Epidemiological Characteristics and Genetic Diversity of Chicken Infectious Anemia Virus (CIAV) in Guangdong Province, China
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Sample Collection and Processing
2.2.1. Sampling Strategy
2.2.2. Data Analysis
2.3. Detection of CIAV Antibodies in Serum Samples
2.4. Sample DNA Extraction and Virus Identification
2.5. Isolation of the Virus
2.6. Indirect Immunofluorescence Assays (IFA)
2.7. Sequence Analysis
3. Results
3.1. CIAV Serological Detection
3.2. PCR-Based CIAV Detection
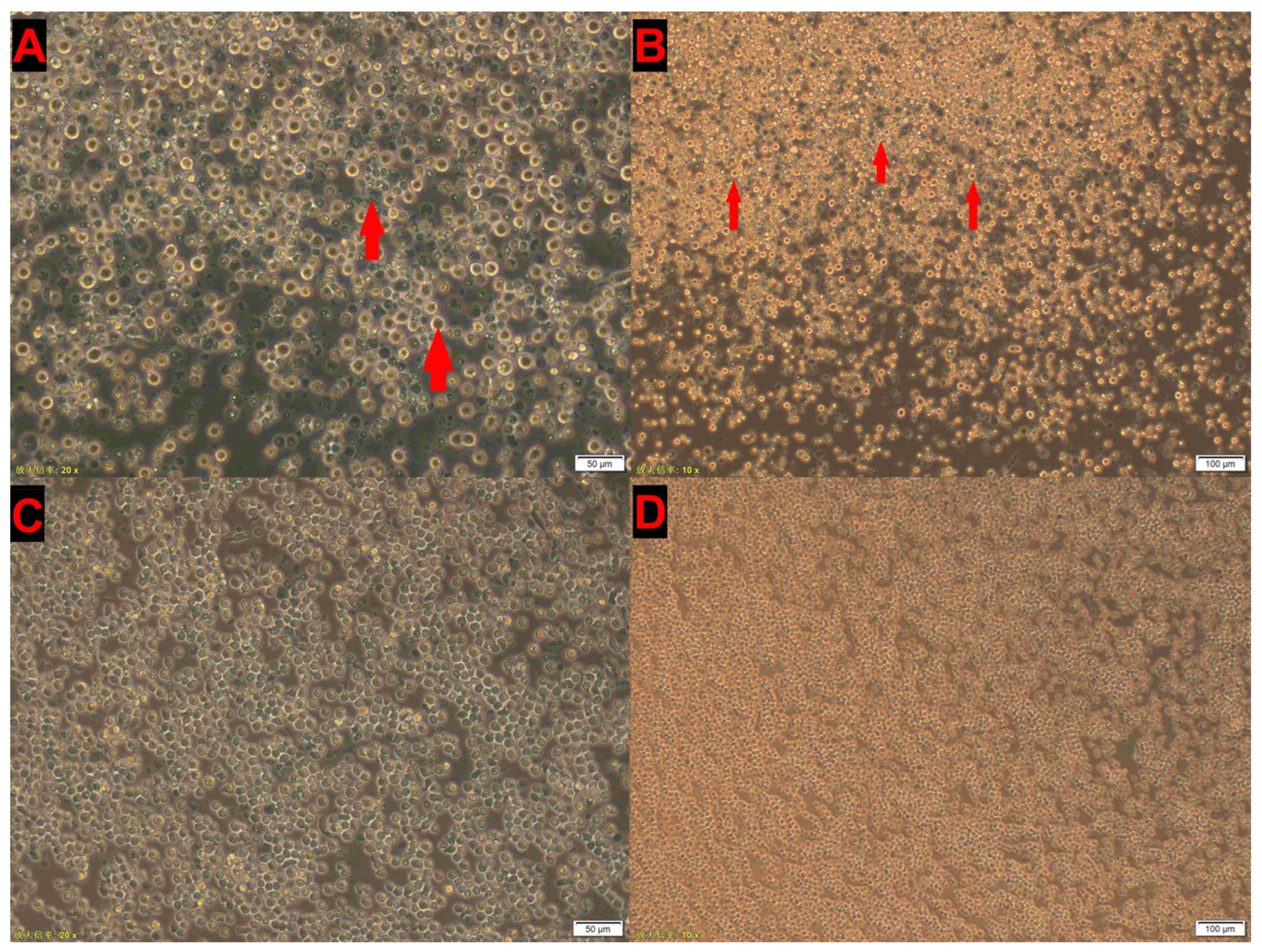
3.3. CIAV Isolation in MSB1 Cells
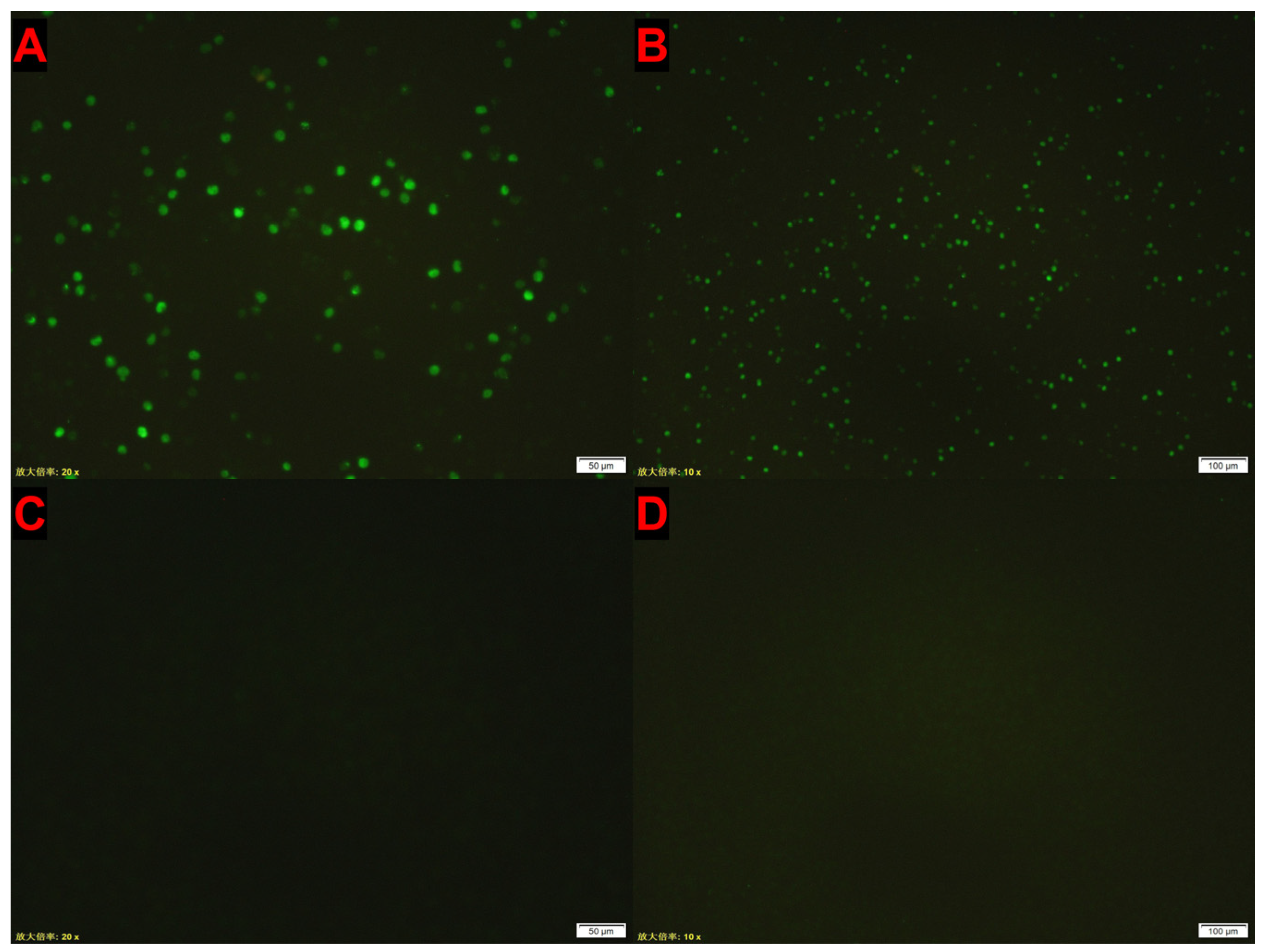
3.4. IFA Detection
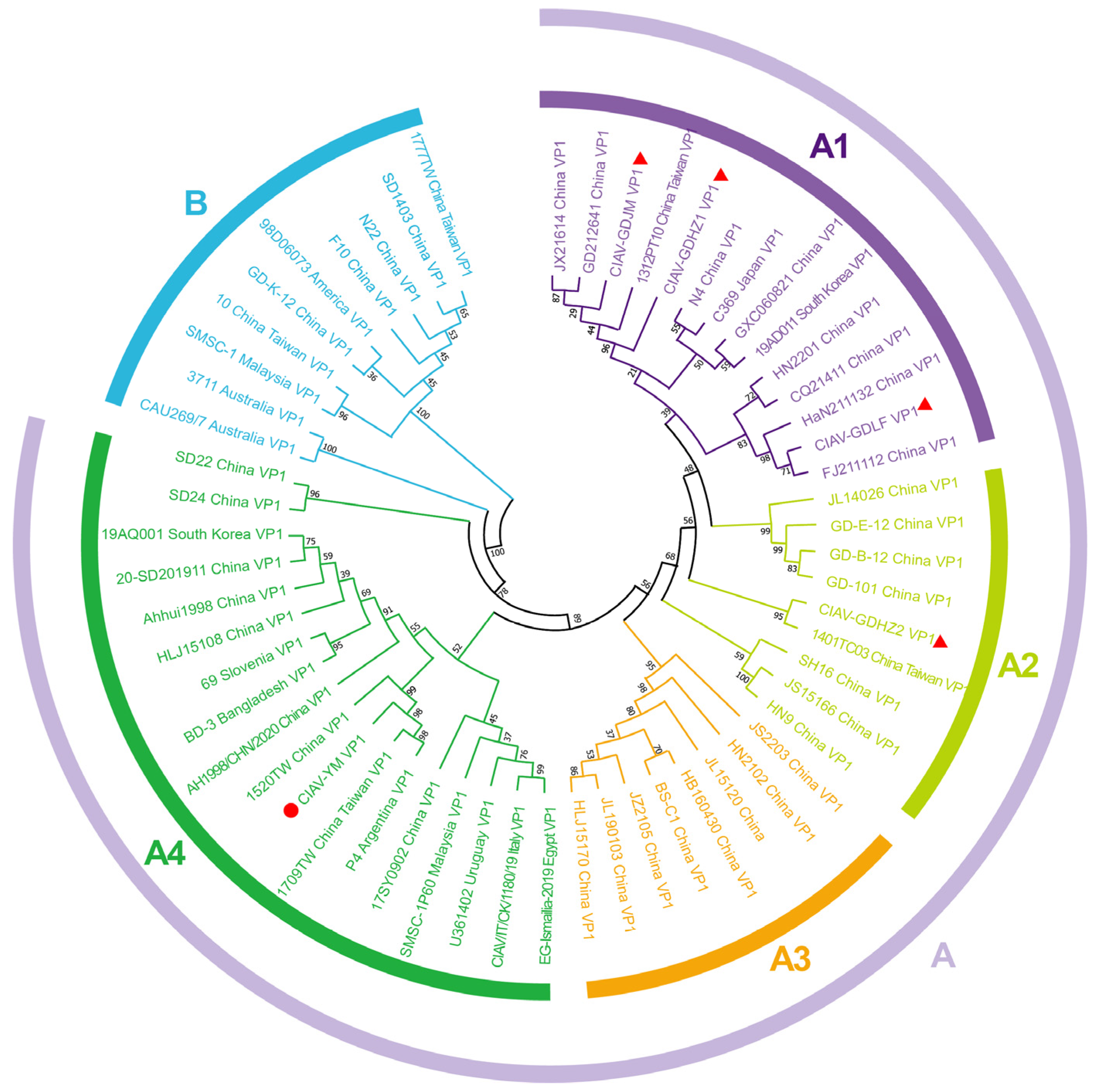
3.5. Genetic Evolution Analysis of CIAV Isolates
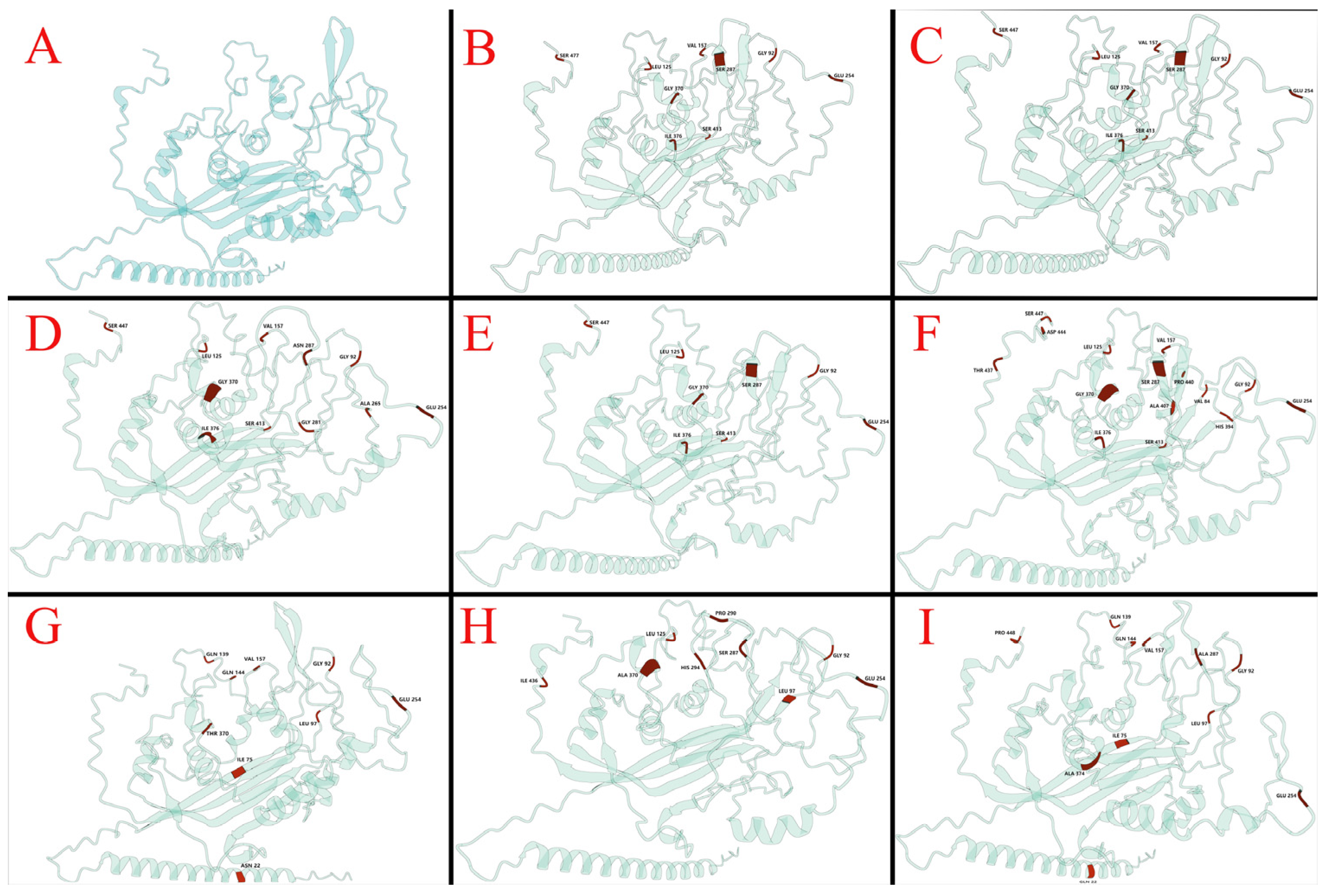
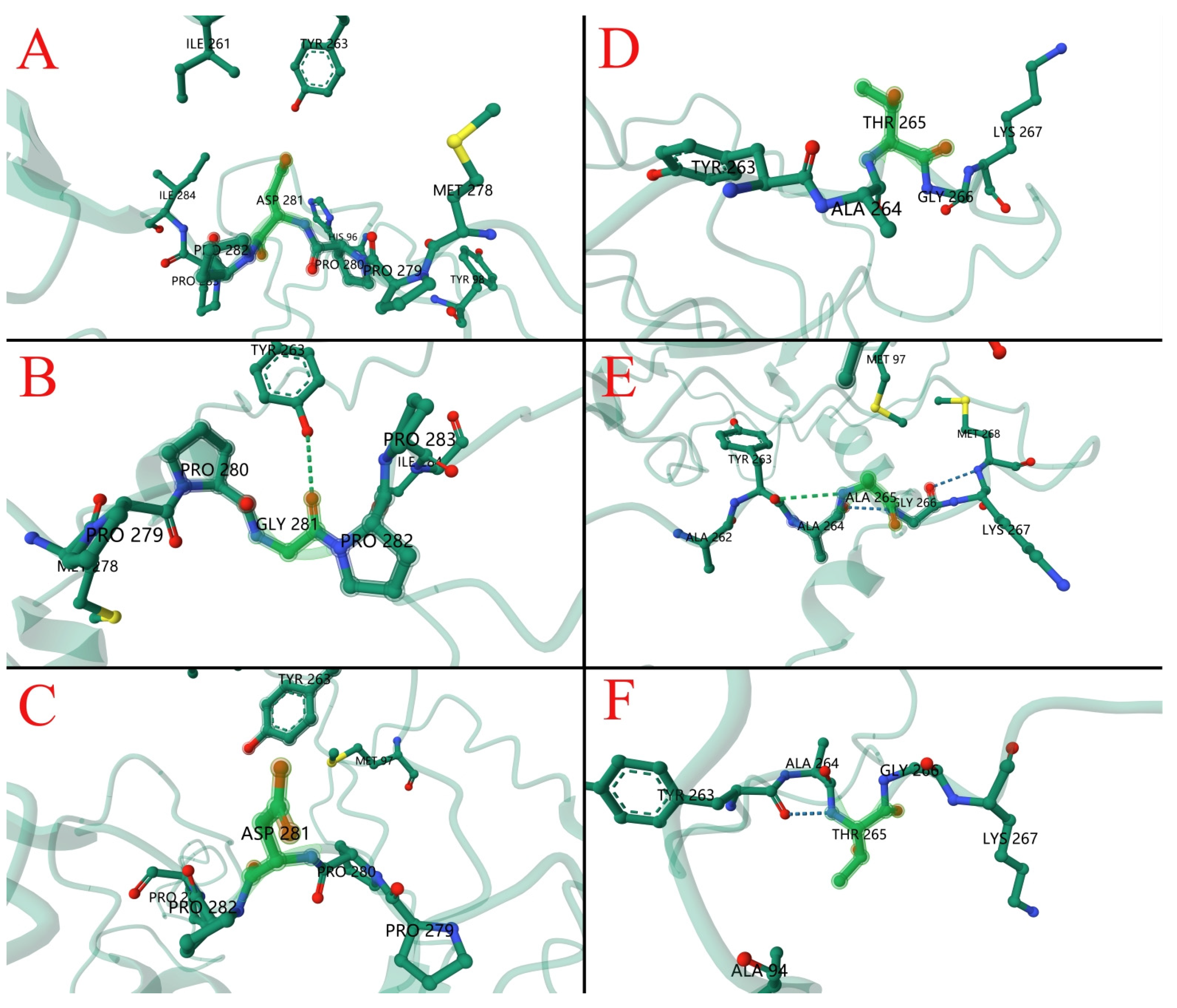
3.6. Amino Acid Sequence and Structure Analysis of CIAV Isolates
4. Discussion
4.1. Serological Analysis of CIAV Strains
4.2. Molecular Detection Method
4.3. Isolation of CIAV Strains
4.4. Phylogenetic Analysis of CIAV Strains
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yuasa, N.; Taniguchi, T.; Yoshida, I. Isolation and Some Characteristics of an Agent Inducing Anemia in Chicks. Avian Dis. 1979, 23, 366–385. [Google Scholar] [CrossRef]
- Cui, X.; Yin, G.; Wu, D.; Feng, J.; Li, F.; Qu, L. Identification of chicken infectious anaemia virus. Chin. J. Prev. Vet. Med. 1992, 6, 3–5. [Google Scholar]
- Ducatez, M.F.; Chen, H.; Guan, Y.; Muller, C.P. Molecular epidemiology of chicken anemia virus (CAV) in southeastern Chinese live birds markets. Avian Dis. 2008, 52, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Deng, X.; Xie, Z.; Zhang, Y.; Xie, Z.; Xie, L.; Luo, S.; Fan, Q.; Zeng, T.; Huang, J.; et al. Molecular characterization of chicken anemia virus in Guangxi Province, southern China, from 2018 to 2020. J. Vet. Sci. 2022, 23, e63. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Wang, Z.; Lei, X.; Lu, J.; Yan, Z.; Qin, J.; Chen, F.; Xie, Q.; Lin, W. Epidemiology, molecular characterization, and recombination analysis of chicken anemia virus in Guangdong province, China. Arch. Virol. 2020, 165, 1409–1417. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Jia, H.; Hu, Y.; Wang, Y.; Cui, Z.; Qi, L.; Zhao, P. Molecular characterization and pathogenicity study of a highly pathogenic strain of chicken anemia virus that emerged in China. Front. Cell. Infect. Microbiol. 2023, 13, 1171622. [Google Scholar] [CrossRef]
- Tseng, T.-Y.; Liu, Y.-C.; Hsu, Y.-C.; Chang, P.-C.; Hsieh, M.-K.; Shien, J.-H.; Ou, S.-C. Preparation of Chicken Anemia Virus (CAV) Virus-Like Particles and Chicken Interleukin-12 for Vaccine Development Using a Baculovirus Expression System. Pathogens 2019, 8, 262. [Google Scholar] [CrossRef]
- Gelderblom, H.; Kling, S.; Lurz, R.; Tischer, I.; von Bülow, V. Morphological characterization of chicken anaemia agent (CAA). Arch. Virol. 1989, 109, 115–120. [Google Scholar] [CrossRef]
- Moeini, H.; Omar, A.R.; Rahim, R.A.; Yusoff, K. Development of a DNA vaccine against chicken anemia virus by using a bicistronic vector expressing VP1 and VP2 proteins of CAV. Comp Immunol. Microbiol. Infect. Dis. 2011, 34, 227–236. [Google Scholar] [CrossRef]
- Peng, Y.; Luo, Y.; Pan, L.; Hou, Y.; Qin, L.; Lan, L.; Ouyang, K.; Chen, Y.; Wei, Z.; Qin, Y.; et al. Immunogenicity analysis based on VP1 and VP2 proteins of bovine enterovirus. Virology 2024, 600, 110260. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Chen, L.; Wang, Q.; Zhou, M.; Zhao, H.; Chi, Z.; Wang, Y.; Chang, S.; Zhao, P. Genomic Characterization of CIAV Detected in Contaminated Attenuated NDV Vaccine: Epidemiological Evidence of Source and Vertical Transmission from SPF Chicken Embryos in China. Front. Vet. Sci. 2022, 9, 930887. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fang, L.; Cui, S.; Fu, J.; Li, X.; Zhang, H.; Cui, Z.; Chang, S.; Shi, W.; Zhao, P. Genomic Characterization of Recent Chicken Anemia Virus Isolates in China. Front. Microbiol. 2017, 8, 401. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ma, L.; Li, T.; Xie, Q.; Wan, Z.; Qin, A.; Ye, J.; Shao, H.; Wang, S. Isolation and genomic characterization of chicken infectious anemia virus in Jiangsu province of China during 2020–2022. Front. Vet. Sci. 2024, 11, 1378120. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Huang, Y.; Wang, L.; Luo, J.; Yan, N.; Zhang, L.; Huang, H.; Zhou, J.; Li, Z.; Xu, C. Genomic Sequence and Pathogenicity of the Chicken Anemia Virus Isolated from Chicken in Yunnan Province, China. Front. Vet. Sci. 2022, 9, 860134. [Google Scholar] [CrossRef]
- Zhang, Q.; Shi, H.; Liu, G.; Yao, Z.; Liu, H.; Wei, L.; Zhang, Y. Serological investigation of infectious anemia in Wenchang chickens in Hainan province. Prog. Vet. Med. 2024, 45, 121–125. [Google Scholar] [CrossRef]
- Bao, W.; Fan, L.; Lu, L.; Wu, Y.; Xu, H.; Zhang, X. Serological investigation of chicken immunosuppressive diseases in some chicken farms in Ningbo City, Zhejiang Province. China Anim. Health Insp. 2017, 34, 29–31. [Google Scholar]
- Li, J.; Lou, Y.; Li, P.; Wang, T.; Lv, Z.; Guo, Z.; Geng, N.; Meng, F.; Liu, S.; Li, N. Retrospective Investigation and Genetic Variation Analysis of Chicken Infectious Anemia in Shandong Province, 2020–2022. Vet. Sci. 2023, 10, 263. [Google Scholar] [CrossRef]
- Xu, S.; Zhang, Z.; Xu, X.; Ji, J.; Yao, L.; Kan, Y.; Xie, Q.; Bi, Y. Molecular Characteristics of Chicken Infectious Anemia Virus in Central and Eastern China from 2020 to 2022. Animals 2023, 13, 2709. [Google Scholar] [CrossRef]
- Shah, P.T.; Bahoussi, A.N.; Cui, X.; Shabir, S.; Wu, C.; Xing, L. Genetic diversity, distribution, and evolution of chicken anemia virus: A comparative genomic and phylogenetic analysis. Front. Microbiol. 2023, 14, 1145225. [Google Scholar] [CrossRef]
- Bhatt, P.; Shukla, S.K.; Mahendran, M.; Dhama, K.; Chawak, M.M.; Kataria, J.M. Prevalence of chicken infectious anaemia virus (CIAV) in commercial poultry flocks of northern India: A serological survey. Transbound. Emerg. Dis. 2011, 58, 458–460. [Google Scholar] [CrossRef]
- Kabir, M.A.A.; Saha, S.; Hossain, M.G.; Khan, K.A.; Islam, M.A.; Rahman, L. Serological survey on the prevalence of chicken infectious anemia virus in broiler breeder and layer farms in some selected areas of Bangladesh. J. Adv. Vet. Anim. Res. 2021, 8, 323–329. [Google Scholar] [CrossRef]
- Hien, N.D.; Tran, D.H.; Bich, T.N.; Khanh, N.P.; Nguyen, L.T. First detection and genetic characterization of chicken infectious anemia virus in the Mekong Delta, Vietnam. Open Vet. J. 2023, 13, 690–696. [Google Scholar] [CrossRef]
- Yuasa, N.; Yoshida, I. Experimental egg transmission of chicken anemia agent. Natl. Inst. Anim. Health Q. 1983, 23, 99–100. [Google Scholar]
- Yuasa, N.; Taniguchi, T.; Goda, M.; Shibatani, M.; Imada, T.; Hihara, H. Isolation of chicken anemia agent with MDCC-MSB1 cells from chickens in the field. Natl. Inst. Anim. Health Q. 1983, 23, 75–77. [Google Scholar]
- Techera, C.; Marandino, A.; Tomás, G.; Grecco, S.; Hernández, M.; Hernández, D.; Panzera, Y.; Pérez, R. Origin, spreading and genetic variability of chicken anaemia virus. Avian Pathol. 2021, 50, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Renshaw, R.W.; Soiné, C.; Weinkle, T.; O’Connell, P.H.; Ohashi, K.; Watson, S.; Lucio, B.; Harrington, S.; Schat, K.A. A hypervariable region in VP1 of chicken infectious anemia virus mediates rate of spread and cell tropism in tissue culture. J. Virol. 1996, 70, 8872–8878. [Google Scholar] [CrossRef]
- Lai, G.H.; Lin, M.K.; Lien, Y.Y.; Cheng, J.H.; Sun, F.C.; Lee, M.S.; Chen, H.J.; Lee, M.S. Characterization of the DNA binding activity of structural protein VP1 from chicken anaemia virus. BMC Vet. Res. 2018, 14, 155. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Imada, T.; Kaji, N.; Mase, M.; Tsukamoto, K.; Tanimura, N.; Yuasa, N. Identification of a genetic determinant of pathogenicity in chicken anaemia virus. J. Gen. Virol. 2001, 82, 1233–1238. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Zhang, B.; Jiang, T. Isolation, Identification and Pathogenicity of a Chicken Infectious Anemia Virus. Chin. J. Vet. Drug 2020, 54, 17–23. [Google Scholar]
- Todd, D.; Scott, A.N.; Ball, N.W.; Borghmans, B.J.; Adair, B.M. Molecular basis of the attenuation exhibited by molecularly cloned highly passaged chicken anemia virus isolates. J. Virol. 2002, 76, 8472–8474. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.; Wu, B.; Sun, B.; Chen, F.; Ji, J.; Ma, J.; Xie, Q. Phylogenetic and molecular characterization of chicken anemia virus in southern China from 2011 to 2012. Sci. Rep. 2013, 3, 3519. [Google Scholar] [CrossRef]

| Primer Name | Sequence (5′–3′) | Amplicon Length (bp) |
|---|---|---|
| CIAV-124-F | TGCCGGTTCTTTAATCACCC | 124 bp |
| CIAV-124-R | ATCCCTCATTCTTAGTGGCAA |
| Primer Name | Sequence (5′ to 3′) | Amplicon Length (bp) |
|---|---|---|
| CIAV-F | GCTCTCCAAGAAGATACTCCA | 723 bp |
| CIAV-R | TATGTTAGGTTCATTGACGCT | |
| CIAV-F684 | AGTAGGTATACGCAAGGCGGT | 684 bp |
| CIAV-R684 | AGCCTCACACTATACGTA | |
| CIAV-F1243 | GCGGTATCGTAGACGAGC | 1243 bp |
| CIAV-R1243 | CCCTTTTCAGGGCTGCG |
| City | Breed | Number of Serum Samples | Number of CIAV Positive Serum Samples | CIAV Positive Rate of Serum Samples (%) | 95% Confidence Interval (CI) | |
|---|---|---|---|---|---|---|
| Lower CI (%) | Upper CI (%) | |||||
| Guangzhou | Yangshan chicken | 51 | 47 | 92.16 | 81.16 | 97.04 |
| Foshan | Ma chicken | 136 | 115 | 84.56 | 77.44 | 89.76 |
| Zhanjiang | Three yellow chicken | 50 | 50 | 100.00 | 92.89 | 100.00 |
| Huizhou | Ma chicken | 98 | 67 | 68.37 | 58.77 | 76.60 |
| Qingyuan | Ma chicken | 82 | 70 | 85.37 | 76.04 | 91.51 |
| Jiangmen | Three yellow chicken | 162 | 117 | 72.22 | 64.70 | 78.64 |
| Zhuhai | Yangshan chicken | 129 | 115 | 89.15 | 82.40 | 93.64 |
| Zhongshan | Three yellow chicken | 52 | 20 | 38.46 | 26.44 | 52.31 |
| Shantou | Three yellow chicken | 26 | 26 | 100 | 87.06 | 100 |
| Farm Name | Breed | Number of Samples | Days-Old of Chickens in the Farm | City | Positive Rate of CAV (Positive Tissue Samples/Total Tissue Samples) | Isolates | Year of Collection |
|---|---|---|---|---|---|---|---|
| Farm A | Three yellow chicken | 12 | 70 | Lufeng | 8.33% (1/12) | CIAV-GDLF | 2018 |
| Farm B | Ma chicken | 10 | 52 | Foshan | 0% (0/10) | 2018 | |
| Farm C | Three yellow chicken | 21 | 65 | Huizhou | 9.52% (2/21) | CIAV-GDHZ1 CIAV-GDHZ2 | 2018 |
| Farm D | Ma chicken | 15 | 67 | Qingyuan | 0% (0/15) | 2018 | |
| Farm E | Three yellow chicken | 17 | 59 | Jiangmen | 5.89% (1/17) | CIAV-GDJM | 2019 |
| Farm F | Yangshan chicken | 13 | 95 | Zhuhai | 0% (0/13) | 2019 | |
| Farm G | Ma chicken | 17 | 83 | Shantou | 0% (0/17) | 2019 |
| Strain | Substitution of the Amino Acid Residues in VP1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 75 | 89 | 125 | 139 | 141 | 144 | 265 | 281 | 287 | 394 | |
| CIAV-YM | V | T | I | K | Q | E | T | D | T | Q |
| CIAV-GDHZ1 | - | - | L | - | - | - | - | - | S | - |
| CIAV-GDHZ2 | - | - | L | - | - | - | - | - | S | - |
| CIAV-JM | - | - | L | - | - | - | A | G | N | - |
| CIAV-LF | - | - | L | - | - | - | - | - | S | - |
| N22 | I | - | - | Q | - | Q | - | - | - | - |
| Ahhui1998 | I | - | - | Q | - | Q | - | - | - | - |
| JL15120 | - | - | L | - | - | - | - | - | S | - |
| 98D06073 | I | - | - | Q | - | Q | - | - | A | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Li, W.; Liu, Y.; Lin, J.; Luo, H.; Wang, Y.; Xu, F.; Liang, Z.; Mei, K.; Huang, S. Epidemiological Characteristics and Genetic Diversity of Chicken Infectious Anemia Virus (CIAV) in Guangdong Province, China. Vet. Sci. 2025, 12, 972. https://doi.org/10.3390/vetsci12100972
Lu Y, Li W, Liu Y, Lin J, Luo H, Wang Y, Xu F, Liang Z, Mei K, Huang S. Epidemiological Characteristics and Genetic Diversity of Chicken Infectious Anemia Virus (CIAV) in Guangdong Province, China. Veterinary Sciences. 2025; 12(10):972. https://doi.org/10.3390/vetsci12100972
Chicago/Turabian StyleLu, Yongkun, Wenjun Li, Yingying Liu, Junjie Lin, Haojian Luo, Yiqiao Wang, Fenfen Xu, Zhaoping Liang, Kun Mei, and Shujian Huang. 2025. "Epidemiological Characteristics and Genetic Diversity of Chicken Infectious Anemia Virus (CIAV) in Guangdong Province, China" Veterinary Sciences 12, no. 10: 972. https://doi.org/10.3390/vetsci12100972
APA StyleLu, Y., Li, W., Liu, Y., Lin, J., Luo, H., Wang, Y., Xu, F., Liang, Z., Mei, K., & Huang, S. (2025). Epidemiological Characteristics and Genetic Diversity of Chicken Infectious Anemia Virus (CIAV) in Guangdong Province, China. Veterinary Sciences, 12(10), 972. https://doi.org/10.3390/vetsci12100972




