A Multiplex PCR Assay for Simultaneous Detection of Giardia duodenalis, Cryptosporidium parvum, Blastocystis spp. and Enterocytozoon bieneusi in Goats
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples and Parasite DNA
2.2. Genomic DNA Extraction
2.3. Primer Design
2.4. Construction of the Standard Plasmids
2.5. Singleplex PCR Assays
2.6. Multiplex PCR Assay
2.7. Sensitivity and Specificity
2.8. Analysis of the Clinical Samples
3. Results
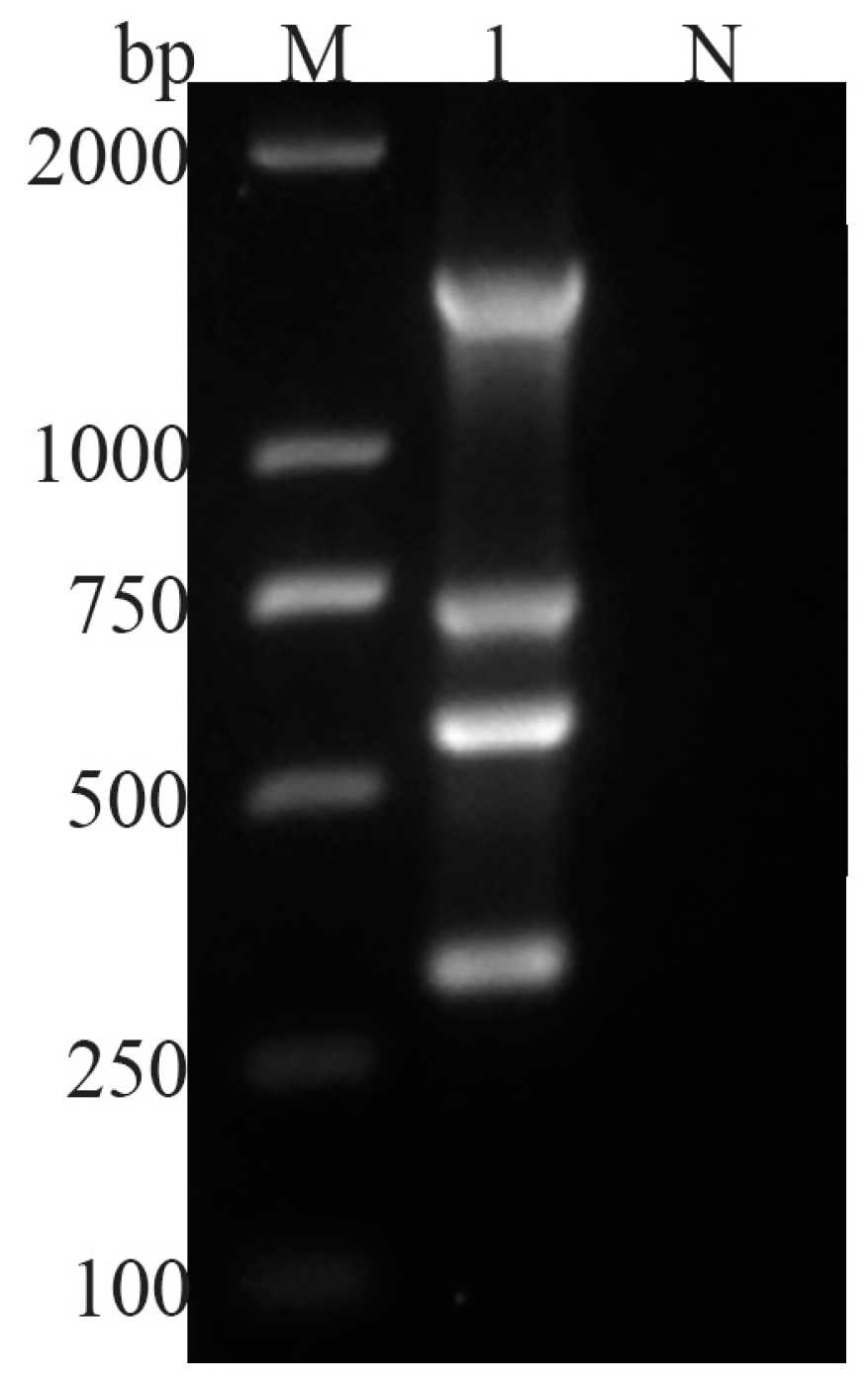
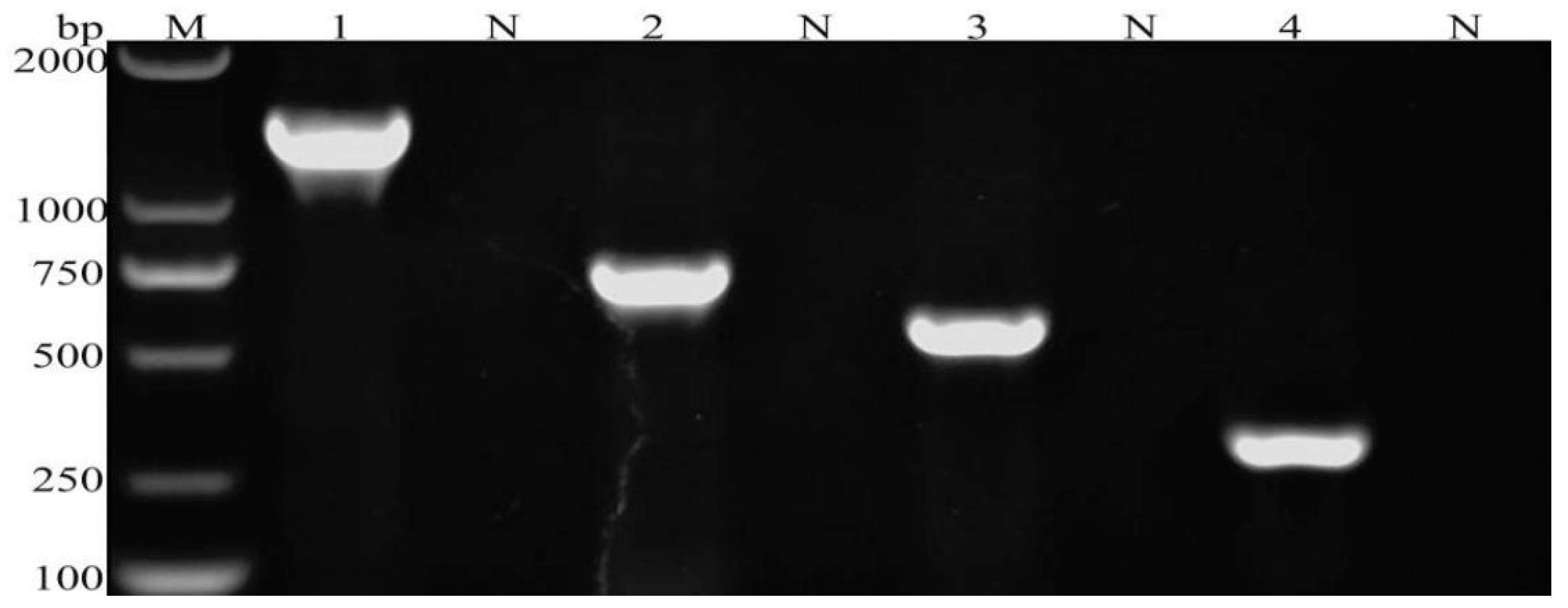
3.1. Multiplex PCR Assay Results
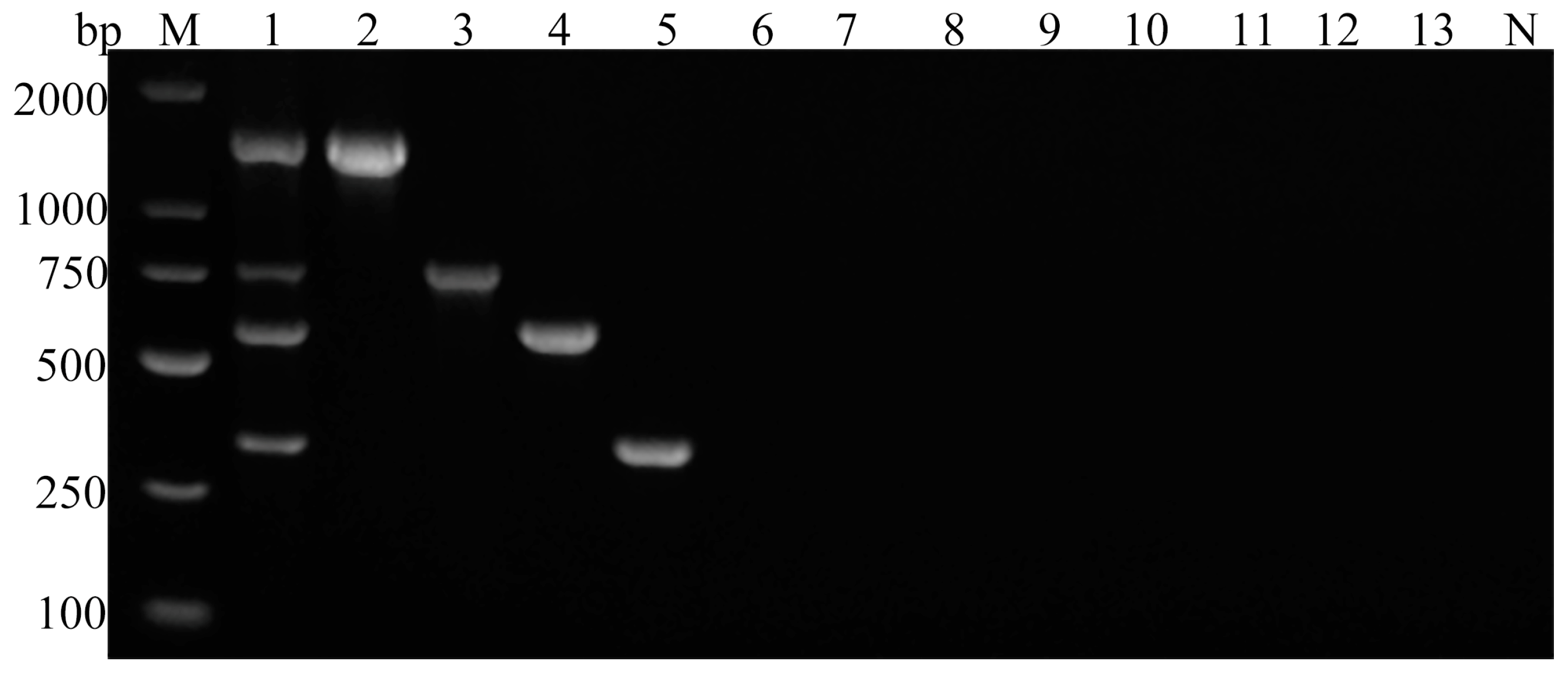
3.2. Sensitivity and Specificity
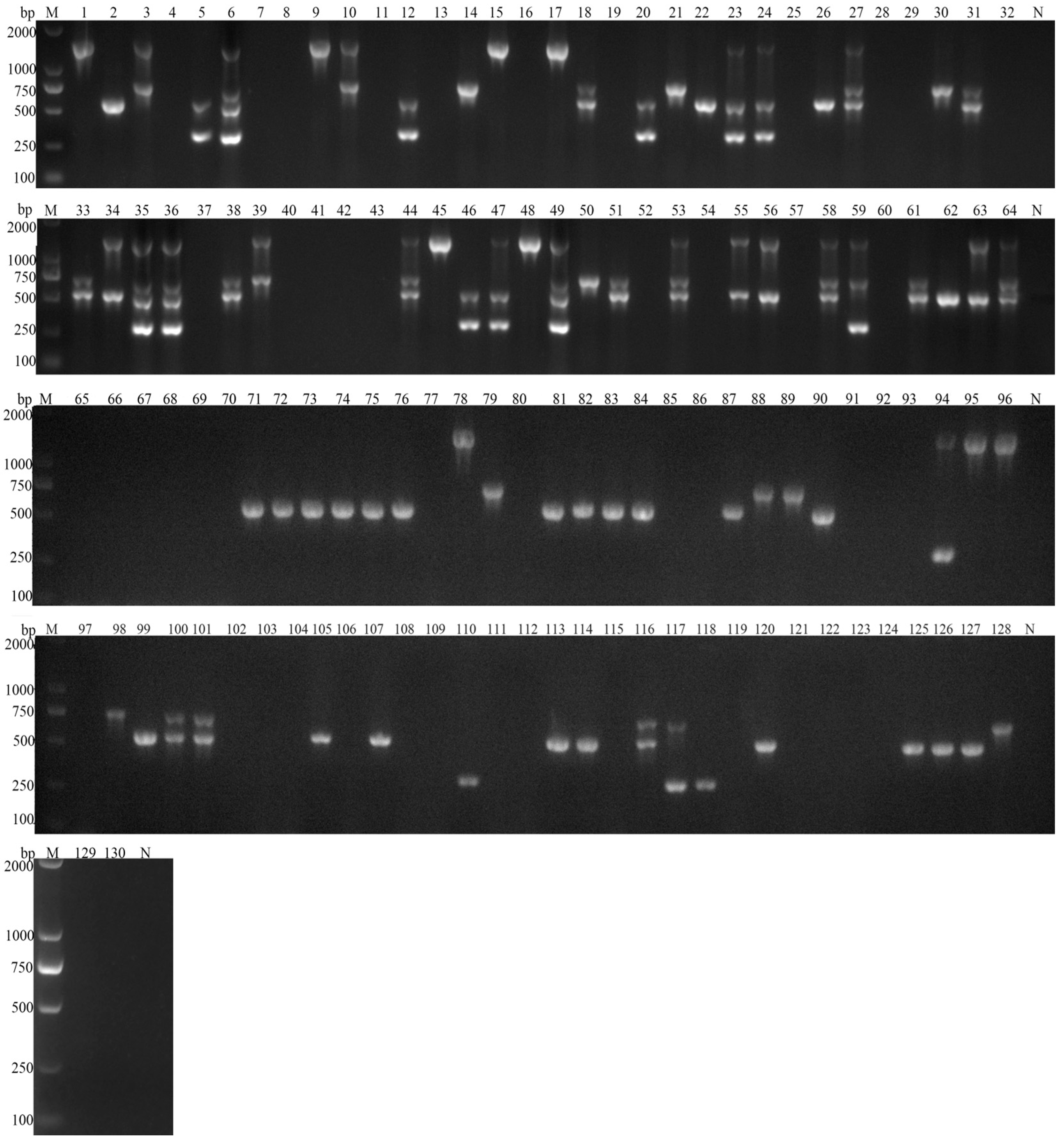
3.3. Field Evaluation of Multiplex PCR Assay
3.4. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feng, Y.; Xiao, L. Zoonotic potential and molecular epidemiology of Giardia species and Giardiasis. Clin. Microbiol. Rev. 2011, 24, 110–140. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Wang, H.; Li, Y.; Mu, X.; Yuan, K.; Wu, A.; Guo, J.; Hong, Y.; Zhang, H. Occurrence and genotypic identification of Blastocystis spp., Enterocytozoon bieneusi, and Giardia duodenalis in Leizhou black goats in Zhanjiang City, Guangdong Province, China. Animals 2023, 13, 2777. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.M.; Keithly, J.S.; Paya, C.V.; LaRusso, N.F. Cryptosporidiosis. N. Engl. J. Med. 2002, 346, 1723–1731. [Google Scholar] [CrossRef]
- Zhang, K.; Zheng, S.; Wang, Y.; Wang, K.; Wang, Y.; Gazizova, A.; Han, K.; Yu, F.; Chen, Y.; Zhang, L. Occurrence and molecular characterization of Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi, and Blastocystis sp. in captive wild animals in zoos in Henan, China. BMC Vet. Res. 2021, 17, 332. [Google Scholar] [CrossRef]
- Savioli, L.; Smith, H.; Thompson, A. Giardia and Cryptosporidium join the neglected diseases initiative. Trends Parasitol. 2006, 22, 203–208. [Google Scholar] [CrossRef]
- Cai, W.; Ryan, U.; Xiao, L.; Feng, Y. Zoonotic Giardiasis: An update. Parasitol. Res. 2021, 120, 4199–4218. [Google Scholar] [CrossRef]
- Golomazou, E.; Mamedova, S.; Eslahi, A.V.; Karanis, P. Cryptosporidium and agriculture: A review. Sci. Total Environ. 2024, 916, 170057. [Google Scholar] [CrossRef] [PubMed]
- Lang, J.; Han, H.; Dong, H.; Qin, Z.; Fu, Y.; Qin, H.; Zhang, J.; Zhao, J.; Li, X.; Zhao, G.; et al. Molecular characterization and prevalence of Cryptosporidium spp. in sheep and goats in western Inner Mongolia, China. Parasitol. Res. 2023, 122, 537–545. [Google Scholar] [CrossRef]
- Liu, J.; Platts-Mills, J.A.; Juma, J.; Kabir, F.; Nkeze, J.; Okoi, C.; Operario, D.J.; Uddin, J.; Ahmed, S.; Alonso, P.L.; et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: A reanalysis of the GEMS case-control study. Lancet 2016, 388, 1291–1301. [Google Scholar] [CrossRef]
- Abarca, N.; Santín, M.; Ortega, S.; Maloney, J.G.; George, N.S.; Molokin, A.; Cardona, G.A.; Dashti, A.; Köster, P.C.; Bailo, B.; et al. Molecular detection and characterization of Blastocystis spp. and Enterocytozoon bieneusi in cattle in Northern Spain. Vet. Sci. 2021, 8, 191. [Google Scholar]
- Diao, N.C.; Zhao, B.; Chen, Y.; Wang, Q.; Chen, Z.Y.; Yang, Y.; Sun, Y.H.; Shi, J.F.; Li, J.M.; Shi, K.; et al. Prevalence of Eimeria spp. among goats in China: A systematic review and meta-analysis. Front. Cell Infect. Microbiol. 2022, 12, 806085. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Cheng, C.; Feng, Q.; Ma, Y.; Hua, E.; Jiang, S.; Hou, Z.; Liu, D.; Yang, A.; Cheng, D.; et al. Prevalence and risk factors associated with gastrointestinal parasites in goats (Capra hircus) and sheep (Ovis aries) from three provinces of China. Front. Microbiol. 2023, 14, 1287835. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, H.; Wang, R.; Zhang, L. Giardia duodenalis infections in humans and other animals in China. Front. Microbiol. 2017, 8, 2004. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, F.; Zhang, Z.; Yang, X.; Ahmad, A.A.; Li, X.; Du, A.; Hu, M. Recent research progress in China on Haemonchus contortus. Front. Microbiol. 2017, 8, 1509. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Feng, Y.; Xiao, L. Diagnosis and molecular typing of Enterocytozoon bieneusi: The significant role of domestic animals in transmission of human microsporidiosis. Res. Vet. Sci. 2020, 133, 251–261. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, X.; Yang, R.; Zhao, W.; Li, N.; Guo, Y.; Xiao, L.; Feng, Y. Molecular characterization of the waterborne pathogens Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi, Cyclospora cayetanensis and Eimeria spp. in wastewater and sewage in Guangzhou, China. Parasit. Vectors 2021, 14, 66. [Google Scholar] [CrossRef]
- Guy, R.A.; Payment, P.; Krull, U.J.; Horgen, P.A. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 2003, 69, 5178–5185. [Google Scholar] [CrossRef]
- Bourli, P.; Eslahi, A.V.; Tzoraki, O.; Karanis, P. Waterborne transmission of protozoan parasites: A review of worldwide outbreaks an update 2017–2022. J. Water Health 2023, 21, 1421–1447. [Google Scholar] [CrossRef]
- Omarova, A.; Tussupova, K.; Berndtsson, R.; Kalishev, M.; Sharapatova, K. Protozoan parasites in drinking water: A system approach for improved water, sanitation and hygiene in developing countries. Int. J. Environ. Res. Public Health 2018, 15, 495. [Google Scholar] [CrossRef]
- Adeyemo, F.E.; Singh, G.; Reddy, P.; Stenström, T.A. Methods for the detection of Cryptosporidium and Giardia: From microscopy to nucleic acid based tools in clinical and environmental regimes. Acta Trop. 2018, 184, 15–28. [Google Scholar] [CrossRef]
- Jahan, N.; Khatoon, R.; Ahmad, S. A comparison of microscopy and enzyme linked immunosorbent assay for diagnosis of Giardia lamblia in human faecal Specimens. J. Clin. Diagn. Res. 2014, 8, DC04–DC06. [Google Scholar] [PubMed]
- Markoulatos, P.; Siafakas, N.; Moncany, M. Multiplex polymerase chain reaction: A practical approach. J. Clin. Lab. Anal. 2002, 16, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Alvarez, S.; Pérez-Roth, E. Multiplex PCR in clinical microbiology. Enferm. Infecc. Microbiol. Clin. 2004, 22, 183. [Google Scholar] [CrossRef] [PubMed]
- Tu, H.L.C.; Nugraheni, Y.R.; Tiawsirisup, S.; Saiwichai, T.; Thiptara, A.; Kaewthamasorn, M. Development of a novel multiplex PCR assay for the detection and differentiation of Plasmodium caprae from Theileria luwenshuni and Babesia spp. in goats. Acta Trop. 2021, 220, 105957. [Google Scholar] [CrossRef]
- Hu, W.; Wu, S.; Yu, X.; Abullahi, A.Y.; Song, M.; Tan, L.; Wang, Z.; Jiang, B.; Li, G. A multiplex PCR for simultaneous detection of three zoonotic parasites Ancylostoma ceylanicum, A. caninum, and Giardia lamblia assemblage A. BioMed Res. Int. 2015, 2015, 406168. [Google Scholar] [CrossRef]
- Fernandez, S.; Pagotto, A.H.; Furtado, M.M.; Katsuyama, A.M.; Madeira, A.M.; Gruber, A. A multiplex PCR assay for the simultaneous detection and discrimination of the seven Eimeria species that infect domestic fowl. Parasitology 2003, 127 Pt 4, 317–325. [Google Scholar] [CrossRef]
- Monaghan, T.F.; Rahman, S.N.; Agudelo, C.W.; Wein, A.J.; Lazar, J.M.; Everaert, K.; Dmochowski, R.R. Foundational statistical principles in medical research: Sensitivity, specificity, positive predictive value, and negative predictive value. Medicina 2021, 57, 503. [Google Scholar] [CrossRef]
- Xu, R.; Zhao, D.Y.; Lu, K.; Li, H.; Hong, Y.; Lin, J.J.; Zhu, C.G. Development of colloidal gold immunochromatographic test strips for diagnosis of Schistosoma japonicum in livestock. Chin. J. Schistosomiasis Control 2015, 27, 474–478. [Google Scholar]
- Rinaldi, L.; Veneziano, V.; Cringoli, G. Dairy goat production and the importance of gastrointestinal strongyle parasitism. Trans. R. Soc. Trop. Med. Hyg. 2007, 101, 745–746. [Google Scholar] [CrossRef]
- Ashraf, A.; Tramboo, S.R.; Maqbool, I.; Allaie, I.M.; Bulbul, K.H.; Shahardar, R.A.; Wani, Z.A.; Sheikh, F.D. Occurrence of GI parasites in ruminants of Kashmir and Ladakh. J. Parasit. Dis. 2022, 46, 196–201. [Google Scholar] [CrossRef]
- Wu, Y.Y.; Chang, Y.K.; Zheng, S.J.; Wang, L.; Li, D.F.; Li, G.; Zhang, S.M.; Zhang, H.Z.; Li, J.K.; Ning, C.S.; et al. Investigation of the prevalence of intestinal parasites in grazing livestock in parts of Tibet. Chin. Vet. Sci. 2017, 38, 1011–1016. [Google Scholar]
- Peng, X.Q.; Tian, G.R.; Ren, G.J.; Yu, Z.Q.; Lok, J.B.; Zhang, L.X.; Wang, X.T.; Song, J.K.; Zhao, G.H. Infection rate of Giardia duodenalis, Cryptosporidium spp. and Enterocytozoon bieneusi in cashmere, dairy and meat goats in China. Infect. Genet. Evol. 2016, 41, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Gomes-Gonçalves, S.; Palmeira, J.D.; Ferreira, H.; Santos-Silva, S.; Mesquita, J.R. Occurrence and phylogenetic analysis of zoonotic Enteropathogenic protist parasites in asymptomatic domestic ruminants from Portugal. Pathogens 2023, 12, 1341. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Zhang, Z.; Zhao, A.; Jing, B.; Guan, G.; Luo, J.; Zhang, L. Distribution and molecular characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi amongst grazing adult sheep in Xinjiang, China. Parasitol. Int. 2019, 71, 80–86. [Google Scholar] [CrossRef]
- Liu, Y.F.; Liu, H.; Li, K.R.; Liu, Y.H. Prevalence and molecular detection of human Cryptosporidium infections: A review. Chin. J. Schisto Control 2024, 36, 105–110. [Google Scholar]
- Yu, F.; Zhang, K.; Wang, Y.; Li, D.; Cui, Z.; Huang, J.; Zhang, S.; Li, X.; Zhang, L. CRISPR/Cas12a-based on-site diagnostics of Cryptosporidium parvum IId-subtype-family from human and cattle stool samples. Parasit. Vectors 2021, 14, 208. [Google Scholar] [CrossRef]
- Madadi, S.; Mahami-Oskouei, M.; Rafeey, M.; Spotin, A.; Aminisani, N.; Mahami-Oskouei, L.; Ghoyounchi, R.; Berahmat, R. Comparative evaluation of Cryptosporidium infection in malnourished and well-nourished children: Parasitic infections are affected by the interaction of nutritional status and socio-demographic characteristics. Comp. Immunol. Microbiol. Infect. Dis. 2020, 68, 101406. [Google Scholar] [CrossRef]
- Tan, L.; Wu, S.; Abdullahi, A.Y.; Yu, X.; Hu, W.; Song, M.; Shi, X.; Li, G. PCR-RFLP method to detect zoonotic and host-specific Giardia duodenalis assemblages in dog stool samples. Parasitol. Res. 2016, 115, 2045–2050. [Google Scholar] [CrossRef]
- Guo, J.; Starr, D.; Guo, H. Classification and review of free PCR primer design software. Bioinformatics 2021, 36, 5263–5268. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Mei, X.; Wei, L.; Wang, J.; Feng, X.; Wang, P.; He, B.; Chang, Y.; Xu, F.; Wang, M.; et al. Prevalence and molecular subtyping of Blastocystis in domestic pigeons in Henan Province, central China. J. Eukaryot. Microbiol. 2022, 69, e12888. [Google Scholar] [CrossRef]
- Twomey, D.F.; Barlow, A.M.; Bell, S.; Chalmers, R.M.; Elwin, K.; Giles, M.; Higgins, R.J.; Robinson, G.; Stringer, R.M. Cryptosporidiosis in two alpaca (Lama pacos) holdings in the South-West of England. Vet. J. 2008, 175, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Fusaro, C.; Bernal, J.E.; Baldiris-Ávila, R.; González-Cuello, R.; Cisneros-Lorduy, J.; Reales-Ruiz, A.; Castro-Orozco, R.; Sarria-Guzmán, Y. Molecular prevalence and subtypes distribution of Blastocystis spp. in humans of Latin America: A systematic review. Trop. Med. Infect. Dis. 2024, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Escalante, L.; Yang, C.; Sulaiman, I.; Escalante, A.A.; Montali, R.J.; Fayer, R.; Lal, A.A. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 1999, 65, 1578–1583. [Google Scholar] [CrossRef]
- Liu, J.B.; Zhao, H.X. Development of PCR diagnostic method for Trichomonas foetus infection based on 18S rRNA. Chin. J. Parasitol. Parasit. Dis. 2022, 40, 682–685. [Google Scholar]
- Liu, X.; Tang, L.; Li, W.; Li, C.; Gu, Y. Prevalence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi from large-scale cattle farms in Anhui Province, China. J. Vet. Med. Sci. 2022, 84, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhang, Y.; Jian, F.; Zhang, L.; Wang, R.; Cao, S.; Wang, X.; Yan, Y.; Ning, C. Development of duplex PCR for simultaneous detection of Theileria spp. and Anaplasma spp. in sheep and goats. Exp. Parasitol. 2017, 176, 1–7. [Google Scholar] [CrossRef]
- Henegariu, O.; Heerema, N.A.; Dlouhy, S.R.; Vance, G.H.; Vogt, P.H. Multiplex PCR: Critical parameters and step-by-step protocol. Biotechniques 1997, 23, 504–511. [Google Scholar] [CrossRef]
- Edwards, M.C.; Gibbs, R.A. Multiplex PCR: Advantages, development, and applications. PCR Methods Appl. 1994, 3, S65–S75. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Z.J.; Yang, J.F.; Chen, Z.; Guan, G.Q.; Ren, Q.Y.; Liu, A.H.; Luo, J.X.; Yin, H.; Li, Y.Q. Multiplex PCR for diagnosis of Theileria uilenbergi, Theileria luwenshuni, and Theileria ovis in small ruminants. Parasitol. Res. 2014, 113, 527–531. [Google Scholar] [CrossRef]

| Parasite | Target (GenBank Accession) | Primer Sequence (5′-3′) | Fragment Size (bp) |
|---|---|---|---|
| G. duodenalis | VSP with INR (XM_001710026.2) | F: CCTGTGCTTAAGTTCCGACG | 1400 |
| R: CGGTACTGCTAGCACATTCC | |||
| C. parvum | SSU rRNA (XM_626998.1) | F: GCTCTTGGACCTTGGAAAACG | 755 |
| R: GCTCGTCCATACCTTCAATCC | |||
| Blastocystis spp. | SSU rRNA (AB071000.1) | F: GCCCTATCAGCTTTGGATGG | 573 |
| R: GAATACCCCCAACTGTCCCT | |||
| E. bieneusi | 18S rRNA (KJ719492.1) | F: CAAGAGTGTCTATGGTGGATGC | 314 |
| R: CGAACACTAAGATTTCCCCGC |
| Parasite | True Positive | False Negative | True Negative | False Positive | Sensitivity (%) | Specificity (%) | Positive Predictive Value (%) | Negative Predictive Value (%) |
|---|---|---|---|---|---|---|---|---|
| G. duodenalis | 30 | 0 | 120 | 0 | 100 | 100 | 100 | 100 |
| E.bieneusi | 16 | 0 | 134 | 0 | 100 | 100 | 100 | 100 |
| C. parvum | 32 | 0 | 118 | 0 | 100 | 100 | 100 | 100 |
| Blastocystis spp. | 54 | 0 | 96 | 0 | 100 | 100 | 100 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, X.; Xu, H.; Mu, X.; Yuan, K.; Li, Y.; Xu, N.; Li, Q.; Zeng, W.; Chen, S.; Hong, Y. A Multiplex PCR Assay for Simultaneous Detection of Giardia duodenalis, Cryptosporidium parvum, Blastocystis spp. and Enterocytozoon bieneusi in Goats. Vet. Sci. 2024, 11, 448. https://doi.org/10.3390/vetsci11090448
Yu X, Xu H, Mu X, Yuan K, Li Y, Xu N, Li Q, Zeng W, Chen S, Hong Y. A Multiplex PCR Assay for Simultaneous Detection of Giardia duodenalis, Cryptosporidium parvum, Blastocystis spp. and Enterocytozoon bieneusi in Goats. Veterinary Sciences. 2024; 11(9):448. https://doi.org/10.3390/vetsci11090448
Chicago/Turabian StyleYu, Xingang, Hui Xu, Xuanru Mu, Kaijian Yuan, Yilong Li, Nuo Xu, Qiaoyu Li, Wenjing Zeng, Shengfeng Chen, and Yang Hong. 2024. "A Multiplex PCR Assay for Simultaneous Detection of Giardia duodenalis, Cryptosporidium parvum, Blastocystis spp. and Enterocytozoon bieneusi in Goats" Veterinary Sciences 11, no. 9: 448. https://doi.org/10.3390/vetsci11090448
APA StyleYu, X., Xu, H., Mu, X., Yuan, K., Li, Y., Xu, N., Li, Q., Zeng, W., Chen, S., & Hong, Y. (2024). A Multiplex PCR Assay for Simultaneous Detection of Giardia duodenalis, Cryptosporidium parvum, Blastocystis spp. and Enterocytozoon bieneusi in Goats. Veterinary Sciences, 11(9), 448. https://doi.org/10.3390/vetsci11090448






