A Simple and Sensitive RT-qPCR Technology for Rapid Detection of Porcine Reproductive and Respiratory Syndrome Virus
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Source
2.2. Virus, Vaccines, and Recombinant Plasmids
2.3. Main Reagents and Instruments
2.4. Nucleic Acid Extraction
2.5. Design and Screening of Specific Primer Probes for PRRSV
2.6. Fully Pre-Mixed RT-qPCR Development of the System and Reaction Procedures
2.7. Optimization of the PRRSV RT-qPCR System
2.8. Comparison of the Fully Pre-Mixed RT-qPCR System with the Commercial Master Mix
2.9. Specific Assay
2.10. Sensitivity Assay
2.11. Inclusive Assay
2.12. Precision Assay
2.13. Anti-Interference Test
2.14. Detection of the Real Samples
3. Results
3.1. Screening of Prime Probes Specific for PRRSV
3.2. Effect of PCR Enhancers for Fully Pre-Mixed RT-qPCR Systems
3.3. Optimization of the PRRSV RT-qPCR Reaction System
3.4. Comparison of the Amplification Effect of Fully Pre-Mixed RT-qPCR System with Commercial Master Mix
3.5. PRRSV RT-qPCR Specificity Test
3.6. The PRRSV RT-qPCR Sensitivity Test and the Establishment of the Standard Curve
3.7. PRRSV RT-qPCR Inclusive Assay
3.8. PRRSV RT-qPCR Precision Assay
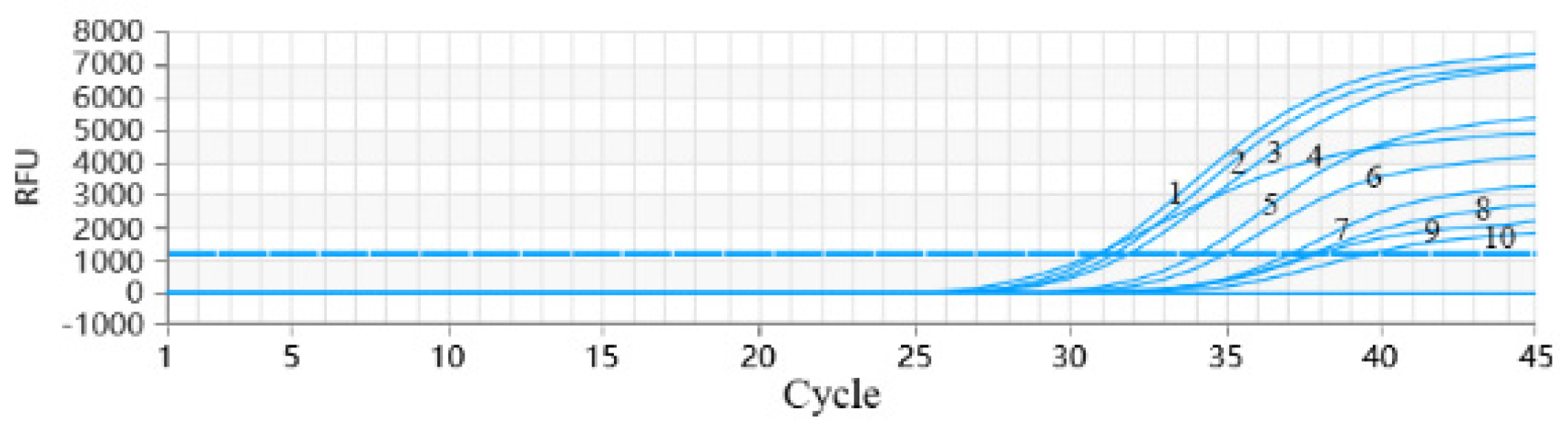
3.9. PRRSV RT-qPCR Anti-Interference Test
3.10. Detection of Real Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Name | Specifications | Production Batch Number | Source |
|---|---|---|---|
| Swine Epidemic Encephalitis Vaccine, live (Strain SA14-14-2) | 10 copies/bottle | 20220721 | Wuhan Keda Biotechnology Co., Ltd. (Wuhan, China) |
| Porcine Parvovirus vaccine, inactivated (Strain WH-1) | 20 mL/bottle | 20220817 | Wuhan Keda Biotechnology Co., Ltd. (Wuhan, China) |
| Swine Influenza Virus Subtype H1N1 Vaccine, inactivated (Strain TJ) | 20 mL/bottle | 20220502 | Wuhan Keda Biotechnology Co., Ltd. (Wuhan, China) |
| Swine Pseudorabies Vaccine, live (Strain HB-98) | 10 copies/bottle | 2212022 | Zhongmu Industry Co., Ltd. (Shanghai, China) |
| Classical Swine Fever Live Vaccine (Cell-Derived) (CVCC AV1412 Strain) | 10 copies/bottle | 2201011 | Zhongmu Industry Co., Ltd. (Shanghai, China) |
| Porcine circovirus disease type 2 vaccine, inactivated (Strain SH) | 20 mL/bottle | 072110006 | PLeiro Biotechnology Engineering Co., Ltd. (Wuhan, China) |
| Swine Transmissible Gastroenteritis, Porcine Epidemic Diarrhea and Porcine Rotavirus (GP5 type) Vaccine, live (Strain huadu + Strain CV777 + Strain NX) | 1 copies/bottle | 2022020 | Harbin Weike Biotechnology Co., Ltd. (Harbin, China) |
| Foot-and-mouth disease type O and A bivalent 3B protein epitope deletion vaccine, inactivated (O/rV-1 Strain + A/rV-2 Strain) | 20mL/bottle | 0AA220618 | Inner Mongolia Biwei Antai Biotechnology Co., Ltd. (Hohhot, China) |
| Haemophilus parasuis Disease Quadrivalent Propolis Vaccine, Inactivated (Type 4 SD02 Strain + Type 5 HN02 Strain + Type 12 GZ01 Strain + Type13 JX03 Strain) | 20 mL/bottle | 202209003 | Shandong Huahong Biotechnology Engineering Co., Ltd. (Binzhou, China) |
| Swine Staphylococcus Bee Propolis Inactivated Vaccine (Swine Staphylococcus serogroup C BHZZ-L1 Strain + Swine Staphylococcus type 2 BHZZ-L4 Strain). | 20 mL/bottle | 202209009 | Shandong Huahong Biotechnology Engineering Co., Ltd. (Binzhou, China) |
| Piglet Enterotoxigenic E. coli Disease Tri-valent Inactivated Vaccine (containing K88, K99, 987P flagellar antigens) | 10 mL/bottle | 202209006 | Shandong Huahong Biotechnology Engineering Co., Ltd. (Binzhou, China) |
| Porcine Mycoplasmal Pneumonia Inactivated Vaccine (Strain J) | 20 mL/bottle | 20220808 | Shandong Huahong Biotechnology Engineering Co., Ltd. (Binzhou, China) |
| Triple Live Vaccine for Classical Swine Fever, Swine Erysipelas and Pasteurellosis in Pigs (Cell-Derived + G4T10 Strain + EO630 Strain) | 10 copies/bottle | 2208037 | Zhongmu Industry Co., Ltd. (Shanghai, China) |
| Porcine Atrophic Rhinitis Inactivated Vaccine (Bordetella pertussis JB5 Strain) | 20 mL/bottle | 20220602 | Wuhan Keda Biotechnology Co., Ltd. (Wuhan, China) |
| Triple-valent Inactivated Vaccine for Porcine Contagious Pleuropneumonia (Serotype 1 9901 Strain, Serotype 2 XT9904 Strain, Serotype 7 GZ9903 Strain) | 20 mL/bottle | 20220706 | Wuhan Keda Biotechnology Co., Ltd. (Wuhan, China) |
| Experimental Group | |||||
|---|---|---|---|---|---|
| Exogenous Dry Perturbation Material | Constituent Content | Actual Dosage (μL) | Endogenous Dry Perturbation Material | Constituent Content | Actual Dosage (μL) |
| Group 1 | Ceftiofur sodium for injection (300 μL) | 100 | Group 6 | Throat swab treatment solution * | 100 |
| Fluphenicol powder (30 μL) | |||||
| Telmicocin pre-mix (30 μL) | Group 7 | Liver tissue treatment solution * | 100 | ||
| normal saline (240 μL) | |||||
| Group 2 | Amoxicillin for injection (30 μL) | 100 | Group 8 | Lung tissue treatment solution * | 100 |
| Dxycycline hydrochloride-soluble powder (150 μL) | |||||
| Tamrocin-soluble powder (150 μL) normal saline (300 μL) | Group 9 | Intestinal tissue treatment solution * | 100 | ||
| Group 3 | Dexamethasone acetate tablets (2.5 μL) | 100 | Group 10 | Stool swab treatment solution * | 100 |
| gentamicin sulphate (30 μL) | |||||
| normal saline (567.5 μL) | Group 11 | Milk treatment solution * | 100 | ||
| Group 4 | Ribavirin granules (10 μL) | 100 | |||
| Amantadine hydrochloride tablets (15 μL) | Group 12 | whole blood * | 100 | ||
| normal saline (575 μL) | |||||
| Group 5 | Feed residue treatment solution * | 100 | Group 13 | mucin (270 μL) normal saline (270 μL) | 100 |
| control group | |||||
| positive control | constituent content | negative control | constituent content | ||
| Group 14 | Virus titers 101.0 TCID50/μL (100 μL) ddH2O (100 μL) | Group 15 | ddH2O (200 μL) | ||
| Method | Target Gene | Sensitivity/ Amplification Efficiency (E:90–110%) | Amplification Time | Add Sample Steps | Specificity |
|---|---|---|---|---|---|
| The method developed in this study | M gene | 3.12 × 100 copies/μL 100 TCID50/μL E: >90% | About 30 min | 1 | High |
| Qiu et al., 2020 [6] | NSP2/ORF5 | 101 copies/μL E: <90% | About 98 min | 5 | High |
| Chen et al., 2019 [55] | M gene/ N gene | 3.9 × 101 copies/μL 5.6 (100.75) TCID50/mL E: >90% | About 160 min | 4 | High |
| Ma et al., 2024 [58] | N gene | 1.33 × 102 copies/μL E: >110% | About 35 min | 4 | High |
| Amer et al., 2023 [57] | ORF4-6 | 101 copies/μL E: >90% | About 49 min | 3 | High |
| Wang et al., 2024 [52] | ORF6 | 2.8 × 101 copies/μL E: <90% | About 65 min | 4 | High |
| Fornyos et al., 2022 [59] | ORF5 | 101 copies/μL E: =90% | About 70 min | 5 | High |
References
- Lunney, J.K.; Benfield, D.A.; Rowland, R.R.R. Porcine reproductive and respiratory syndrome virus: An update on an emerging and re-emerging viral disease of swine. Virus Res. 2010, 154, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lunney, J.K.; Fang, Y.; Ladinig, A.; Chen, N.; Li, Y.; Rowland, B.; Renukaradhya, G.J. Porcine Reproductive and Respiratory Syndrome Virus (PRRSV): Pathogenesis and Interaction with the Immune System. Annu. Rev. Anim. Biosci. 2016, 4, 129–154. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Kim, S.; Park, C.; Kang, I.; Park, K.H.; Ham, H.J.; Chae, C. Effect of vaccination with a porcine reproductive and respiratory syndrome subunit vaccine on sow reproductive performance in endemic farms. Vet. Rec. 2018, 182, 602. [Google Scholar] [CrossRef]
- Neumann, E.J.; Kliebenstein, J.B.; Johnson, C.D.; Mabry, J.W.; Bush, E.J.; Seitzinger, A.H.; Green, A.L.; Zimmerman, J.J. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J. Am. Vet. Med. Assoc. 2005, 227, 385–392. [Google Scholar] [CrossRef]
- Zimmerman, J.J.; Yoon, K.J.; Wills, R.W.; Swenson, S.L. General overview of PRRSV: A perspective from the United States. Vet. Microbiol. 1997, 55, 187–196. [Google Scholar] [CrossRef]
- Qiu, W.; Meng, K.; Liu, Y.; Zhang, Y.; Wang, Z.; Chen, Z.; Yang, J.; Sun, W.; Guo, L.; Ren, S.; et al. Simultaneous detection of classical PRRSV, highly pathogenic PRRSV and NADC30-like PRRSV by TaqMan probe real-time PCR. J. Virol. Methods 2020, 282, 113774. [Google Scholar] [CrossRef] [PubMed]
- Shin, G.-E.; Park, J.-Y.; Lee, K.-K.; Ko, M.-K.; Ku, B.-K.; Park, C.-K.; Jeoung, H.-Y. Genetic diversity of porcine reproductive and respiratory syndrome virus and evaluation of three one-step real-time RT-PCR assays in Korea. BMC Vet. Res. 2022, 18, 327. [Google Scholar] [CrossRef] [PubMed]
- Fiers, J.; Tignon, M.; Cay, A.B.; Simons, X.; Maes, D. Porcine Reproductive and Respiratory Syndrome virus (PRRSv): A Cross-Sectional Study on ELISA Seronegative, Multivaccinated Sows. Viruses 2022, 14, 1944. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Ye, C.; Chang, X.-B.; Jiang, C.-G.; Wang, S.-J.; Cai, X.-H.; Tong, G.-Z.; Tian, Z.-J.; Shi, M.; An, T.-Q.; et al. Importation and Recombination Are Responsible for the Latest Emergence of Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus in China. J. Virol. 2015, 89, 10712–10716. [Google Scholar] [CrossRef]
- Wills, R.W.; Zimmerman, J.J.; Yoon, K.-J.; Swenson, S.L.; Hoffman, L.J.; McGinley, M.J.; Hill, H.T.; Platt, K.B. Porcine reproductive and respiratory syndrome virus: Routes of excretion. Vet. Microbiol. 1997, 57, 69–81. [Google Scholar] [CrossRef]
- Yun, S.-I.; Lee, Y.-M. Overview: Replication of porcine reproductive and respiratory syndrome virus. J. Microbiol. 2013, 51, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Flores-Contreras, E.A.; Carrasco-González, J.A.; Linhares, D.C.L.; Corzo, C.A.; Campos-Villalobos, J.I.; Henao-Díaz, A.; Melchor-Martínez, E.M.; Iqbal, H.M.N.; González-González, R.B.; Parra-Saldívar, R.; et al. Emergent Molecular Techniques Applied to the Detection of Porcine Viruses. Vet. Sci. 2023, 10, 609. [Google Scholar] [CrossRef]
- Dee, S.; Shah, A.; Cochrane, R.; Clement, T.; Singrey, A.; Edler, R.; Spronk, G.; Niederwerder, M.; Nelson, E. Use of a demonstration project to evaluate viral survival in feed: Proof of concept. Transbound. Emerg. Dis. 2020, 68, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Dee, S.; Shah, A.; Jones, C.; Singrey, A.; Hanson, D.; Edler, R.; Spronk, G.; Niederwerder, M.; Nelson, E. Evidence of viral survival in representative volumes of feed and feed ingredients during long-distance commercial transport across the continental United States. Transbound. Emerg. Dis. 2021, 69, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Dee, N.; Havas, K.; Shah, A.; Singrey, A.; Spronk, G.; Niederwerder, M.; Nelson, E.; Dee, S. Evaluating the effect of temperature on viral survival in plant-based feed during storage. Transbound. Emerg. Dis. 2022, 69, e2105–e2110. [Google Scholar] [CrossRef] [PubMed]
- Arruda, A.G.; Tousignant, S.; Sanhueza, J.; Vilalta, C.; Poljak, Z.; Torremorell, M.; Alonso, C.; Corzo, C.A. Aerosol Detection and Transmission of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV): What Is the Evidence, and What Are the Knowledge Gaps? Viruses 2019, 11, 712. [Google Scholar] [CrossRef] [PubMed]
- Hou, F.-H.; Chia, M.-Y.; Lee, Y.-H.; Liao, J.-W.; Lee, W.-C. A comparably high virulence strain of porcine reproductive and respiratory syndrome virus isolated in Taiwan. Comp. Immunol. Microbiol. Infect. Dis. 2019, 65, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-H.; Kaewprom, K.; Wang, S.-Y.; Lin, C.-F.; Yang, C.-Y.; Chiou, M.-T.; Lin, C.-N. Outbreak of Porcine Reproductive and Respiratory Syndrome Virus 1 in Taiwan. Viruses 2020, 12, 316. [Google Scholar] [CrossRef]
- Fang, Y.; Treffers, E.E.; Li, Y.; Tas, A.; Sun, Z.; van der Meer, Y.; de Ru, A.H.; van Veelen, P.A.; Atkins, J.F.; Snijder, E.J.; et al. Efficient −2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc. Natl. Acad. Sci. USA 2012, 109, E2920–E2928. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Zhao, J.; Chen, L.; Chang, H.-T.; Li, Y.-T.; Liu, H.-Y.; Wang, C.-Q.; Yang, X. Complete Genome Sequence of a Novel Porcine Reproductive and Respiratory Syndrome Virus That Emerged in China. Genome Announc. 2015, 3, e00702-15. [Google Scholar] [CrossRef]
- Brar, M.S.; Shi, M.; Murtaugh, M.P.; Leung, F.C.C. Evolutionary diversification of type 2 porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 2015, 96, 1570–1580. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.Q.; Harrach, B.; Harrison, R.L.; Knowles, N.J.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Mushegian, A.R.; et al. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2016). Arch. Virol. 2016, 161, 2921–2949. [Google Scholar] [CrossRef]
- Montaner-Tarbes, S.; del Portillo, H.A.; Montoya, M.; Fraile, L. Key Gaps in the Knowledge of the Porcine Respiratory Reproductive Syndrome Virus (PRRSV). Front. Vet. Sci. 2019, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.-C.; Xiong, J.-Y.; Ye, C.; Chang, X.-B.; Guo, J.-C.; Jiang, C.-G.; Zhang, G.-H.; Tian, Z.-J.; Cai, X.-H.; Tong, G.-Z.; et al. Genotypic and geographical distribution of porcine reproductive and respiratory syndrome viruses in mainland China in 1996–2016. Vet. Microbiol. 2017, 208, 164–172. [Google Scholar] [CrossRef]
- Shi, M.; Lam, T.T.-Y.; Hon, C.-C.; Hui, R.K.-H.; Faaberg, K.S.; Wennblom, T.; Murtaugh, M.P.; Stadejek, T.; Leung, F.C.-C. Molecular epidemiology of PRRSV: A phylogenetic perspective. Virus Res. 2010, 154, 7–17. [Google Scholar] [CrossRef]
- Chen, N.; Liu, Q.; Qiao, M.; Deng, X.; Chen, X.; Sun, M. Whole genome characterization of a novel porcine reproductive and respiratory syndrome virus 1 isolate: Genetic evidence for recombination between Amervac vaccine and circulating strains in mainland China. Infect. Genet. Evol. 2017, 54, 308–313. [Google Scholar] [CrossRef]
- Sun, Q.; Xu, H.; An, T.; Cai, X.; Tian, Z.; Zhang, H. Recent Progress in Studies of Porcine Reproductive and Respiratory Syndrome Virus 1 in China. Viruses 2023, 15, 1528. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Bo, K.; Wang, X.; Tang, B.; Yang, B.; Jiang, W.; Jiang, P. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China. Vet. J. 2007, 174, 577–584. [Google Scholar] [CrossRef]
- Wu, Q.; Li, Z.; Zhang, G.; Niu, J.; Zeng, X.; Sun, B.; Ma, J. Genetic diversity and phylogenetic analysis of porcine reproductive and respiratory syndrome virus in southern China from 2007 to 2014. J. Vet. Sci. 2017, 18, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Zhao, P.; Dong, J.; Liu, Y.; Zhang, L.; Liang, P.; Wang, L.; Song, C. Genetic characterization of 11 porcine reproductive and respiratory syndrome virus isolates in South China from 2014 to 2015. Virol. J. 2017, 14, 139. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Zeng, M.; Zhao, M.; Huang, L. Research Progress on the detection methods of porcine reproductive and respiratory syndrome virus. Front. Microbiol. 2023, 14, 1097905. [Google Scholar] [CrossRef] [PubMed]
- Espy, M.J.; Uhl, J.R.; Sloan, L.M.; Buckwalter, S.P.; Jones, M.F.; Vetter, E.A.; Yao, J.D.C.; Wengenack, N.L.; Rosenblatt, J.E.; Cockerill, F.R.; et al. Real-Time PCR in Clinical Microbiology: Applications for Routine Laboratory Testing. Clin. Microbiol. Rev. 2006, 19, 165–256. [Google Scholar] [CrossRef]
- Chen, Y.; Shi, K.; Liu, H.; Yin, Y.; Zhao, J.; Long, F.; Lu, W.; Si, H. Development of a multiplex qRT-PCR assay for detection of African swine fever virus, classical swine fever virus and porcine reproductive and respiratory syndrome virus. J. Vet. Sci. 2021, 22, e87. [Google Scholar] [CrossRef] [PubMed]
- Long, F.; Chen, Y.; Shi, K.; Yin, Y.; Feng, S.; Si, H. Development of a Multiplex Crystal Digital RT-PCR for Differential Detection of Classical, Highly Pathogenic, and NADC30-like Porcine Reproductive and Respiratory Syndrome Virus. Animals 2023, 13, 594. [Google Scholar] [CrossRef]
- Cheng, D.; Zhao, J.-J.; Li, N.; Sun, Y.; Zhou, Y.-J.; Zhu, Y.; Tian, Z.-J.; Tu, C.; Tong, G.-Z.; Qiu, H.-J. Simultaneous detection of Classical swine fever virus and North American genotype Porcine reproductive and respiratory syndrome virus using a duplex real-time RT-PCR. J. Virol. Methods 2008, 151, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Tian, Y.; Zhou, D.; Duan, Z.; Guo, R.; Liu, Z.; Yuan, F.; Liu, W. A Multiplex RT-PCR Assay to Detect and Discriminate Porcine Reproductive and Respiratory Syndrome Viruses in Clinical Specimens. Viruses 2017, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Xi, J.; Chen, X.; Hu, S.; Chen, N.; Qiao, S.; Wan, S.; Bao, D. The development of a sensitive droplet digital PCR for quantitative detection of porcine reproductive and respiratory syndrome virus. Int. J. Biol. Macromol. 2017, 104, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.L.; Chai, L.Y.; Tian, R.B.; Zhao, Y.; Chen, H.Y.; Wang, Z.Y. Simultaneous detection of porcine reproductive and respiratory syndrome virus and porcine circovirus 3 by SYBR Green I-based duplex real-time PCR. Mol. Cell. Probes 2020, 49, 5. [Google Scholar] [CrossRef] [PubMed]
- Kolmodin, L.A.; Birch, D.E. Polymerase chain reaction. Basic principles and routine practice. Methods Mol. Biol. 2002, 192, 3–18. [Google Scholar] [PubMed]
- Sakhabutdinova, A.R.; Chemeris, A.V.; Garafutdinov, R.R. Enhancement of PCR efficiency using mono- and disaccharides. Anal. Biochem. 2020, 606, 113858. [Google Scholar] [CrossRef]
- Karunanathie, H.; Kee, P.S.; Ng, S.F.; Kennedy, M.A.; Chua, E.W. PCR enhancers: Types, mechanisms, and applications in long-range PCR. Biochimie 2022, 197, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, X.; Zou, R.; Xue, Y.; Lou, X.; He, M. Bovine Thrombin Enhances the Efficiency and Specificity of Polymerase Chain Reaction. Biotechniques 2018, 57, 289–294. [Google Scholar] [CrossRef]
- Li, X.; Galliher-Beckley, A.; Pappan, L.; Trible, B.; Kerrigan, M.; Beck, A.; Hesse, R.; Blecha, F.; Nietfeld, J.C.; Rowland, R.R.; et al. Comparison of Host Immune Responses to Homologous and Heterologous Type II Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Challenge in Vaccinated and Unvaccinated Pigs. Biomed. Res. Int. 2014, 2014, 416727. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.J. Heterogeneity of porcine reproductive and respiratory syndrome virus: Implications for current vaccine efficacy and future vaccine development. Vet. Microbiol. 2000, 74, 309–329. [Google Scholar] [CrossRef] [PubMed]
- Weesendorp, E.; Morgan, S.; Stockhofe-Zurwieden, N.; Popma-De Graaf, D.J.; Graham, S.P.; Rebel, J.M.J. Comparative analysis of immune responses following experimental infection of pigs with European porcine reproductive and respiratory syndrome virus strains of differing virulence. Vet. Microbiol. 2013, 163, 1–12. [Google Scholar] [CrossRef]
- Guo, B.; Lager, K.M.; Henningson, J.N.; Miller, L.C.; Schlink, S.N.; Kappes, M.A.; Kehrli, M.E.; Brockmeier, S.L.; Nicholson, T.L.; Yang, H.-C.; et al. Experimental infection of United States swine with a Chinese highly pathogenic strain of porcine reproductive and respiratory syndrome virus. Virology 2013, 435, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Ruan, S.; Ren, W.; Yu, B.; Yu, X.; Wu, H.; Li, W.; Jiang, Y.; He, Q. Development and Implementation of a Quadruple RT-qPCR Method for the Identification of Porcine Reproductive and Respiratory Syndrome Virus Strains. Viruses 2023, 15, 1946. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Liang, W.; Wang, X.; Chen, H.; Fan, J.; Song, W.; Hua, L.; Tang, X.; Chen, H.; Peng, Z.; et al. Epidemiological and genetic characteristics of porcine reproduction and respiratory syndrome virus 2 in mainland China, 2017–2018. Arch. Virol. 2020, 165, 1621–1632. [Google Scholar] [CrossRef]
- Zhang, Q.; Yang, F.; Gao, J.; Zhang, W.; Xu, X. Development of multiplex TaqMan qPCR for simultaneous detection and differentiation of eight common swine viral and bacterial pathogens. Braz. J. Microbiol. 2021, 53, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Munguía-Ramírez, B.; Armenta-Leyva, B.; Henao-Díaz, A.; Ye, F.; Baum, D.H.; Giménez-Lirola, L.G.; Zimmerman, J.J. Evaluation of a Porcine Endogenous Reference Gene (Internal Sample Control) in a Porcine Reproductive and Respiratory Syndrome Virus RT-qPCR. Vet. Sci. 2023, 10, 381. [Google Scholar] [CrossRef] [PubMed]
- Tu, T.; Pang, M.; Jiang, D.; Zhou, Y.; Wu, X.; Yao, X.; Luo, Y.; Yang, Z.; Ren, M.; Lu, A.; et al. Development of a Real-Time TaqMan RT-PCR Assay for the Detection of NADC34-like Porcine Reproductive and Respiratory Syndrome Virus. Vet. Sci. 2023, 10, 279. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhu, H.; Zhan, C.; Chen, P.; Wu, B.; Peng, Z.; Qian, P.; Cheng, G. Establishment and Application of a Quadruplex Real-Time Reverse-Transcription Polymerase Chain Reaction Assay for Differentiation of Porcine Reproductive and Respiratory Syndrome Virus, Porcine Circovirus Type 2, Porcine Circovirus Type 3, and Streptococcus suis. Microorganisms 2024, 12, 427. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-Y.; Deng, M.-C.; Wang, F.-I.; Tsai, H.-J.; Yang, C.-H.; Chang, C.; Huang, Y.-L. The application of a duplex reverse transcription real-time PCR for the surveillance of porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. J. Virol. Methods 2014, 201, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.-H.; Chen, X.-Z.; Hu, D.-M.; Yu, X.-L.; Wang, L.-L.; Han, W.; Wu, J.-J.; Cao, Z.; Wang, C.-B.; Zhang, Q.; et al. Rapid differential detection of classical and highly pathogenic North American Porcine Reproductive and Respiratory Syndrome virus in China by a duplex real-time RT-PCR. J. Virol. Methods 2009, 161, 192–198. [Google Scholar] [CrossRef]
- Chen, N.; Ye, M.; Xiao, Y.; Li, S.; Huang, Y.; Li, X.; Tian, K.; Zhu, J. Development of universal and quadruplex real-time RT-PCR assays for simultaneous detection and differentiation of porcine reproductive and respiratory syndrome viruses. Transbound. Emerg. Dis. 2019, 66, 2271–2278. [Google Scholar] [CrossRef]
- Shi, K.; Chen, Y.; Yin, Y.; Long, F.; Feng, S.; Liu, H.; Qu, S.; Si, H. A Multiplex Crystal Digital PCR for Detection of African Swine Fever Virus, Classical Swine Fever Virus, and Porcine Reproductive and Respiratory Syndrome Virus. Front. Vet. Sci. 2022, 9, 926881. [Google Scholar] [CrossRef] [PubMed]
- Amer, H.M.; Chae, H.; Roh, H.S.; Jo, Y.M.; Kim, W.G.; Chae, J.B.; Shin, S.-U.; Kang, J.W. Development of a one-step reverse transcription-quantitative polymerase chain reaction assay for the detection of porcine reproductive and respiratory syndrome virus. PLoS ONE 2023, 18, e0293042. [Google Scholar]
- Ma, Y.; Shi, K.; Chen, Z.; Shi, Y.; Zhou, Q.; Mo, S.; Wei, H.; Hu, L.; Mo, M. Simultaneous Detection of Porcine Respiratory Coronavirus, Porcine Reproductive and Respiratory Syndrome Virus, Swine Influenza Virus, and Pseudorabies Virus via Quadruplex One-Step RT-qPCR. Pathogens 2024, 13, 341. [Google Scholar] [CrossRef]
- Fornyos, K.; Szabó, I.; Lebhardt, K.; Bálint, Á. Development of a Farm-Specific Real-Time Quantitative RT-PCR Assay for the Detection and Discrimination of Wild-Type Porcine Reproductive Respiratory Syndrome Virus and the Vaccine Strain in a Farm under Eradication. Acta Vet. Hung. 2022, 70, 254–261. [Google Scholar] [CrossRef]





| Primer and Probe | Sequence of 5′-3′ | Fragment Length (bp) |
|---|---|---|
| PRRSV-F1 | 5′-TACATTCTGGCCCCTGCCCA-3′ | 196 bp |
| PRRSV-R1 | 5′-CCTCACCACTTGGAACAATTTATAC-3′ | |
| PRRSV-P1 | 6-FAM-TGCCACCCAACACGAGGC-MGB | |
| PRRSV-F2 | 5′-GGAATGGCCAGCCAGTCAA-3′ | 122 bp |
| PRRSV-R2 | 5′-TTCTTTTTAGGCCTCTTCGGGGTAA-3′ | |
| PRRSV-P2 | 6-FAM-AGCTGTGCCAAATGCTG-MGB |
| Enhancer | Ct Value | Ct Mean | SD | ||
|---|---|---|---|---|---|
| Control | 17.160 | 16.762 | 17.379 | 17.100 | 0.255 |
| BT | 16.293 | 15.902 | 16.027 | 16.074 | 0.163 |
| gp32 | 14.102 | 13.746 | 13.879 | 13.909 | 0.146 |
| DMSO | 14.176 | 14.121 | 14.436 | 14.244 | 0.137 |
| DTT | 16.395 | 16.277 | 16.057 | 16.243 | 0.146 |
| Sucrose | 16.285 | 16.739 | 16.012 | 16.345 | 0.299 |
| Total | 14.488 | 13.908 | 13.871 | 14.089 | 0.244 |
| Project | Parameter | Ct Average Value (3.12 × 105/3.12 × 104/ 3.12 × 103 copies/μL) | Amplification Efficiency/R2 |
|---|---|---|---|
| Primer concentration (μmol/L) | 0.2 | 22.66/25.92/29.45 | 97.006%/R2 = 1.000 |
| 0.4 | 22.96/25.99/29.76 | 99.580%/R2 = 0.997 | |
| 0.6 | 23.40/26.04/30.13 | 98.303%/R2 = 0.985 | |
| 0.8 | 23.18/26.27/29.68 | 102.977%/R2 = 0.999 | |
| Probe concentration (μmol/L) | 0.1 | 22.60/25.29/29.09 | 102.862%/R2 = 0.992 |
| 0.2 | 22.53/24.89/28.96 | 104.790%/R2 = 0.977 | |
| 0.3 | 22.75/25.73/29.61 | 98.619%/R2 = 0.993 | |
| Annealing temperature (°C) | 58 | 23.69/26.05/28.61 | 97.676%/R2 = 0.998 |
| 60 | 23.33/25.58/29.43 | 112.743%/R2 = 0.978 | |
| 62 | 23.64/26.19/29.68 | 114.306%/R2 = 0.991 |
| RT-qPCR System | Amplification Efficiency | R2 | Repeatability | |||||
|---|---|---|---|---|---|---|---|---|
| 3.12 × 105 | 3.12 × 104 | 3.12 × 103 | ||||||
| Ct Mean | CV (%) | Ct Mean | CV (%) | Ct Mean | CV (%) | |||
| A | 101.840% | 0.999 | 21.715 | 0.407% | 25.344 | 0.458% | 28.813 | 0.671% |
| B | 83.677% | 0.960 | 24.203 | 4.086% | 29.324 | 3.240% | 31.770 | 0.243% |
| C | 90.050% | 0.997 | 23.789 | 1.436% | 27.434 | 6.242% | 30.574 | 5.276% |
| Concentration (copies/µL) | Intra-Batch Assay | Inter-Batch Assay | ||
|---|---|---|---|---|
| Ct Mean ± SD | CV (%) | Ct Mean ± SD | CV (%) | |
| 3.12 × 105 | 21.43 ± 0.08 | 0.150% | 21.54 ± 0.08 | 0.369% |
| 3.12 × 104 | 25.32 ± 0.07 | 0.268% | 25.38 ± 0.06 | 0.248% |
| 3.12 × 103 | 28.86 ± 0.02 | 0.081% | 28.84 ± 0.06 | 0.201% |
| Dates | Viral Titers (TCID50/μL) | Xi’an Gentier 96R | ABl 7500 | ABl StepOne Plus | SD | CV % |
|---|---|---|---|---|---|---|
| Ct Value | ||||||
| 6.24 | 103.0 | 21.99/21.85/21.90 | 21.48/21.45/21.39 | 21.02/21.11/21.29 | 0.35 | 1.61% |
| 6.25 | 21.76/21.62/21.66 | 22.08/21.82/21.96 | 21.11/21.02/21.30 | 0.37 | 1.72% | |
| 6.27 | 21.61/21.62/21.80 | 21.79/21.72/21.90 | 21.20/21.22/21.10 | 0.30 | 1.40% | |
| 6.28 | 21.40/21.42/21.49 | 21.72/21.82/21.64 | 21.21/21.02/21.24 | 0.26 | 1.21% | |
| 6.29 | 21.61/21.58/21.61 | 21.66/21.55/21.66 | 20.98/21.01/21.10 | 0.29 | 1.37% | |
| 6.24 | 102.0 | 25.81/25.62/25.44 | 24.80/24.69/24.87 | 24.67/24.44/24.64 | 0.49 | 1.95% |
| 6.25 | 25.09/25.10/25.20 | 25.97/25.68/25.85 | 24.54/24.42/24.35 | 0.61 | 2.47% | |
| 6.27 | 24.76/24.89/24.78 | 25.17/25.23/25.23 | 24.70/24.90/24.64 | 0.23 | 0.94% | |
| 6.28 | 24.74/24.71/24.86 | 25.35/25.36/25.10 | 25.02/25.10/24.87 | 0.24 | 0.97% | |
| 6.29 | 24.91/25.01/24.88 | 25.23/25.50/25.31 | 26.00/25.80/25.81 | 0.42 | 1.65% | |
| 6.24 | 101.0 | 28.81/28.78/28.71 | 28.68/28.67/28.42 | 28.22/28.22/28.22 | 0.25 | 0.89% |
| 6.25 | 28.49/28.46/28.61 | 28.46/28.50/28.40 | 28.13/28.14/28.23 | 0.17 | 0.60% | |
| 6.27 | 28.37/28.41/28.30 | 28.41/28.43/28.50 | 28.09/28.11/28.09 | 0.16 | 0.57% | |
| 6.28 | 28.20/28.17/28.20 | 28.72/28.60/28.66 | 28.26/28.52/28.25 | 0.22 | 0.79% | |
| 6.29 | 28.28/28.21/28.35 | 29.48/29.48/29.40 | 29.28/29.31/29.31 | 0.55 | 1.92% | |
| Experimental Group | Control Group | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Endogenous Interfering Substances (1–5) | Exogenous Interfering Substances (6–13) | Positive | Negative | ||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| T1 | 28.09 | 28.35 | 28.42 | 28.41 | 29.06 | 28.36 | 29.50 | 29.01 | 28.88 | 29.21 | 32.40 | 32.05 | 30.93 | 28.48 | -- |
| 28.87 | 28.77 | 28.39 | 28.50 | 28.63 | 28.57 | 28.86 | 29.50 | 28.52 | 29.36 | 32.84 | 32.62 | 31.01 | 28.25 | -- | |
| T2 | 29.04 | 28.58 | 27.83 | 28.55 | 29.01 | 28.48 | 29.83 | 28.62 | 29.47 | 29.80 | 33.01 | 31.76 | 31.30 | 27.95 | -- |
| 28.46 | 28.94 | 21.43 | 29.77 | 29.31 | 28.48 | 29.39 | 29.37 | 28.70 | 29.50 | 32.55 | 32.29 | 30.52 | 28.67 | -- | |
| Clinical Samples (247) | PRRSV RT-qPCR | Control Kits |
|---|---|---|
| Samples (known PRRSV positivity 30) | 30/30 | 30/30 |
| Samples (known PRRSV negative 10) | 0/10 | 0/10 |
| Blood serum samples (93) | 11/93 | 11/93 |
| Swab samples (114) | 3/114 | 0/114 |
| Positive detection rate | 14/207 (6.8%) | 11/207 (5.3%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, H.; Xiao, X.; Sun, Y.; Chen, Y.; Zhang, Y.; Li, P.; Jin, H.; Li, Y.; Yin, R. A Simple and Sensitive RT-qPCR Technology for Rapid Detection of Porcine Reproductive and Respiratory Syndrome Virus. Vet. Sci. 2025, 12, 26. https://doi.org/10.3390/vetsci12010026
Zhao H, Xiao X, Sun Y, Chen Y, Zhang Y, Li P, Jin H, Li Y, Yin R. A Simple and Sensitive RT-qPCR Technology for Rapid Detection of Porcine Reproductive and Respiratory Syndrome Virus. Veterinary Sciences. 2025; 12(1):26. https://doi.org/10.3390/vetsci12010026
Chicago/Turabian StyleZhao, Hongri, Xingyu Xiao, Yajuan Sun, Yang Chen, Yongzhe Zhang, Peng Li, Hui Jin, Ying Li, and Rui Yin. 2025. "A Simple and Sensitive RT-qPCR Technology for Rapid Detection of Porcine Reproductive and Respiratory Syndrome Virus" Veterinary Sciences 12, no. 1: 26. https://doi.org/10.3390/vetsci12010026
APA StyleZhao, H., Xiao, X., Sun, Y., Chen, Y., Zhang, Y., Li, P., Jin, H., Li, Y., & Yin, R. (2025). A Simple and Sensitive RT-qPCR Technology for Rapid Detection of Porcine Reproductive and Respiratory Syndrome Virus. Veterinary Sciences, 12(1), 26. https://doi.org/10.3390/vetsci12010026





