Transcriptomic Analysis of Metformin’s Effect on Bovine Viral Diarrhea Virus Infection
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell and BVDV Information
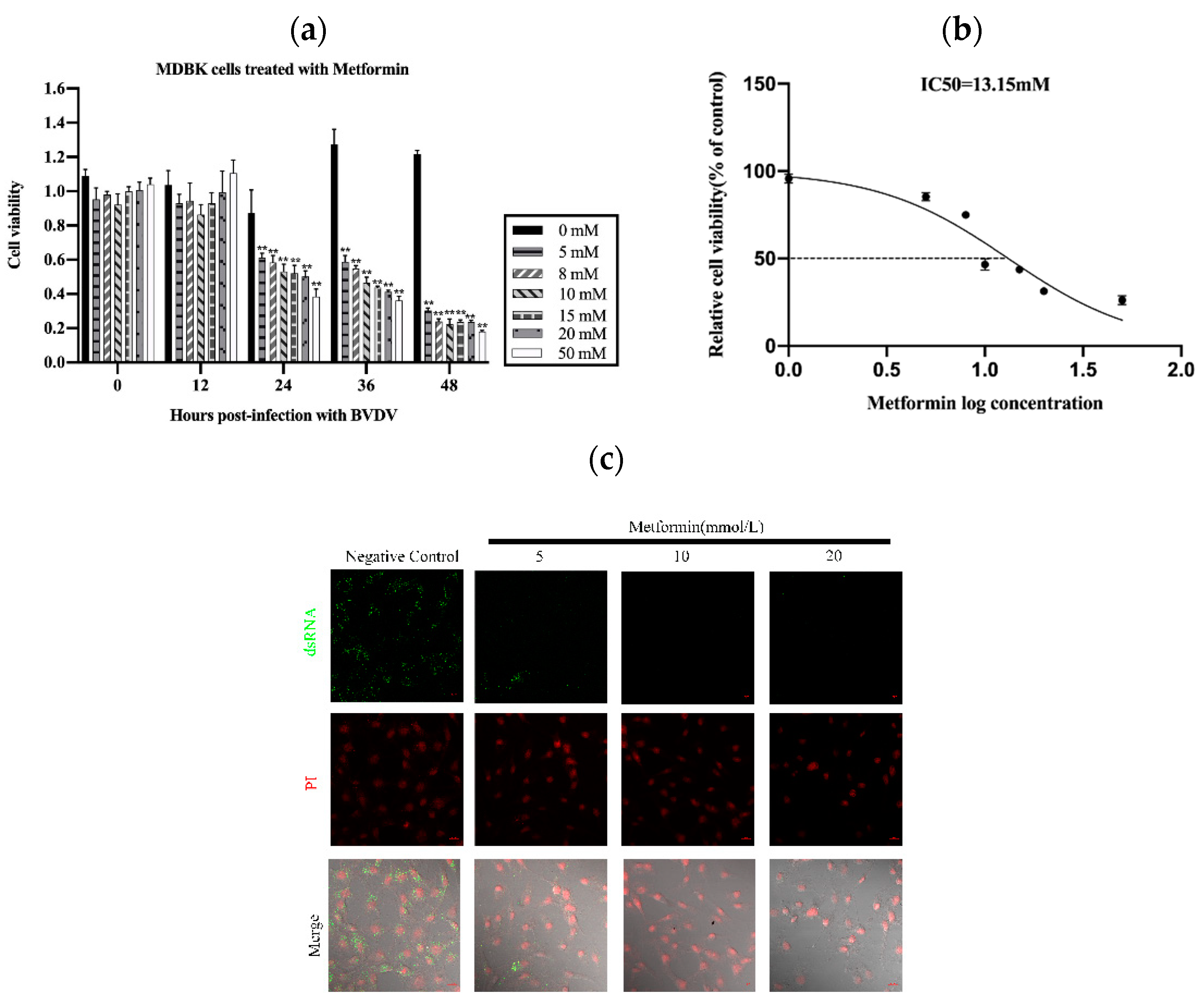
2.2. Cytotoxicity Analysis
2.3. BVDV dsRNA Detection
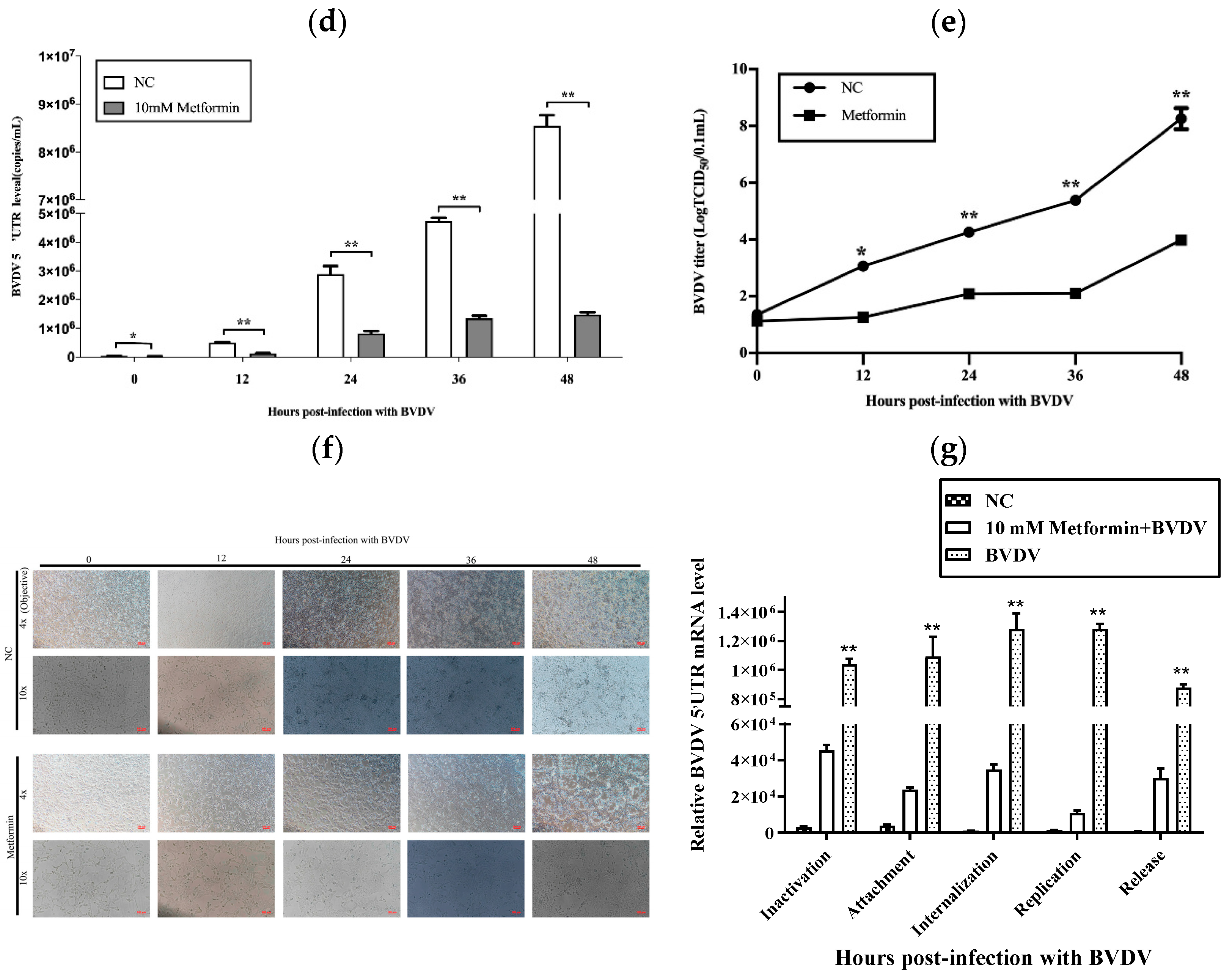
2.4. Quantitative Real-Time PCR (qPCR)
2.5. Viral Titer
2.6. Virus Replication Cycle Assay
2.7. Sample Collection
2.8. Preparation of Samples for Transcriptomic Sequencing
2.9. Quality Control and Read Mapping to the Reference Genome
2.10. Quantification of Gene Expression Levels
2.11. Differential Expression Analysis
2.12. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis
2.13. Gene Set Enrichment Analysis (GSEA)
2.14. Protein–Protein Interaction (PPI) Analysis
2.15. Validation of the Transcriptome Sequencing Data
2.16. Transcriptome Profile Submission
2.17. Statistical Analysis
3. Results
3.1. Met Served as a Novel Inhibitor of BVDV Replication In Vitro
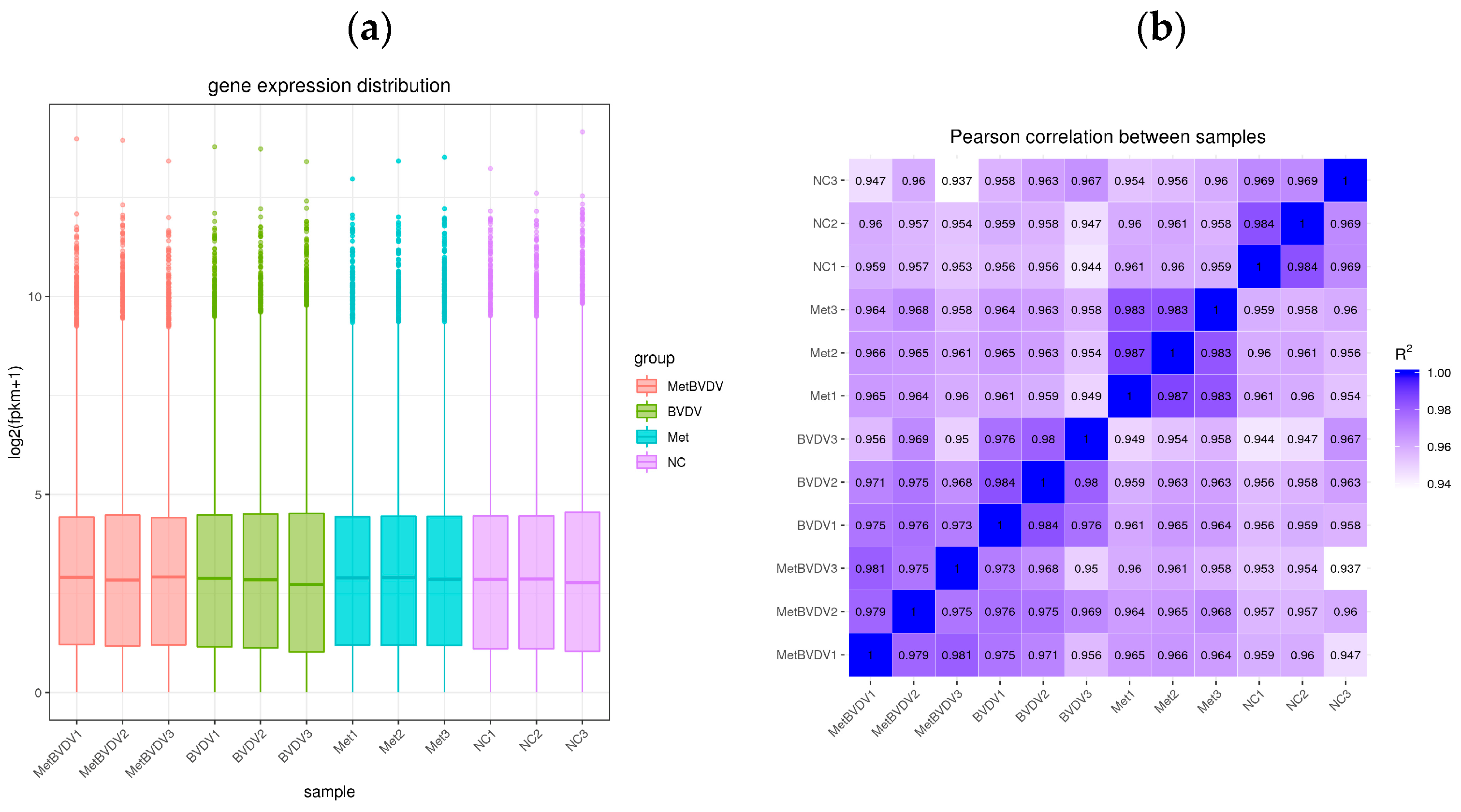
3.2. Evaluation of Transcriptomic Sequencing Data
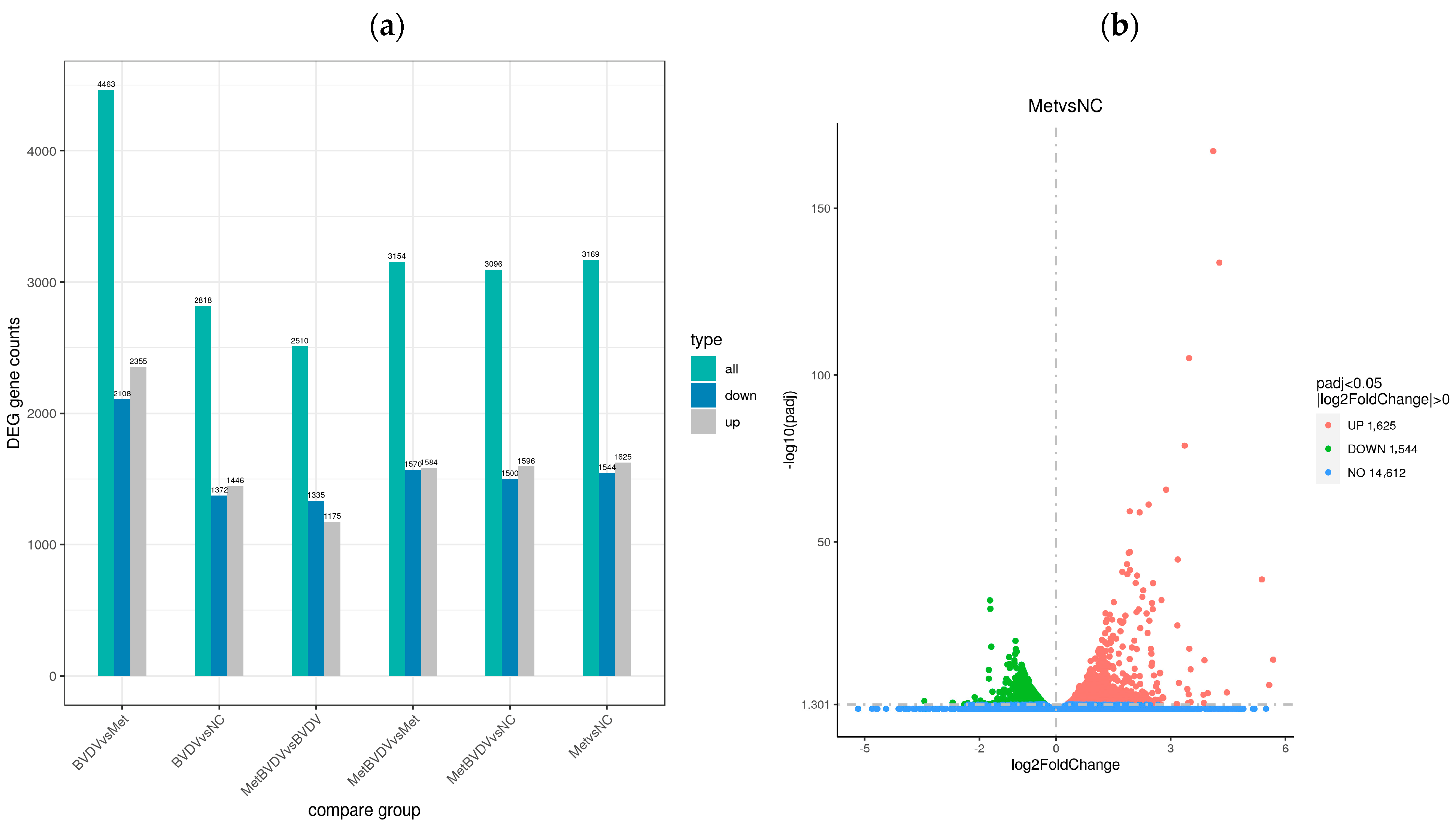
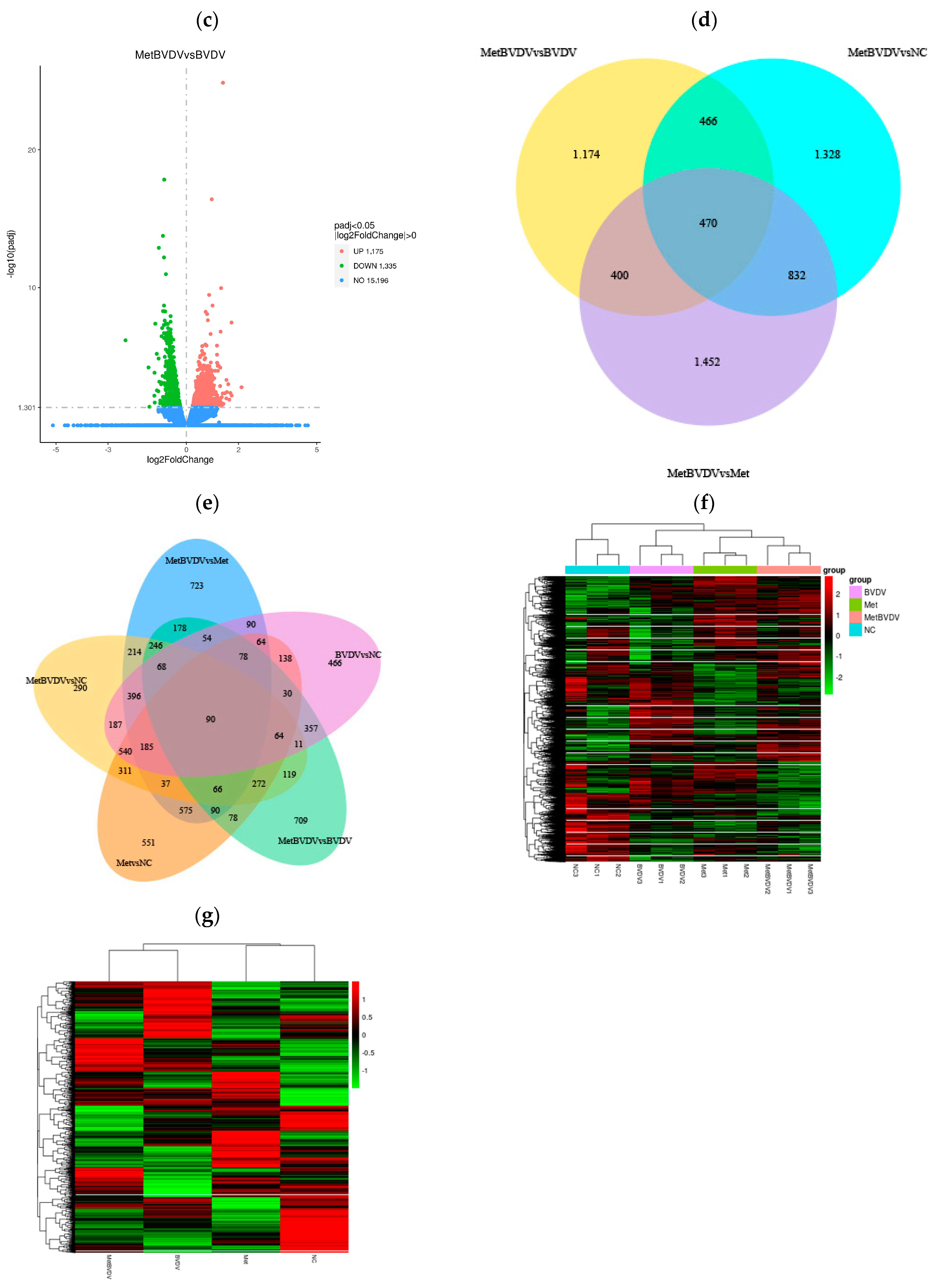
3.3. Differential Gene Expression Analysis
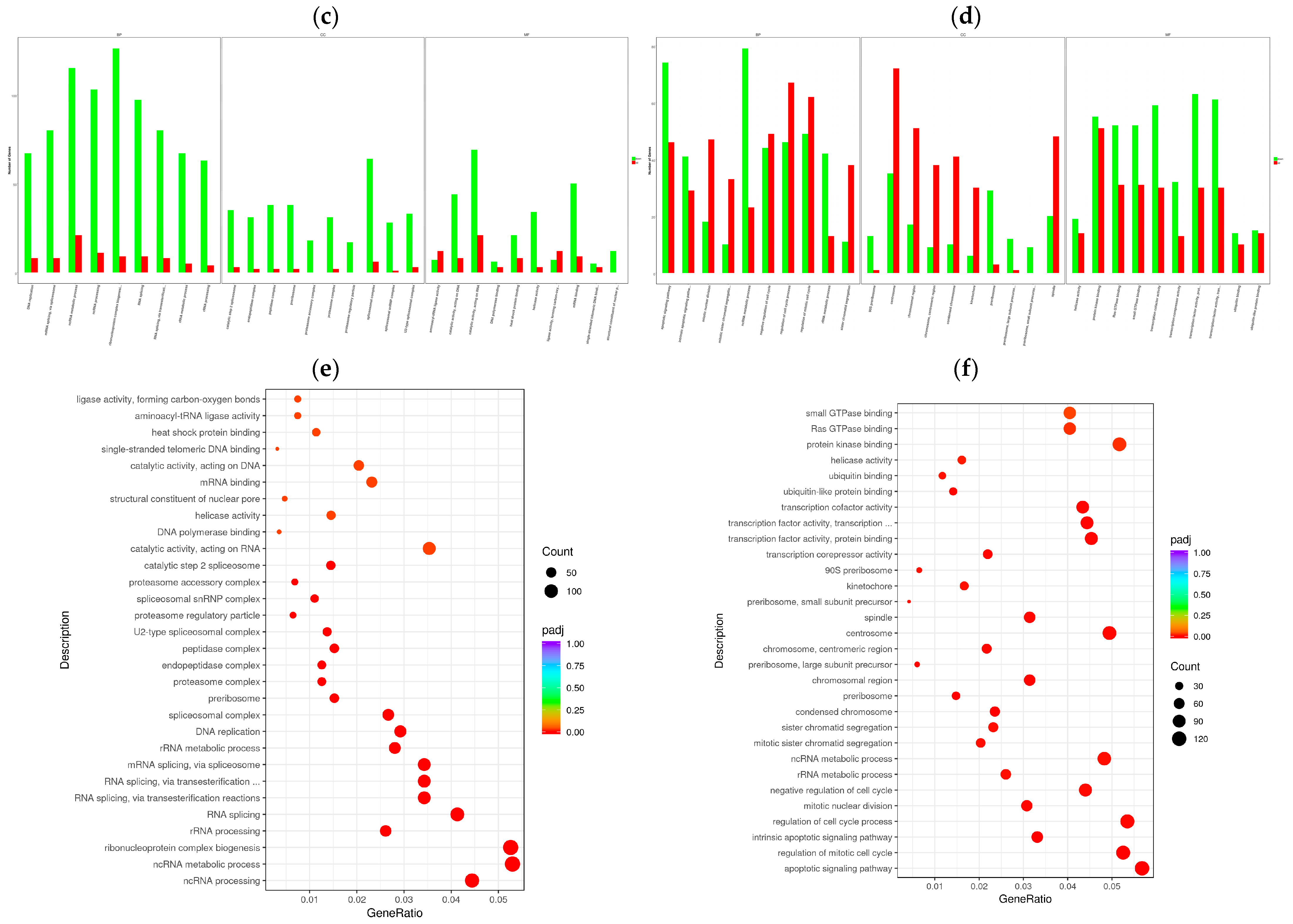
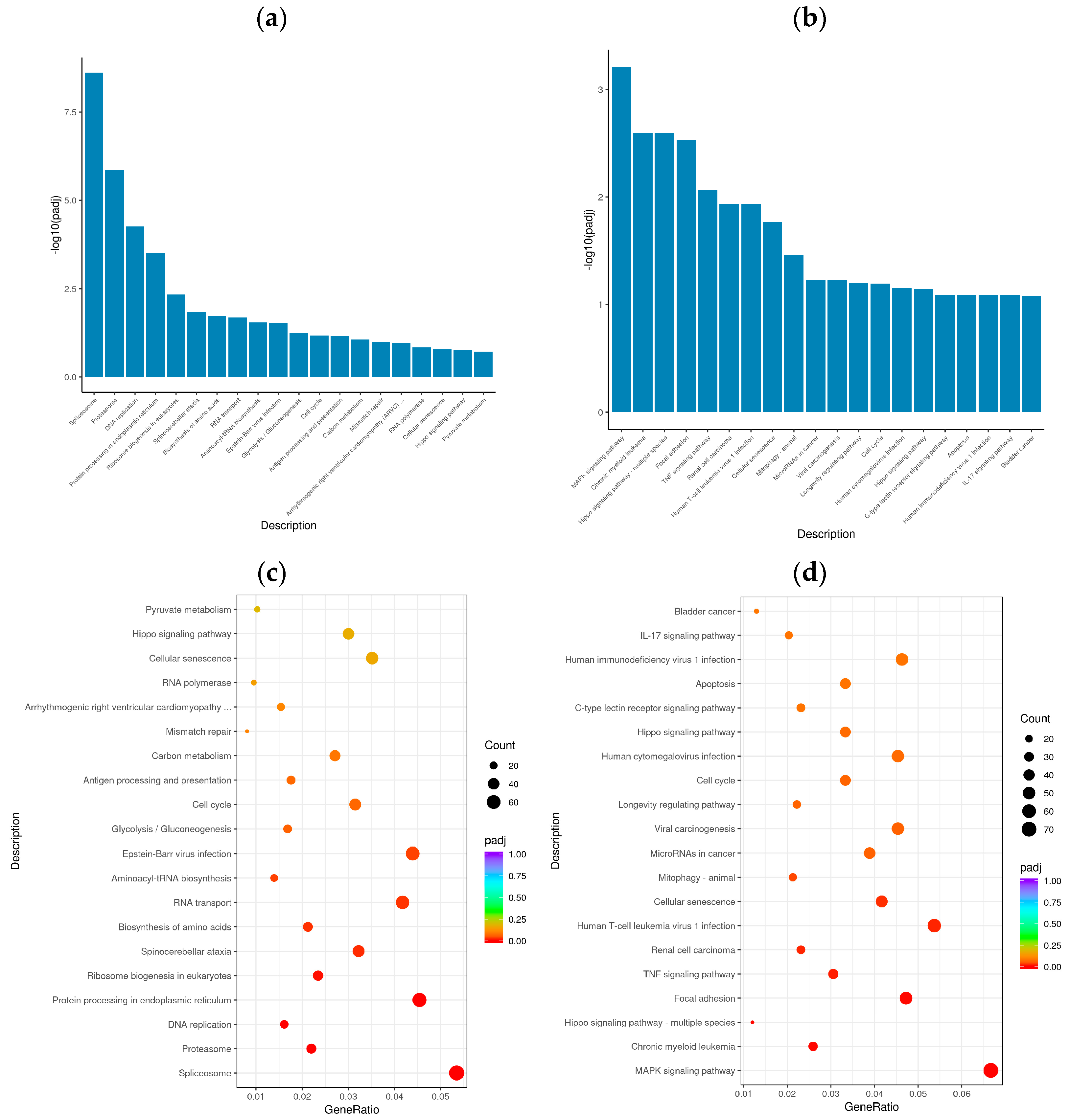
3.4. Enrichment Analysis of GO Terms and KEGG Pathways
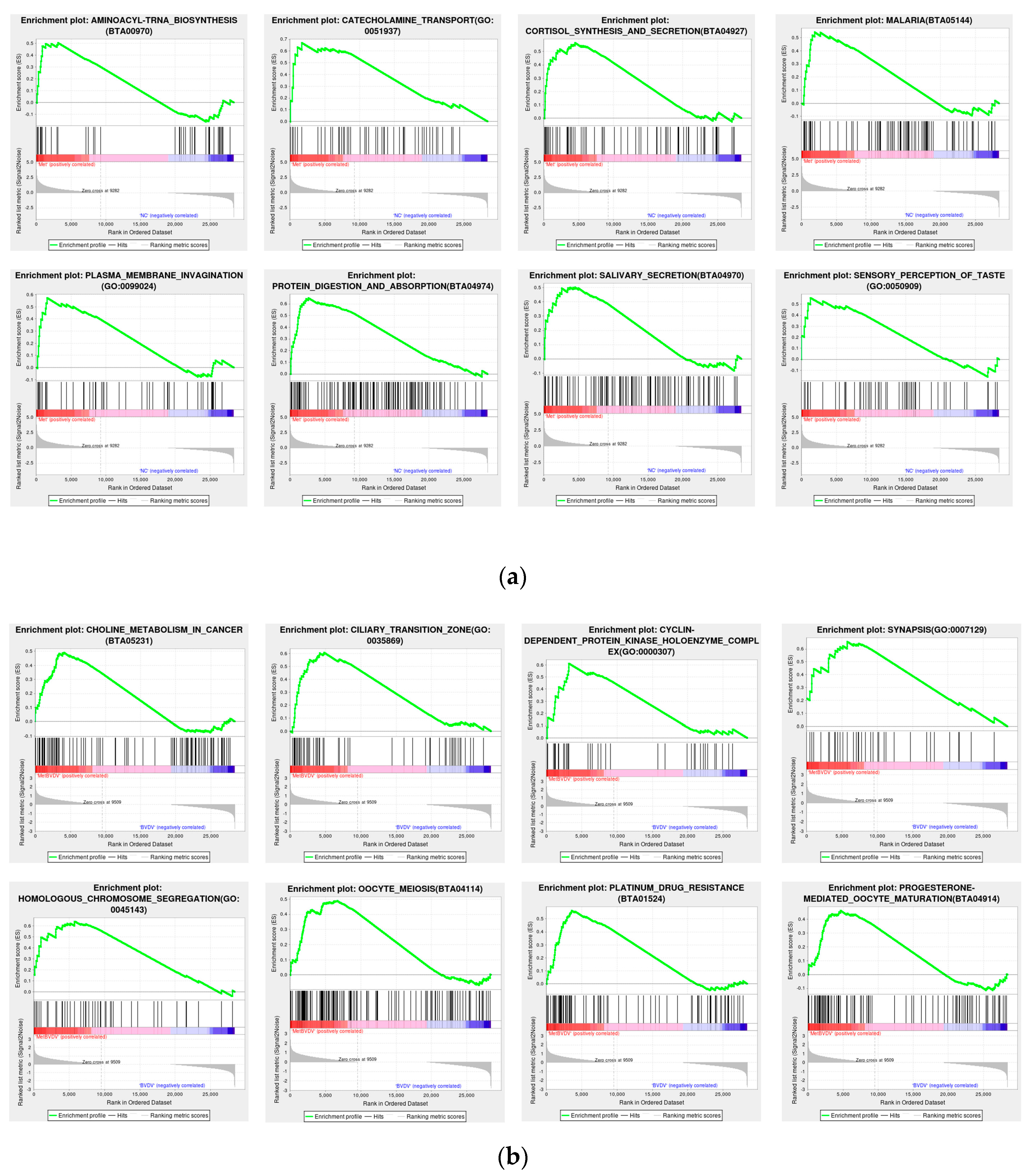
3.5. GSEA
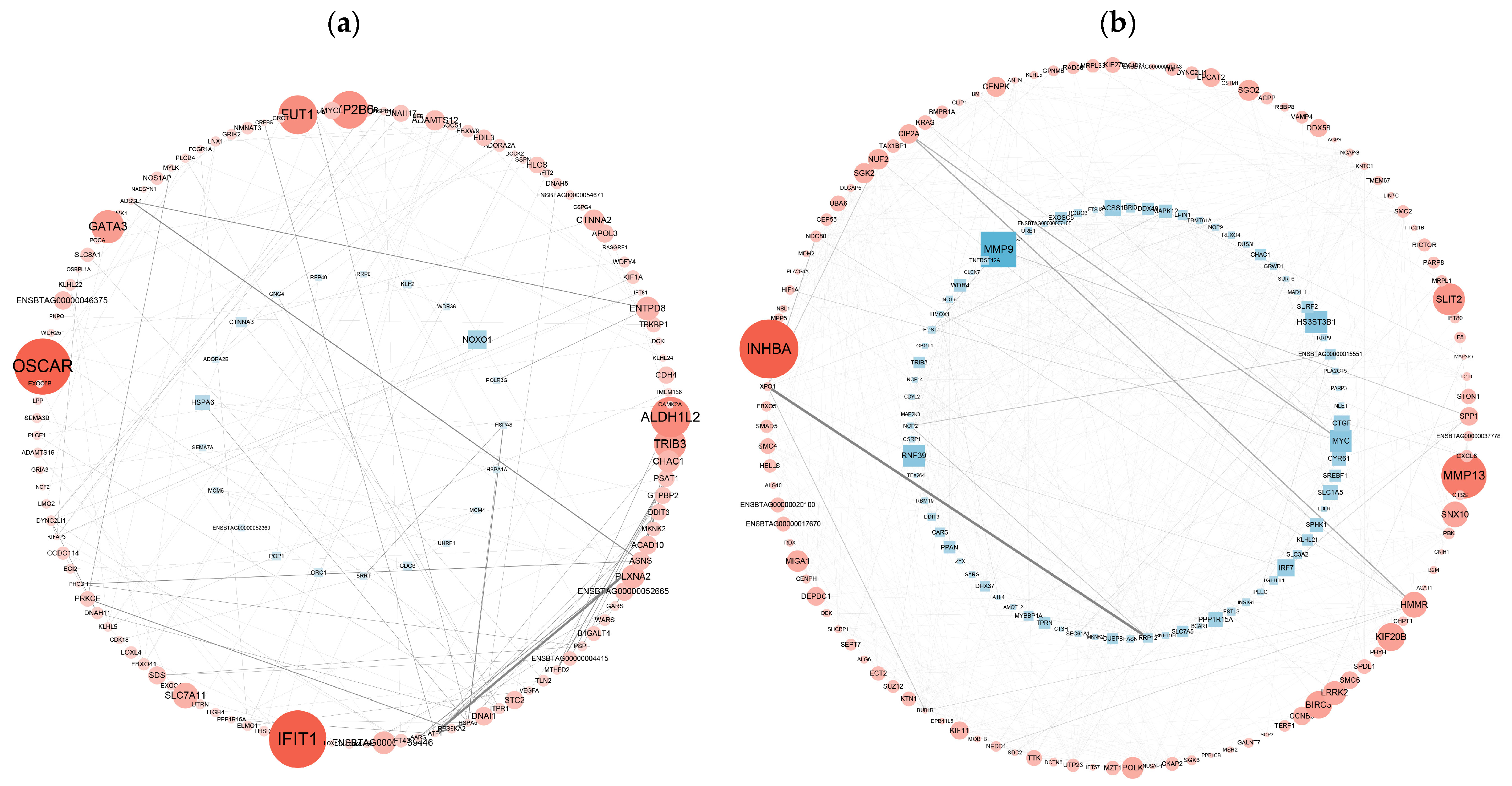
3.6. PPI Network Analysis of DEGs
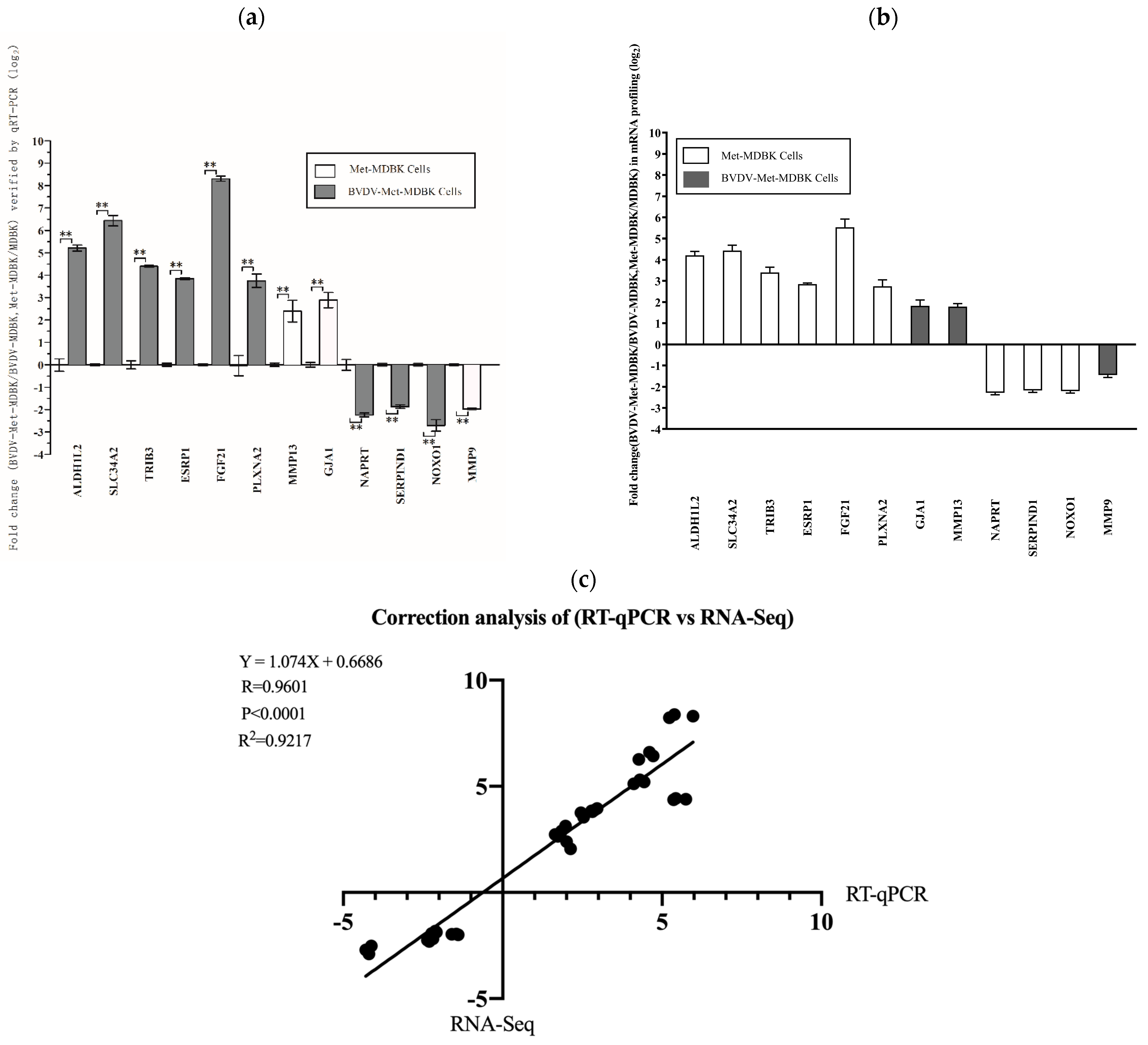
3.7. qPCR Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Evans, C.A.; Pinior, B.; Larska, M.; Graham, D.; Schweizer, M.; Guidarini, C.; Decaro, N.; Ridpath, J.; Gates, M.C. Global knowledge gaps in the prevention and control of bovine viral diarrhoea (BVD) virus. Transbound. Emerg. Dis. 2019, 66, 640–652. [Google Scholar] [CrossRef] [PubMed]
- Bürgi, N.; Josi, C.; Bürki, S.; Schweizer, M.; Pilo, P. Mycoplasma bovis co-infection with bovine viral diarrhea virus in bovine macrophages. Vet. Res. 2018, 49, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, J.; Qiu, Z.; Zhang, K.; Liang, F.; Zhou, Q.; Wang, L.; Li, J. Prevalence characteristic of BVDV in some large scale dairy farms in Western China. Front. Vet. Sci. 2022, 9, 961337. [Google Scholar] [CrossRef] [PubMed]
- Richter, V.; Lebl, K.; Baumgartner, W.; Obritzhauser, W.; Käsbohrer, A.; Pinior, B. A systematic worldwide review of the direct monetary losses in cattle due to bovine viral diarrhoea virus infection. Vet. J. 2017, 220, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wang, H.; Zhang, X.; Shi, K.; Li, J.; Zong, Y.; Du, R. Reaearch progress on drugs against BVDV. Heilongjiang Anim. Sci. Vet. Med. 2019, 9, 34–38. [Google Scholar]
- Oğuzoğlu, T.C.; Muz, D.; Timurkan, M.Ö.; Koç, B.T.; Özşahin, E.; Burgu, I.; Akça, Y.; Demirbağ, Z. Expression and production of recombinant proteins from immunodominant E gene regions of bovine viral diarrhoea virus 1 (BVDV-1) turkish field strains for prophylactic purpose. Rev. Med. Vet. 2017, 168, 183–191. [Google Scholar]
- Zhu, J.; Wang, C.; Zhang, L.; Zhu, T.; Li, H.; Wang, Y.; Xue, K.; Qi, M.; Peng, Q.; Chen, Y.; et al. Isolation of BVDV-1a, 1m, and 1v strains from diarrheal calf in China and identification of its genome sequence and cattle virulence. Front. Vet. Sci. 2022, 9, 1008107. [Google Scholar] [CrossRef]
- Bonora, E. Reduction in the incidence of type 2 diabetes mellitus with lifestyle intervention or metformin. Lendocrinologo 2002, 346, 393–403. [Google Scholar] [CrossRef]
- Anderson, J.J.A.; Couper, J.J.; Giles, L.C.; Leggett, C.E.; Gent, R.; Coppin, B.; Peña, A.S. Effect of Metformin on vascular function in children with type 1 diabetes: A 12 month randomized controlled trial. J. Clin. Endocrinol. Metab. 2017, 102, 4448–4456. [Google Scholar] [CrossRef]
- Romero-Gómez, M.; Diago, M.; Andrade, R.J.; Calleja, J.L.; Salmerón, J.; Fernández-Rodríguez, C.M. Treatment of insulin resistance with metformin in naïve genotype 1 chronic hepatitis C patients receiving peginterferon alfa-2a plus ribavirin. Hepatology 2009, 50, 1702–1708. [Google Scholar] [CrossRef]
- Tsai, W.L.; Chang, T.H.; Sun, W.C.; Chan, H.H.; Wu, C.C.; Hsu, P.I.; Cheng, J.S.; Yu, M.L. Metformin monophosphate-activated protein kinase. Oncotarget 2017, 8, 91928–91937. [Google Scholar] [CrossRef] [PubMed]
- Meireles, C.G.; Pereira, S.A.; Valadares, L.P.; Rêgo, D.F.; Simeoni, L.A.; Guerra, E.N.S.; Lofrano-Porto, A. Effects of metformin on endometrial cancer: Systematic review and meta-analysis. Gynecol. Oncol. 2017, 147, 167–180. [Google Scholar] [CrossRef]
- Campbell, J.M.; Bellman, S.M.; Stephenson, M.D.; Lisy, K. Metformin reduces all-cause mortality and diseases of ageing independent of its effect on diabetes control: A systematic review and meta-analysis. Ageing Res. Rev. 2017, 40, 31–44. [Google Scholar] [CrossRef]
- Long, F.; Zhang, M.; Yang, X.; Liang, X.; Su, L.; An, T.; Zhang, G.; Zeng, Z.; Liu, Y.; Chen, W.; et al. The Antimalaria Drug Artesunate Inhibits Porcine Reproductive and Respiratory Syndrome Virus Replication by Activating AMPK and Nrf2/HO-1 Signaling Pathways. J. Virol. 2022, 96, e01487-21. [Google Scholar] [CrossRef]
- Chen, W.C.; Hossen, M.; Liu, W.; Yen, C.H.; Huang, C.H.; Hsu, Y.C.; Lee, J.C. Grape Seed Proanthocyanidins Inhibit Replication of the Dengue Virus by Targeting NF-kB and MAPK-Mediated Cyclooxygenase-2 Expression. Viruses 2023, 15, 884. [Google Scholar] [CrossRef]
- Li, S.; Hu, X.; Tian, R.; Chen, J.; Li, Z.; Zhao, X.; Kuang, L.; Duo, L. RNA-Seq-based transcriptomic profiling of primary interstitial cells of Cajal in response to bovine viral diarrhea virus infection. Vet. Res. Commun. 2019, 43, 143–153. [Google Scholar] [CrossRef]
- Xu, Y.; Duan, S.; Ye, W.; Zheng, Z.; Zhang, J.; Gao, Y.; Ye, S. SLC34A2 promotes cell proliferation by activating STX17-mediated autophagy in esophageal squamous cell carcinoma. Thorac. Cancer 2024, 15, 1369–1384. [Google Scholar] [CrossRef]
- Nanni, M.; Ranieri, D.; Persechino, F.; Torrisi, M.R.; Belleudi, F. The aberrant expression of the mesenchymal variant of FGFR2 in the epithelial context inhibits autophagy. Cells 2019, 8, 653. [Google Scholar] [CrossRef]
- Qi, Y.; Liu, Y.; Rong, W. RNA-Seq and its applications: A new technology for transcriptomics. Hereditas 2011, 33, 1191–1202. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Berk, M.; Chung, Y.M.; Cui, D.; Zhu, Z.; Chakraborty, A.A.; Sharifi, N. Chronic hypoxia stabilizes 3βHSD1 via autophagy suppression. Cell Rep. 2024, 43, 113575. [Google Scholar] [CrossRef]
- De, L.A.; Maiello, M.R.; D’Alessio, A.; Pergameno, M.; Normanno, N. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: Role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin. Ther. Targets 2012, 16 (Suppl. S2), S17–S27. [Google Scholar] [CrossRef]
- Bussey, K.J. Integrating data on DNA copy number with gene expression levels and drug sensitivities in the NCI-60 cell line panel. Mol. Cancer Ther. 2006, 372, 853–867. [Google Scholar] [CrossRef]
- Purcell, M.K.; Kurath, G.; Garver, K.A.; Herwig, R.P.; Winton, J.R. Quantitative expression profiling of immune response genes in rainbow trout following infectious haematopoietic necrosis virus (IHNV) infection or DNA vaccination. Fish Shellfish. Immunol. 2004, 17, 447–462. [Google Scholar] [CrossRef]
- Kim, S.J.; Syed, G.H.; Siddiqui, A. Hepatitis C Virus Induces the Mitochondrial Translocation of Parkin and Subsequent Mitophagy. PLoS Pathog. 2013, 9, e1003285. [Google Scholar] [CrossRef] [PubMed]
- Jassey, A.; Liu, C.H.; Changou, C.A.; Richardson, C.D.; Hsu, H.Y.; Lin, L.T. Hepatitis C virus non-structural protein 5A (NS5A) disrupts mitochondrial dynamics and induces mitophagy. Cells 2019, 8, 290. [Google Scholar] [CrossRef]
- Kim, S.J.; Syed, G.H.; Khan, M.; Chiu, W.W.; Sohail, M.A.; Gish, R.G. Hepatitis C virus triggers mitochondrial fission and attenuates apoptosis to promote viral persistence. Proc. Natl. Acad. Sci. USA 2014, 111, 6413–6418. [Google Scholar] [CrossRef]
- Agnello, V.; Ábel, G.; Elfahal, M.; Knight, G.B.; Zhang, Q.-X. Hepatitis C Virus and Other Flaviviridae Viruses Enter Cells via Low Density Lipoprotein Receptor. Proc. Natl. Acad. Sci. USA 1999, 96, 12766–12771. [Google Scholar] [CrossRef]
- Buckwold, V.E.; Beer, B.E.; Donis, R.O. Bovine Viral Diarrhea Virus as a Surrogate Model of Hepatitis C Virus for the Evaluation of Antiviral Agents. Antivir. Res. 2003, 60, 1–15. [Google Scholar] [CrossRef]
- Zitzmann, N.; Mehta, A.S.; Carrouée, S.; Butters, T.D.; Platt, F.M.; McCauley, J.; Blumberg, B.S.; Dwek, R.A.; Block, T.M. Imino Sugars Inhibit the Formation and Secretion of Bovine Viral Diarrhea Virus, a Pestivirus Model of Hepatitis C Virus: Implications for the Development of Broad Spectrum Anti-Hepatitis Virus Agents. Proc. Natl. Acad. Sci. USA 1999, 96, 11878–11882. [Google Scholar] [CrossRef]
- Kim, S.J.; Jang, J.Y.; Kim, E.J.; Cho, E.K.; Ahn, D.G.; Kim, C.; Park, H.S.; Jeong, S.W.; Lee, S.H.; Kim, S.G.; et al. Ginsenoside Rg3 restores hepatitis C virus-induced aberrant mitochondrial dynamics and inhibits virus propagation. Hepatology 2017, 66, 758–771. [Google Scholar] [CrossRef]
- Liang, X.; Ji, X.; Ran, D. Isolation and identification of bovine viral diarrhea virus TC strain and sequence analysis of E0 gene. Anim. Husb. Vet. Med. 2016, 48, 36–40. [Google Scholar]
- Farfan-Morales, C.N.; Cordero-Rivera, C.D.; Osuna-Ramos, J.F.; Monroy Muñoz, I.E.; De Jesús-González, L.A.; Muñoz-Medina, J.E.; Hurtado Monzón, A.M.; Reyes-Ruiz, J.M.; Del Angel, R. The antiviral effect of metformin on zika and dengue virus infection. Sci. Rep. 2021, 11, 8743. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J. Muench H A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]

| Gene ID | Gene Name | GenBank | Primers (5′→3′) |
|---|---|---|---|
| gene1545 | SLC34A2 | NM_174661.2 | CTGCGTCTTCCAAGGGATTG |
| CCCAGCTACCTTTCCTCCAA | |||
| gene11624 | FGF21 | XM_024979245.1 | TTCCTTGAGGAGGACGCTTT |
| CCTTGCACATGGACTCACAG | |||
| gene6326 | ALDH1L2 | NM_001191391.1 | TTTCTGCCACAGAACAAGGC |
| ATGGGTGGGATGAGGGAAAG | |||
| gene17007 | TRIB3 | NM_001076103.1 | GGAGAACCTGGAAGATGCCT |
| GGCCAGCATGGTAAAGAGTG | |||
| gene1173 | PLXNA2 | NM_001206657.2 | GGAACCTGACGGAGGTAGAG |
| CTAGCCCAAACCAGTCCTGA | |||
| gene21838 | ESRP1 | NM_001193002.1 | ACGGAGGACTGCAAAGAAGA |
| TTAAACTGTCGGAGCGCTTG | |||
| gene1835 | GJA1 | NM_174068.2 | GCCTTCTTGCTGATCCAGTG |
| GCCGAGAAAGGAAACAGTCC | |||
| gene15059 | MMP13 | NM_174389.2 | AACGCCAGACAAATGTGACC |
| CTTCAACCTGCTGAGGATGC | |||
| gene14642 | NAPRT | XM_027560533.1 | AGGTGAACGTCATTGGCATC |
| CCAGGCAATGTCTGCTTCTC | |||
| gene13973 | SERPIND1 | NM_001105046.2 | GCCCTTCCTGTTCCTCATCT |
| AGCTCTTGGTGGTTGTCAGA | |||
| gene20369 | NOXO1 | XM_027527024.1 | GGCCCTTCTCCAACATCTCT |
| GGCCTGGTAGGGTACAAAGT | |||
| gene20676 | MMP9 | NM_174744.2 | CACGCACGACATCTTTCAGT |
| TCACGTAGCCCACATAGTCC | |||
| GAPDH | XM_014482068.1 | AAGGTCGGAGTGAACGGATT | |
| CGTTCTCTGCCTTGACTGTG |
| Sample | Library | Raw_Reads | Clean_Reads | Clean_Bases | Error_Rate | Q20 | Q30 | GC_pct |
|---|---|---|---|---|---|---|---|---|
| BVDV1 | FRAS210001071-1r | 42743418 | 41200374 | 6.18 G | 0.02 | 98.09 | 94.76 | 54.58 |
| BVDV2 | FRAS210001072-1r | 42083470 | 40540156 | 6.08 G | 0.03 | 97.57 | 93.62 | 54.9 |
| BVDV3 | FRAS210001073-1r | 44209666 | 42370430 | 6.36 G | 0.02 | 97.98 | 94.58 | 55.77 |
| Met1 | FRAS210001074-1r | 54087394 | 52340500 | 7.85 G | 0.02 | 97.91 | 94.31 | 53.54 |
| Met2 | FRAS210001075-1r | 47426060 | 45846420 | 6.88 G | 0.03 | 97.88 | 94.31 | 53.91 |
| Met3 | FRAS210001076-1r | 43909684 | 41876892 | 6.28 G | 0.02 | 97.95 | 94.45 | 53.66 |
| MetBVDV1 | FRAS210001077-1r | 42367072 | 40441680 | 6.07 G | 0.02 | 98.02 | 94.63 | 53.28 |
| MetBVDV2 | FRAS210001078-1r | 40551430 | 39255780 | 5.89 G | 0.02 | 98.01 | 94.56 | 53.88 |
| MetBVDV3 | FRAS210001079-1r | 41821636 | 40469088 | 6.07 G | 0.02 | 98.11 | 94.74 | 53.05 |
| NC1 | FRAS210001080-1r | 41858954 | 39774782 | 5.97 G | 0.02 | 98.02 | 94.55 | 53.09 |
| NC2 | FRAS210001081-1r | 43160394 | 41215010 | 6.18 G | 0.02 | 98.01 | 94.56 | 53.15 |
| NC3 | FRAS210001082-1r | 45915446 | 44639786 | 6.7 G | 0.02 | 97.94 | 94.42 | 54.53 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; He, Y.; Chen, J.; Ran, D.; Yue, J.; Fu, Q.; Shi, H. Transcriptomic Analysis of Metformin’s Effect on Bovine Viral Diarrhea Virus Infection. Vet. Sci. 2024, 11, 376. https://doi.org/10.3390/vetsci11080376
Li Z, He Y, Chen J, Ran D, Yue J, Fu Q, Shi H. Transcriptomic Analysis of Metformin’s Effect on Bovine Viral Diarrhea Virus Infection. Veterinary Sciences. 2024; 11(8):376. https://doi.org/10.3390/vetsci11080376
Chicago/Turabian StyleLi, Zeyu, Yuanxiu He, Junzhen Chen, Duoliang Ran, Jianbo Yue, Qiang Fu, and Huijun Shi. 2024. "Transcriptomic Analysis of Metformin’s Effect on Bovine Viral Diarrhea Virus Infection" Veterinary Sciences 11, no. 8: 376. https://doi.org/10.3390/vetsci11080376
APA StyleLi, Z., He, Y., Chen, J., Ran, D., Yue, J., Fu, Q., & Shi, H. (2024). Transcriptomic Analysis of Metformin’s Effect on Bovine Viral Diarrhea Virus Infection. Veterinary Sciences, 11(8), 376. https://doi.org/10.3390/vetsci11080376





