Data-Driven Early Warning Approach for Antimicrobial Resistance Prediction–Anomaly Detection Based on High-Level Indicators
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Overall Early Warning Framework
2.2. Identification of Environmental Indicators
2.3. Data Collection and Sources
2.4. Feature Engineering and Data Preparation
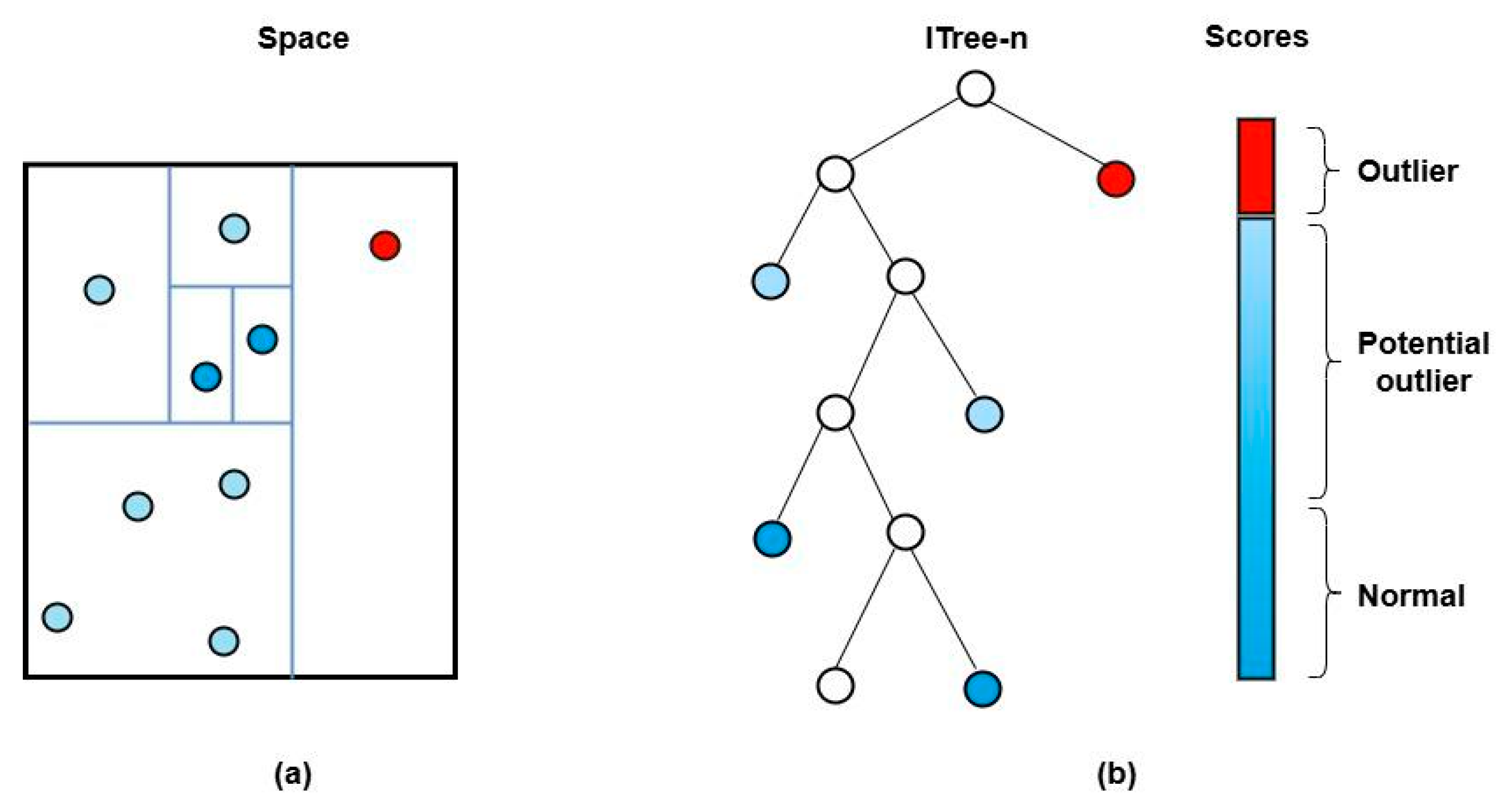
2.5. Anomaly Detection Using iForest
- n_estimators = 100: Number of isolation trees in the ensemble.
- max_samples = “auto”: The number of samples used to build each tree (default: 256).
- contamination = 0.03: The proportion of outliers in the dataset (3%).
- random_state = 42: Seed for reproducibility.
2.6. t-SNE Analysis for Anomaly Score Validation
2.7. SHAP-Based Model Interpretation
3. Results
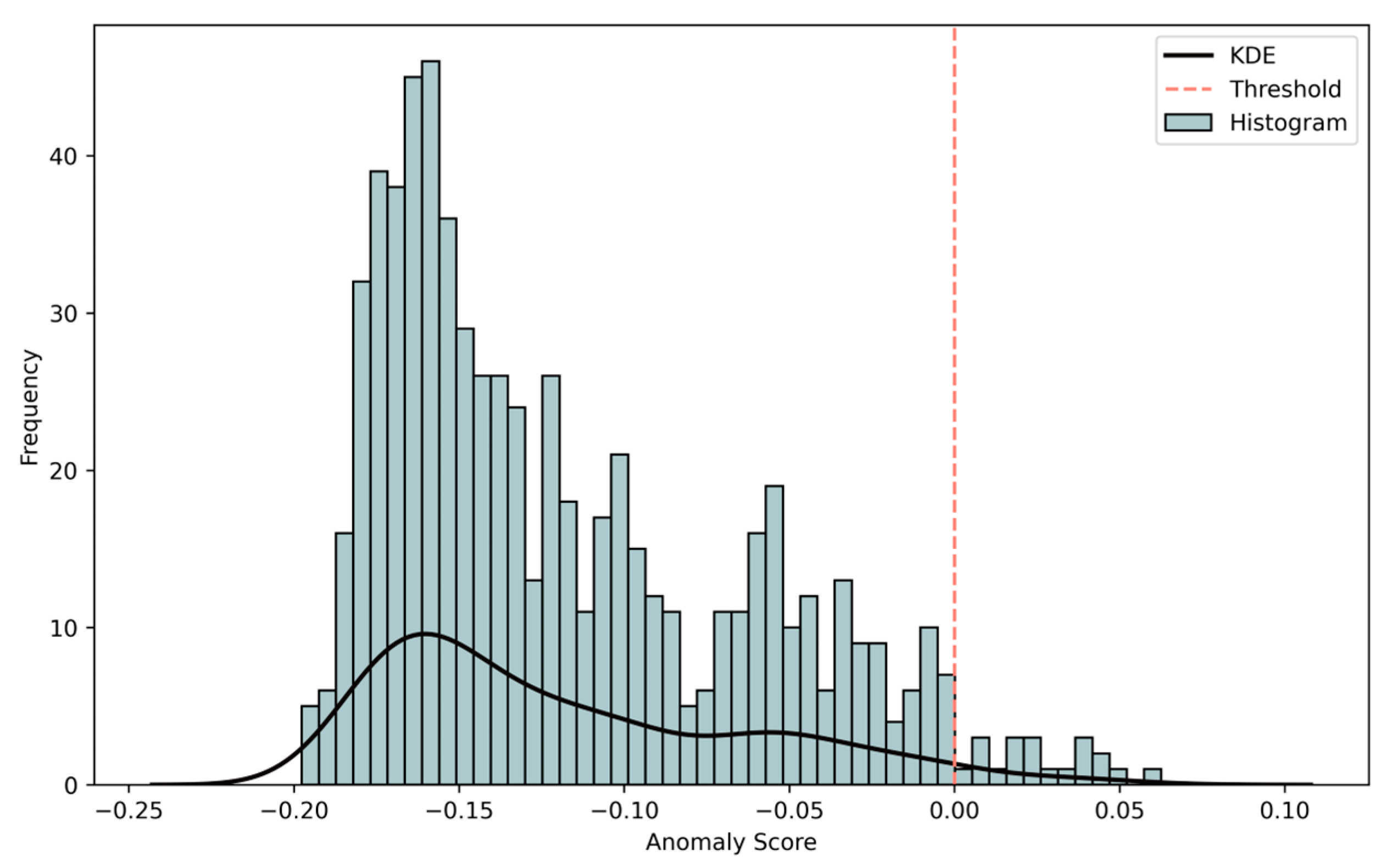
3.1. Anomaly Detection via iForest
3.2. Validation with t-SNE
3.3. SHAP-Based Feature Interpretation
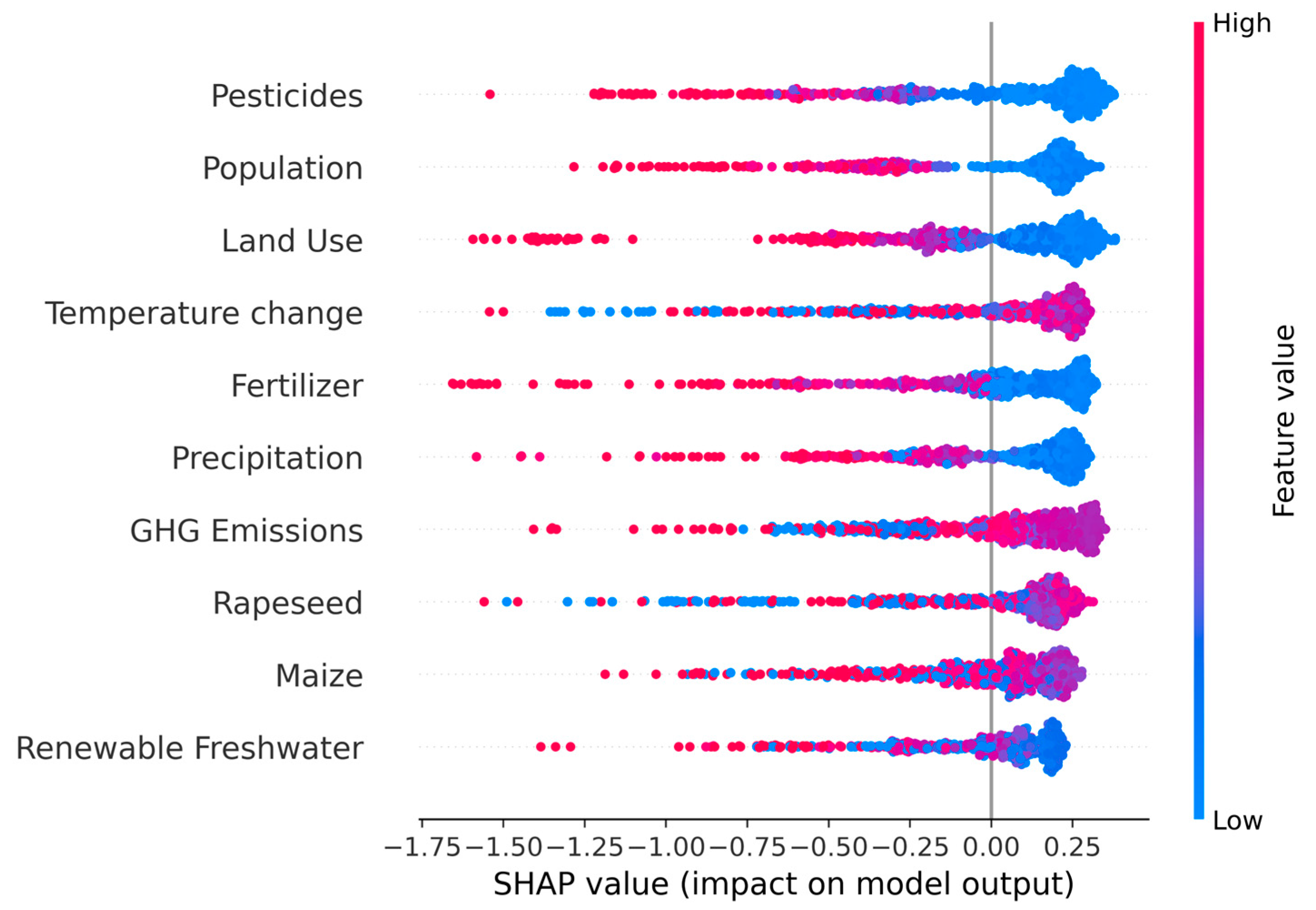
3.3.1. Global Feature Interpretation
3.3.2. Local SHAP Distribution and Directionality of Effect
3.3.3. Feature-Level Interpretation Using SHAP Value Ranges
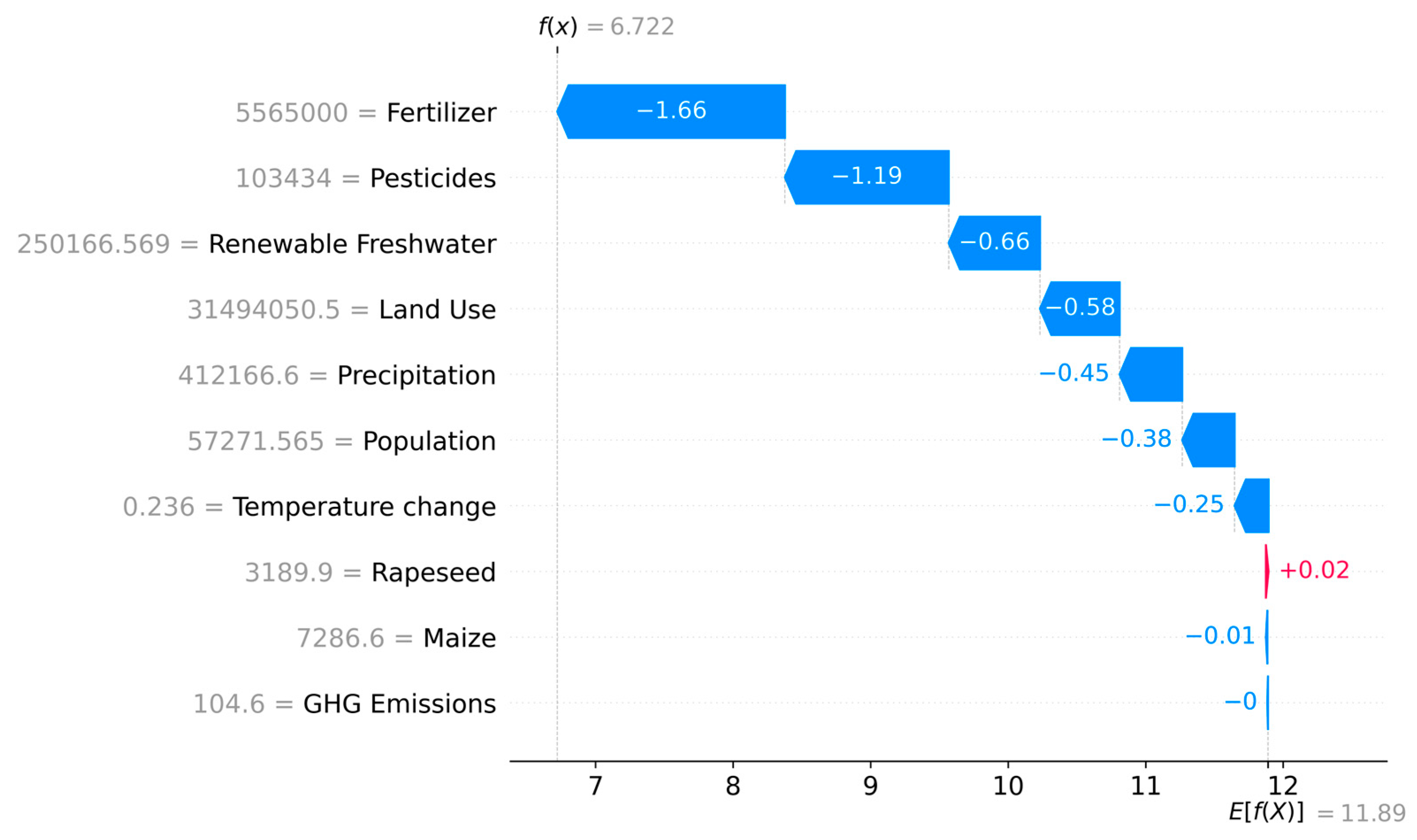
3.3.4. Waterfall SHAP Analysis of Anomalous Samples
4. Discussion
Limitations and Future Directions
- 1.
- Quantify Probabilistic Relationships: Model the conditional probabilities and the strength of influence each environmental driver has on AMR outcomes.
- 2.
- Incorporate Uncertainty: Explicitly account for uncertainty inherent in complex environmental and biological systems.
- 3.
- Develop Predictive Capability: Create a probabilistic model for predicting AMR risk based on environmental inputs, moving toward a true early-warning system.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial resistance |
| ARGs | Antimicrobial resistance genes |
| GHG | Greenhouse gas |
| iForest | Isolation Forest |
| KDE | Kernel Density Estimate |
| MDR | Multidrug-resistant |
| MICE | Multivariate Imputation by Chained Equations |
| MIC | Minimum inhibitory concentration |
| SHAP | Shapley Additive Explanations |
| t-SNE | t-distributed Stochastic Neighbor Embedding |
| WHO | World Health Organization |
References
- Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 7 July 2025).
- Marco-Fuertes, A.; Marin, C.; Lorenzo-Rebenaque, L.; Vega, S.; Montoro-Dasi, L. Antimicrobial Resistance in Companion Animals: A New Challenge for the One Health Approach in the European Union. Vet. Sci. 2022, 9, 208. [Google Scholar] [CrossRef] [PubMed]
- Kerek, Á.; Szabó, Á.; Barnácz, F.; Csirmaz, B.; Kovács, L.; Jerzsele, Á. Antimicrobial Resistance in Commensal Bacteria from Large-Scale Chicken Flocks in the Dél-Alföld Region of Hungary. Vet. Sci. 2025, 12, 691. [Google Scholar] [CrossRef]
- Rhouma, M.; Archambault, M.; Butaye, P. Antimicrobial Use and Resistance in Animals from a One Health Perspective. Vet. Sci. 2023, 10, 319. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.-F. Antibiotic Resistance in the Environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Singer, A.C.; Shaw, H.; Rhodes, V.; Hart, A. Review of Antimicrobial Resistance in the Environment and Its Relevance to Environmental Regulators. Front. Microbiol. 2016, 7, 1728. [Google Scholar] [CrossRef]
- Huijbers, P.M.C.; Flach, C.-F.; Larsson, D.G.J. A Conceptual Framework for the Environmental Surveillance of Antibiotics and Antibiotic Resistance. Environ. Int. 2019, 130, 104880. [Google Scholar] [CrossRef]
- Zhang, Y.; Kuang, F.; Liu, C.; Ma, K.; Liu, T.; Zhao, M.; Lv, G.; Huang, H. Contamination and Health Risk Assessment of Multiple Mycotoxins in Edible and Medicinal Plants. Toxins 2023, 15, 209. [Google Scholar] [CrossRef]
- Sible, C.N.; Kent, A.D.; Margenot, A.J.; Below, F.E. Long-Term Continuous Maize: Impacts on the Soil Microbiome and Implications for Residue Management. Soil Sci. Soc. Am. J. 2024, 88, 1109–1126. [Google Scholar] [CrossRef]
- Wang, P.; Xie, W.; Ding, L.; Zhuo, Y.; Gao, Y.; Li, J.; Zhao, L. Effects of Maize–Crop Rotation on Soil Physicochemical Properties, Enzyme Activities, Microbial Biomass and Microbial Community Structure in Southwest China. Microorganisms 2023, 11, 2621. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Lupwayi, N.; Marc, S.-A.; Siddique, K.H.M.; Bainard, L.D. Anthropogenic Drivers of Soil Microbial Communities and Impacts on Soil Biological Functions in Agroecosystems. Glob. Ecol. Conserv. 2021, 27, e01521. [Google Scholar] [CrossRef]
- Graham, M.E.; Wilson, B.A.; Ramkumar, D.; Rosencranz, H.; Ramkumar, J. Unseen Drivers of Antimicrobial Resistance: The Role of Industrial Agriculture and Climate Change in This Global Health Crisis. Challenges 2025, 16, 22. [Google Scholar] [CrossRef]
- Kurenbach, B.; Marjoshi, D.; Amábile-Cuevas, C.F.; Ferguson, G.C.; Godsoe, W.; Gibson, P.; Heinemann, J.A. Sublethal Exposure to Commercial Formulations of the Herbicides Dicamba, 2,4-Dichlorophenoxyacetic Acid, and Glyphosate Cause Changes in Antibiotic Susceptibility in Escherichia Coli and Salmonella Enterica Serovar Typhimurium. mBio 2015, 6, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Ke, M.; Zhang, Q.; Zhang, F.; Lu, T.; Sun, L.; Qian, H. Response of Microbial Antibiotic Resistance to Pesticides: An Emerging Health Threat. Sci. Total Environ. 2022, 850, 158057. [Google Scholar] [CrossRef] [PubMed]
- MacFadden, D.R.; McGough, S.F.; Fisman, D.; Santillana, M.; Brownstein, J.S. Antibiotic Resistance Increases with Local Temperature. Nat. Clim. Change 2018, 8, 510–514. [Google Scholar] [CrossRef]
- Burnham, J.P. Climate Change and Antibiotic Resistance: A Deadly Combination. Ther. Adv. Infect. Dis. 2021, 8, 2049936121991374. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, X.; Jin, J.; Guo, B.; Qiu, G.; Wang, X.; Zhou, H.; Li, H. Effects of Nitrogen Fertilization on Antibiotic Resistance Gene Spread from Soil to Floodwater in Paddy Fields. Environ. Res. 2025, 274, 121345. [Google Scholar] [CrossRef]
- Leonard, A.F.C.; Zhang, L.; Balfour, A.J.; Garside, R.; Gaze, W.H. Human Recreational Exposure to Antibiotic Resistant Bacteria in Coastal Bathing Waters. Environ. Int. 2015, 82, 92–100. [Google Scholar] [CrossRef]
- Grenni, P.; Ancona, V.; Barra Caracciolo, A. Ecological Effects of Antibiotics on Natural Ecosystems: A Review. Microchem. J. 2018, 136, 25–39. [Google Scholar] [CrossRef]
- Fu, C.; Qin, Y.; Xiang, Q.; Qiao, M.; Zhu, Y. pH Drives the Spatial Variation of Antibiotic Resistance Gene Profiles in Riparian Soils at a Watershed Scale. Environ. Pollut. 2023, 326, 121486. [Google Scholar] [CrossRef]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global Food Demand and the Sustainable Intensification of Agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef]
- Liu, F.T.; Ting, K.M.; Zhou, Z.-H. Isolation Forest. In Proceedings of the 2008 Eighth IEEE International Conference on Data Mining, Pisa, Italy, 15–19 December 2008; pp. 413–422. [Google Scholar]
- Hilal, W.; Gadsden, S.A.; Yawney, J. Financial Fraud: A Review of Anomaly Detection Techniques and Recent Advances. Expert Syst. Appl. 2022, 193, 116429. [Google Scholar] [CrossRef]
- Ten, C.-W.; Hong, J.; Liu, C.-C. Anomaly Detection for Cybersecurity of the Substations. IEEE Trans. Smart Grid 2011, 2, 865–873. [Google Scholar] [CrossRef]
- Flach, M.; Gans, F.; Brenning, A.; Denzler, J.; Reichstein, M.; Rodner, E.; Bathiany, S.; Bodesheim, P.; Guanche, Y.; Sippel, S.; et al. Multivariate Anomaly Detection for Earth Observations: A Comparison of Algorithms and Feature Extraction Techniques. Earth Syst. Dyn. 2017, 8, 677–696. [Google Scholar] [CrossRef]
- Binetti, M.S.; Uricchio, V.F.; Massarelli, C. Isolation Forest for Environmental Monitoring: A Data-Driven Approach to Land Management. Environments 2025, 12, 116. [Google Scholar] [CrossRef]
- van Buuren, S.; Groothuis-Oudshoorn, K. Mice: Multivariate Imputation by Chained Equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Geng, G.; Wang, P.; Sun, L.; Wen, H. Enhanced Isolation Forest-Based Algorithm for Unsupervised Anomaly Detection in Lidar SLAM Localization. World Electr. Veh. J. 2025, 16, 209. [Google Scholar] [CrossRef]
- Cieslak, M.C.; Castelfranco, A.M.; Roncalli, V.; Lenz, P.H.; Hartline, D.K. T-Distributed Stochastic Neighbor Embedding (t-SNE): A Tool for Eco-Physiological Transcriptomic Analysis. Mar. Genom. 2020, 51, 100723. [Google Scholar] [CrossRef]
- Mitchell, R.; Frank, E.; Holmes, G. GPUTreeShap: Massively Parallel Exact Calculation of SHAP Scores for Tree Ensembles 2022. PeerJ Comput. Sci. 2022, 8, e880. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From Local Explanations to Global Understanding with Explainable AI for Trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef]



| Indicator | Data Link | Variables | Temporal Coverage | Geographical Coverage | Use |
|---|---|---|---|---|---|
| Rapeseed Yield | https://www.fao.org/faostat/en/#data/QCL, accessed on 1 March 2025 | Production volume, harvested area, yield (tonnes/ha) | 1961–2023 (annual) | Global (country-level) | Biofuel feedstock and edible oil trends analysis |
| Maize Yield | https://www.fao.org/faostat/en/#data/QCL accessed on 1 March 2025 | Production volume, harvested area, yield (tonnes/ha) | 1961–2023 (annual) | Global (country-level) | Agricultural productivity and food security trends |
| Mean Surface Temperature Change | https://www.fao.org/faostat/en/#data/ET, accessed on 1 March 2025 | Temperature change relative to 1951–1980 baseline | 1961–2023 (annual, monthly, seasonal) | Global (country-level) | Long-term climatic trend assessment |
| Fertilizer Consumption (N, P, K) | https://www.fao.org/faostat/en/#data/RFN accessed on 1 March 2025 | Fertilizer use by nutrient type (tonnes) | 1961–2023 (annual) | Global (country-level) | Agricultural intensification and environmental impact analysis |
| Pesticide Use | https://www.fao.org/faostat/en/#data/RP, accessed on 1 March 2025 | Insecticides, herbicides, fungicides | 1990–2023 (annual) | Global (country-level) | Agrochemical use monitoring and environmental risk assessment |
| Precipitation | [ten00001] Water resources: long-term annual average | Annual precipitation (mm, million m3) | ~1990–present (varies by country) | EU, EFTA, candidate countries | Water availability and climate variability assessment |
| Renewable Freshwater Resources | [ten00003] Fresh water abstraction by source per capita-m3 per capita | Internal/external flows, total renewable freshwater, per capita | ~1990–present (varies by country) | EU, EFTA, candidate countries | Water sustainability and resource pressure analysis |
| Agricultural Land Use | https://ourworldindata.org/land-use, accessed on 1 March 2025 | Arable land, permanent crops, pastures (hectares) | 10,000 BCE–2023 CE (annual) | Global | Long-term land use change and agricultural expansion analysis |
| Population | https://www.fao.org/faostat/en/#data/OA, accessed on 1 March 2025 | Sex, urban/rural status, projections | 1950–2100 (annual) | Global (UN M49 classification) | Demographic analysis for food demand and environmental impact |
| Greenhouse Gas Emissions | [sdg_13_10] Domestic net greenhouse gas emissions | CO2, CH4, N2O, PFCs, HFCs, SF6, NF3 | 1990–latest (some data back to 1985) | EU Member States | Emission trends, mitigation, and policy evaluation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Csorba, S.; Vribék, K.; Farkas, M.; Süth, M.; Strang, O.; Zentai, A.; Farkas, Z. Data-Driven Early Warning Approach for Antimicrobial Resistance Prediction–Anomaly Detection Based on High-Level Indicators. Vet. Sci. 2025, 12, 935. https://doi.org/10.3390/vetsci12100935
Csorba S, Vribék K, Farkas M, Süth M, Strang O, Zentai A, Farkas Z. Data-Driven Early Warning Approach for Antimicrobial Resistance Prediction–Anomaly Detection Based on High-Level Indicators. Veterinary Sciences. 2025; 12(10):935. https://doi.org/10.3390/vetsci12100935
Chicago/Turabian StyleCsorba, Szilveszter, Krisztián Vribék, Máté Farkas, Miklós Süth, Orsolya Strang, Andrea Zentai, and Zsuzsa Farkas. 2025. "Data-Driven Early Warning Approach for Antimicrobial Resistance Prediction–Anomaly Detection Based on High-Level Indicators" Veterinary Sciences 12, no. 10: 935. https://doi.org/10.3390/vetsci12100935
APA StyleCsorba, S., Vribék, K., Farkas, M., Süth, M., Strang, O., Zentai, A., & Farkas, Z. (2025). Data-Driven Early Warning Approach for Antimicrobial Resistance Prediction–Anomaly Detection Based on High-Level Indicators. Veterinary Sciences, 12(10), 935. https://doi.org/10.3390/vetsci12100935








