang Z, Lei X, Cao S, et al. Comparative Analysis of Bacterial Diversity and Composition in Oral Fluid from Pigs of Different Ages and Water Pipe Wall Biofilms. Veterinary Sciences. 2025; 12(11):1022. https://doi.org/10.3390/vetsci12111022
Chicago/Turabian StyleRen, Qinghai, Wenlong Lu, Tingting Zhang, Shengkai Hao, Jiawen Wang, Xinrui Xu, Fei Wang, Zetong Huang, Xiaojing Lei, Shengliang Cao, and et al. 2025. "Comparative Analysis of Bacterial Diversity and Composition in Oral Fluid from Pigs of Different Ages and Water Pipe Wall Biofilms" Veterinary Sciences 12, no. 11: 1022. https://doi.org/10.3390/vetsci12111022
APA StyleRen, Q., Lu, W., Zhang, T., Hao, S., Wang, J., Xu, X., Wang, F., Huang, Z., Lei, X., Cao, S., Chen, D., & Li, Y. (2025). Comparative Analysis of Bacterial Diversity and Composition in Oral Fluid from Pigs of Different Ages and Water Pipe Wall Biofilms. Veterinary Sciences, 12(11), 1022. https://doi.org/10.3390/vetsci12111022





