Pseudomonas aestus Isolation from the Nasal Cavity of a Cat with Chronic Rhinitis
Abstract
Simple Summary
Abstract
1. Introduction
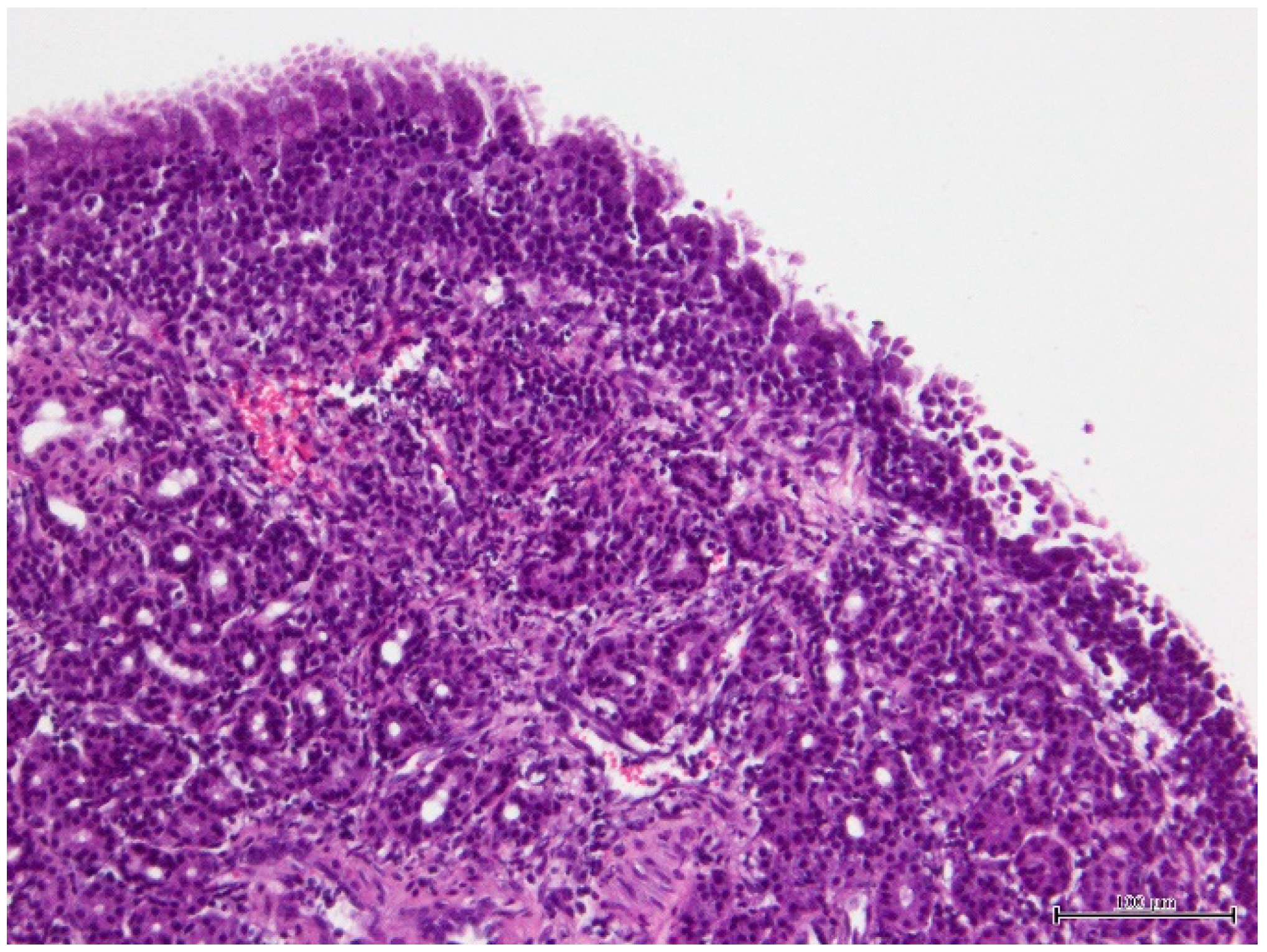
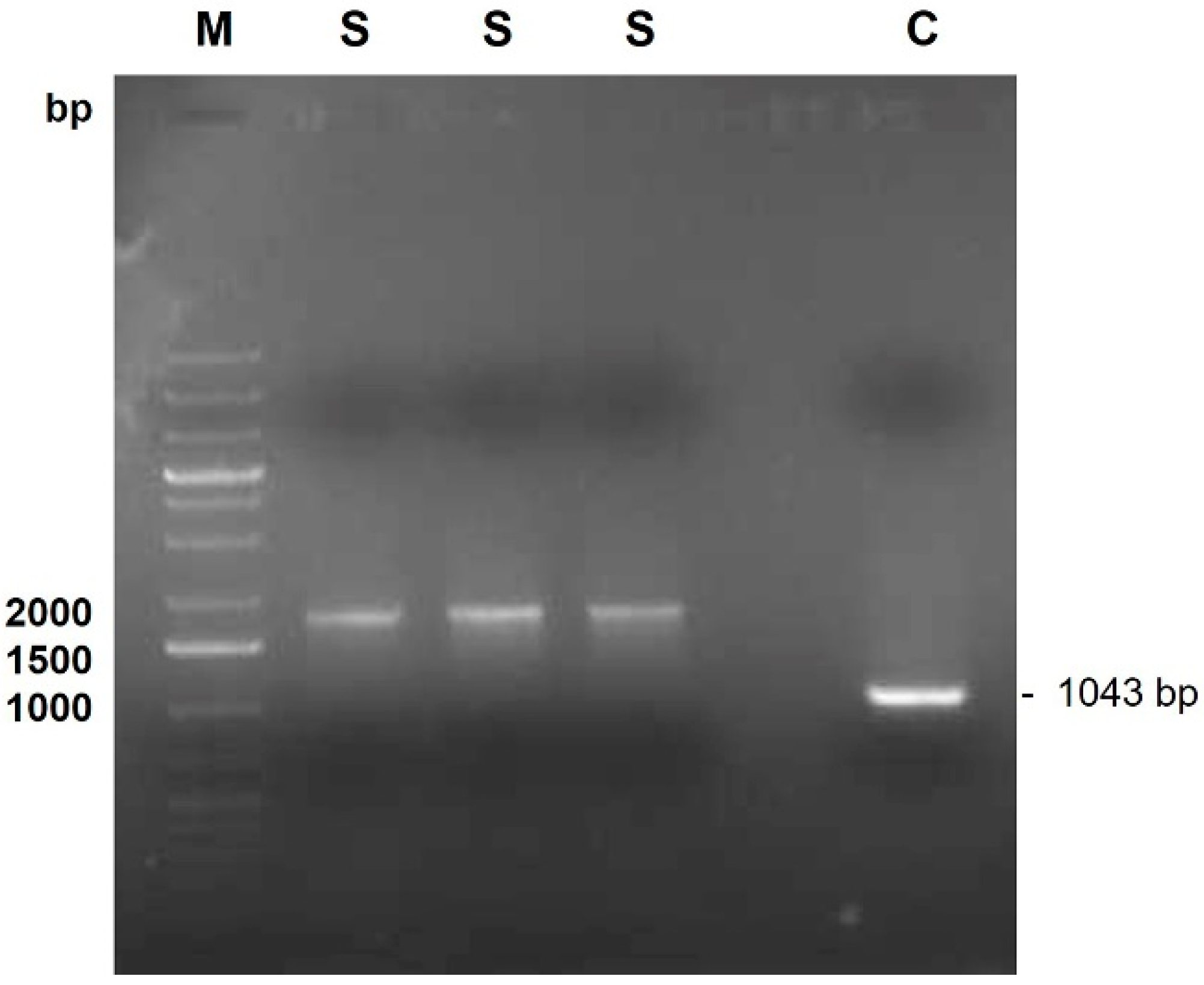
2. Case Presentation
3. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef]
- Peix, A.; Ramírez-Bahena, M.-H.; Velázquez, E. Historical Evolution and Current Status of the Taxonomy of Genus Pseudomonas. Infect. Genet. Evol. 2009, 9, 1132–1147. [Google Scholar] [CrossRef] [PubMed]
- Lalucat, J.; Mulet, M.; Gomila, M.; García-Valdés, E. Genomics in Bacterial Taxonomy: Impact on the Genus Pseudomonas. Genes 2020, 11, 139. [Google Scholar] [CrossRef]
- LPSN Genus Pseudomonas. Available online: https://lpsn.dsmz.de/genus/pseudomonas (accessed on 23 July 2024).
- Sharma, D.; Pakravan, N.; Pritchard, J.C.; Hartmann, F.A.; Young, K.M. Mucoid Pseudomonas aeruginosa Infection in a Cat with Severe Chronic Rhinosinusitis. Vet. Clin. Pathol. 2019, 48, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Markey, B.; Leonard, F.; Archambault, M.; Cullinane, A.; Maguire, D. Pseudomonas, Burkholderia and Stenotrophomonas Species. In Clinical Veterinary Microbiology; Markey, B., Leonard, F., Archambault, M., Cullinane, A., Maguire, D., Eds.; Elsevier: Edinburgh, UK, 2013; pp. 275–288. [Google Scholar]
- Dégi, J.; Moțco, O.A.; Dégi, D.M.; Suici, T.; Mareș, M.; Imre, K.; Cristina, R.T. Antibiotic Susceptibility Profile of Pseudomonas aeruginosa Canine Isolates from a Multicentric Study in Romania. Antibiotics 2021, 10, 846. [Google Scholar] [CrossRef]
- Silva, L.C.A.d.; Pessoa, D.A.d.N.; Maia, L.Â.; Matos, R.A.T.; Macêdo, M.M.d.S. Systemic Infection by Pseudomonas aeruginosa in a Dog. Acta Sci. Vet. 2016, 44, 1–5. [Google Scholar] [CrossRef]
- Polkowska, I.; Sobczyńska-Rak, A.; Golyńska, M. Analysis of Gingival Pocket Microflora and Biochemical Blood Parameters in Dogs Suffering from Periodontal Disease. In Vivo 2014, 28, 1085–1090. [Google Scholar]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-Resistant Pseudomonas Aeruginosa: Clinical Impact and Complex Regulation of Chromosomally Encoded Resistance Mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef]
- Geraldes, C.; Verdial, C.; Cunha, E.; Almeida, V.; Tavares, L.; Oliveira, M.; Gil, S. Evaluation of a Biocide Used in the Biological Isolation and Containment Unit of a Veterinary Teaching Hospital. Antibiotics 2021, 10, 639. [Google Scholar] [CrossRef]
- Tacconelli, E. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Vasconcellos, R.L.F.; Santos, S.N.; Zucchi, T.D.; Silva, F.S.P.; Souza, D.T.; Melo, I.S. Pseudomonas aestus sp. nov., a Plant Growth-Promoting Bacterium Isolated from Mangrove Sediments. Arch. Microbiol. 2017, 199, 1223–1229. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Bischof, J.; Byrne, S.K.; Radomski, C.; Davies, J.E.; Av-Gay, Y.; Vandamme, P. DNA-Based Diagnostic Approaches for Identification of Burkholderia cepacia Complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia Genomovars I and III. J. Clin. Microbiol. 2000, 38, 3165–3173. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D. The Condensed Protocols From Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 2006. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Perneel, M.; Heyrman, J.; Adiobo, A.; Maeyer, K.D.; Raaijmakers, J.M.; De Vos, P.; Höfte, M. Characterization of CMR5c and CMR12a, Novel Fluorescent Pseudomonas Strains from the Cocoyam Rhizosphere with Biocontrol Activity. J. Appl. Microbiol. 2007, 103, 1007–1020. [Google Scholar] [CrossRef] [PubMed]
- Girard, L.; Lood, C.; Höfte, M.; Vandamme, P.; Rokni-Zadeh, H.; van Noort, V.; Lavigne, R.; De Mot, R. The Ever-Expanding Pseudomonas Genus: Description of 43 New Species and Partition of the Pseudomonas putida Group. Microorganisms 2021, 9, 1766. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.B.; Weistein, M.P.; Eliopoulos, G.M.; Jenkins, S.G.; Lewis II, J.S.; Limbago, B.; Mathers, A.J.; Mazzulli, T.; Patel, R.; Ritcher, S.S.; et al. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; CLSI: Wayne, PA, USA, 2017; ISBN 1562387855. [Google Scholar]
- Watts, J.L.; Shryock, T.R.; Apley, M.; Bade, D.J.; Brown, S.D.; Gray, J.T.; Heine, H.; Hunter, R.P.; Mevius, D.J.; Papich, M.G.; et al. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals: Approved Standard, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008; Volume 28. [Google Scholar]
- Magiorakos, A.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F. Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2011, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Reed, N. Chronic Rhinitis in the Cat: An Update. Vet. Clin. Small Anim. Pract. 2020, 50, 311–329. [Google Scholar]
- Michiels, L.; Snaps, F.; Hansen, P.; Clercx, C. A Retrospective Study of Non-Specific Rhinitis in 22 Cats and the Value of Nasal Cytology and Histopathology. J. Feline Med. Surg. 2003, 5, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Reed, N.; Gunn-Moore, D. Nasopharyngeal Disease in Cats—2. Specific Conditions and Their Management Practical Relevance. J. Feline Med. Surg. 2012, 14, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.R.; Foley, J.E.; De Cock, H.E.V.; Clarke, H.E.; Maggs, D.J. Assessment of Infectious Organisms Associated with Chronic Rhinosinusitis in Cats. J. Am. Vet. Med. Assoc. 2005, 227, 579–585. [Google Scholar] [CrossRef] [PubMed]
| Antimicrobial Class | Antimicrobial Agent | Disk Content | Inhibition Halo Diameter (mm) | Classification |
|---|---|---|---|---|
| Aminoglycosides | Gentamicin | 10 µg | 17 | Susceptible |
| Tobramycin | 10 µg | 14 | Intermediate | |
| Amikacin | 30 µg | 16 | Intermediate | |
| Carbapenems | Imipenem | 10 µg | 8 | Resistant |
| Meropenem | 10 µg | 6 | Resistant | |
| Cephalosporins | Ceftazidime | 30 µg | 21 | Susceptible |
| Cefepime | 30 µg | 6 | Resistant | |
| Antipseudomonal penicillin | Piperacillin | 100 µg | 22 | Susceptible |
| Antipseudomonal penicillin + β-lactamase inhibitor | Piperacillin/tazobactam | 100/10 µg | 21 | Susceptible |
| Monobactam | Aztreonam | 30 µg | 6 | Resistant |
| Fluoroquinolones | Ciprofloxacin | 5 µg | 20 | Intermediate |
| Enrofloxacin | 5 µg | 19 | Intermediate | |
| Marbofloxacin | 5 µg | 18 | Intermediate | |
| Ofloxacin | 5 µg | 14 | Intermediate |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abreu, R.; Mouro, S.; Guerreiro, J.F.; Sousa, S.A.; Leitão, J.H.; Pissarra, H.; Cunha, E.; Tavares, L.; Oliveira, M. Pseudomonas aestus Isolation from the Nasal Cavity of a Cat with Chronic Rhinitis. Vet. Sci. 2024, 11, 382. https://doi.org/10.3390/vetsci11080382
Abreu R, Mouro S, Guerreiro JF, Sousa SA, Leitão JH, Pissarra H, Cunha E, Tavares L, Oliveira M. Pseudomonas aestus Isolation from the Nasal Cavity of a Cat with Chronic Rhinitis. Veterinary Sciences. 2024; 11(8):382. https://doi.org/10.3390/vetsci11080382
Chicago/Turabian StyleAbreu, Raquel, Sofia Mouro, Joana F. Guerreiro, Sílvia A. Sousa, Jorge H. Leitão, Hugo Pissarra, Eva Cunha, Luís Tavares, and Manuela Oliveira. 2024. "Pseudomonas aestus Isolation from the Nasal Cavity of a Cat with Chronic Rhinitis" Veterinary Sciences 11, no. 8: 382. https://doi.org/10.3390/vetsci11080382
APA StyleAbreu, R., Mouro, S., Guerreiro, J. F., Sousa, S. A., Leitão, J. H., Pissarra, H., Cunha, E., Tavares, L., & Oliveira, M. (2024). Pseudomonas aestus Isolation from the Nasal Cavity of a Cat with Chronic Rhinitis. Veterinary Sciences, 11(8), 382. https://doi.org/10.3390/vetsci11080382









