Development and Clinical Application of a Molecular Assay for Four Common Porcine Enteroviruses
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses and Clinical Samples
2.2. Primers and Probes
2.3. RNA Extraction and Reverse Transcription
2.4. Construction of Recombinant Plasmids for Standard Curves
2.5. Reaction Conditions of the Singleplex Real-Time PCR
2.6. Optimization of Reaction Conditions for Multiplex Real-Time PCR
2.7. Specificity of Multiplex Real-Time RT-PCR
2.8. Sensitivity of Multiplex Real-Time PCR
2.9. Repeatability of Multiplex Real-Time PCR
2.10. Clinical Sample Detection
3. Results
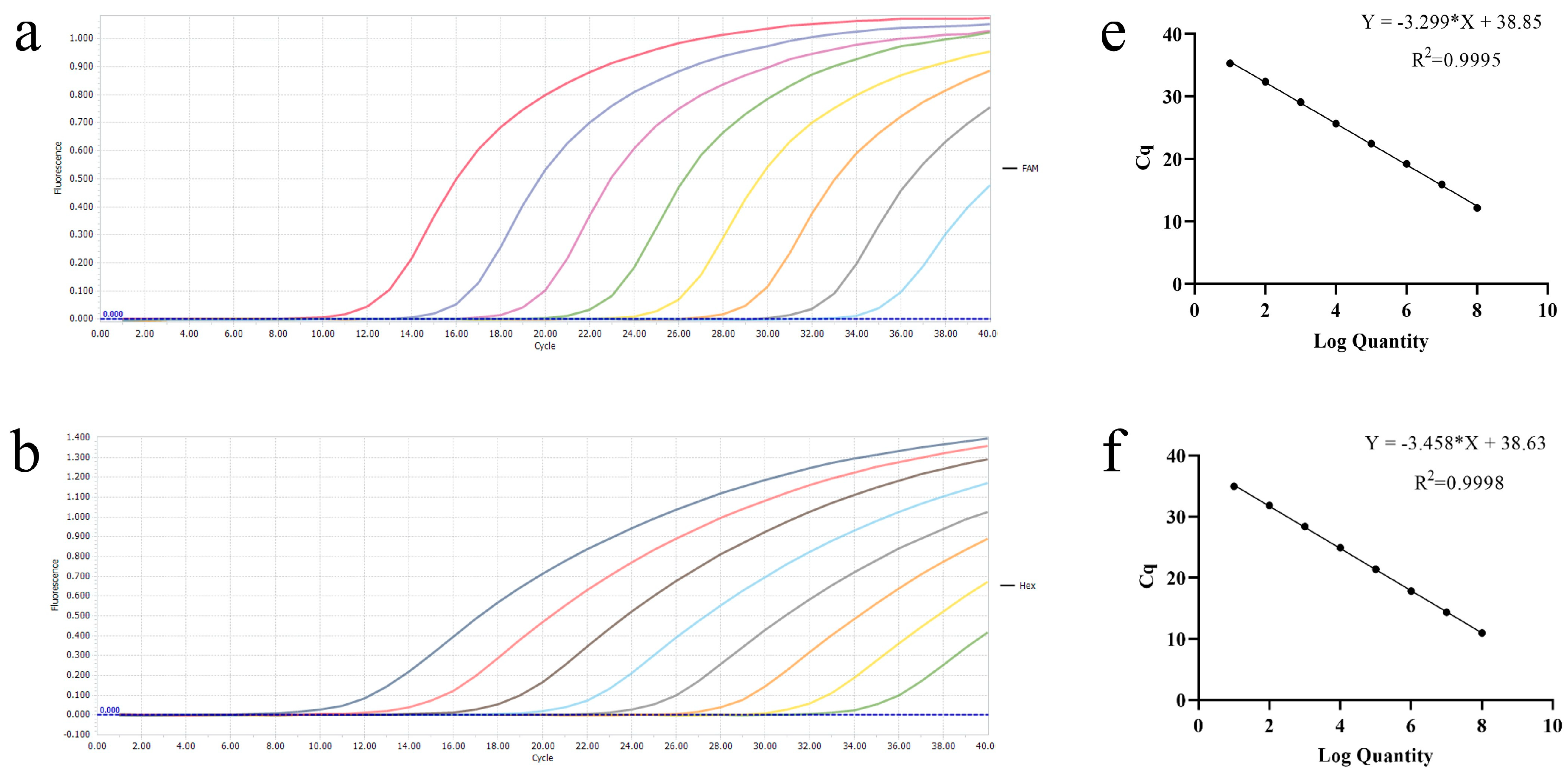
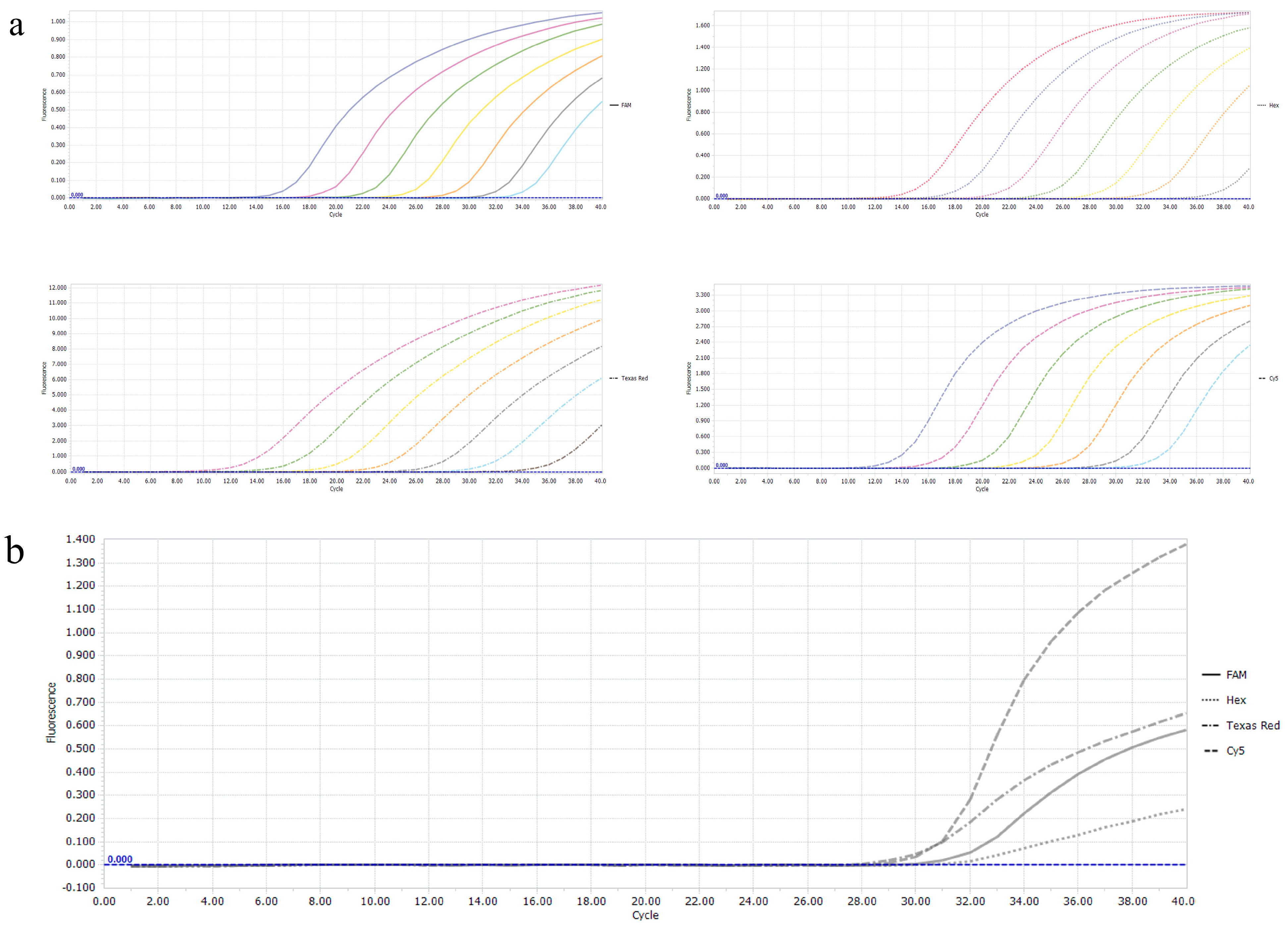
3.1. Establishment of Single Real-Time PCR Standard Curves
3.2. Establishment and Optimization of the Multiplex Real-Time PCR Reaction Method
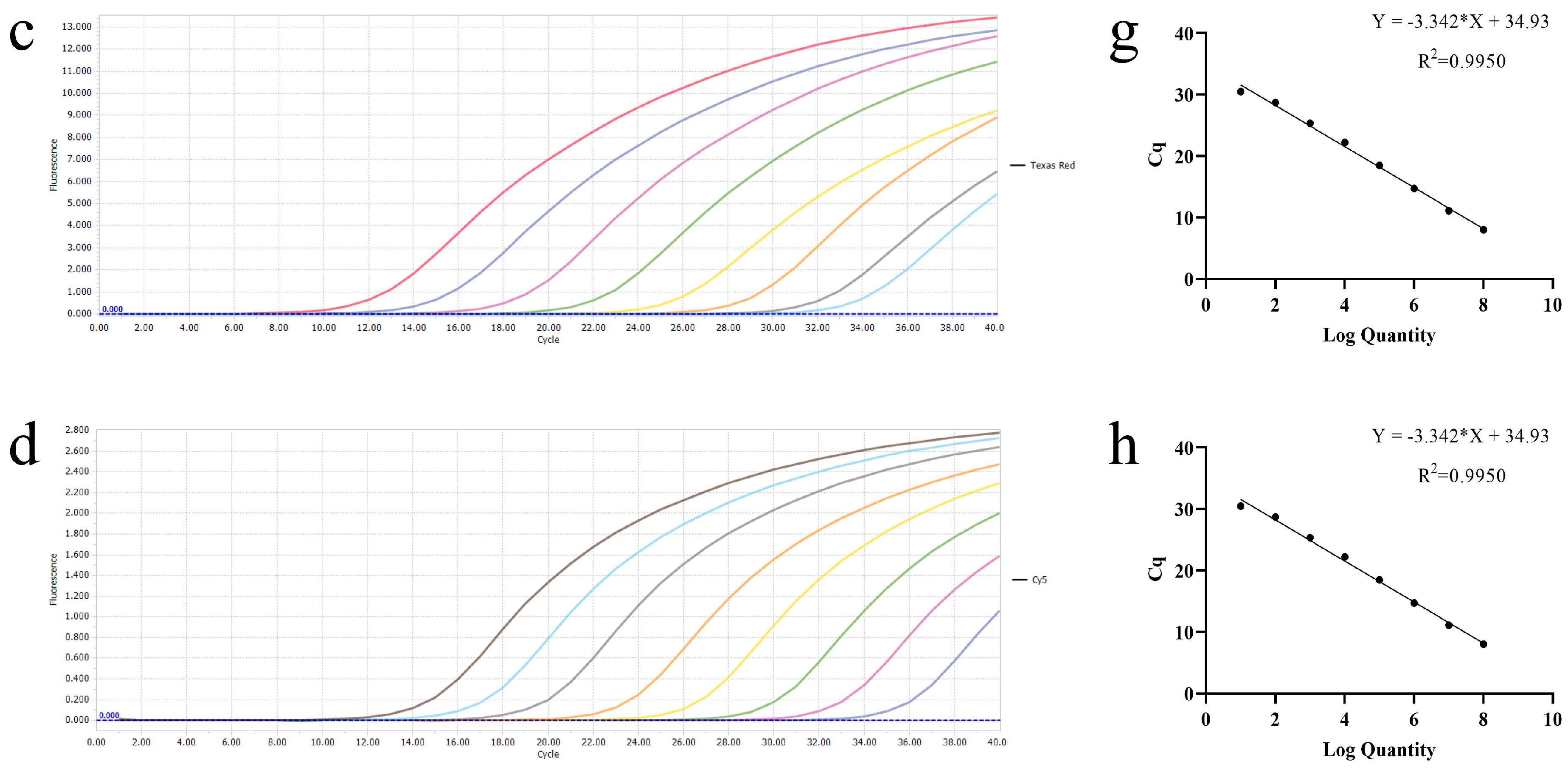
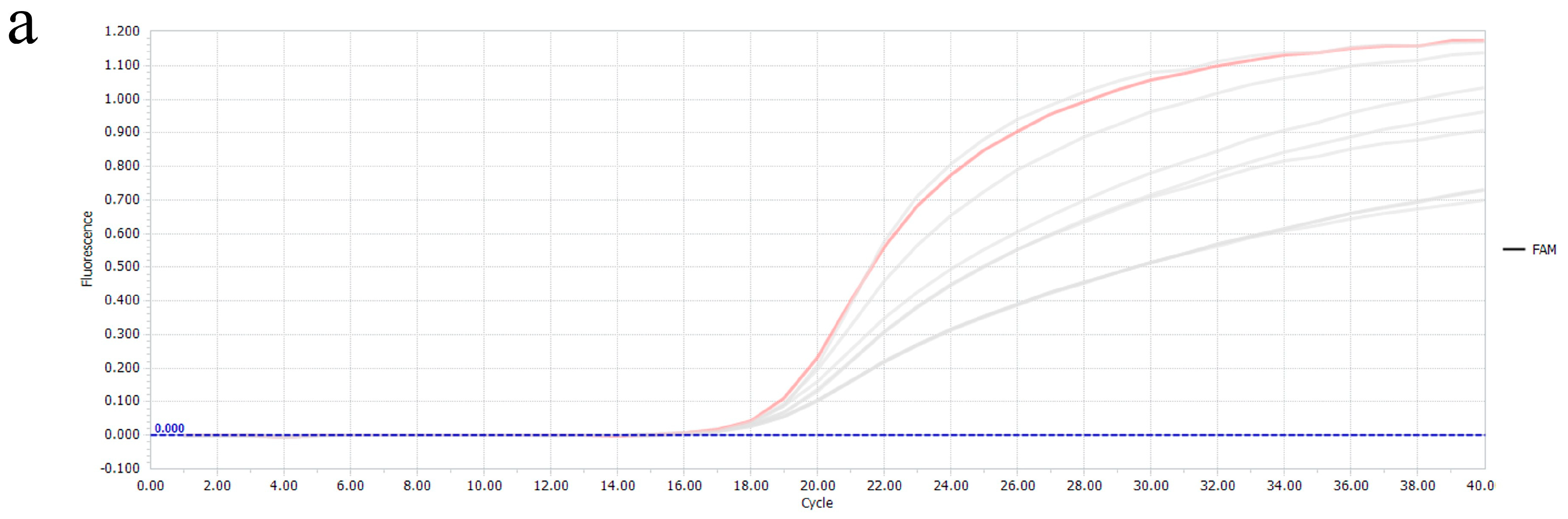
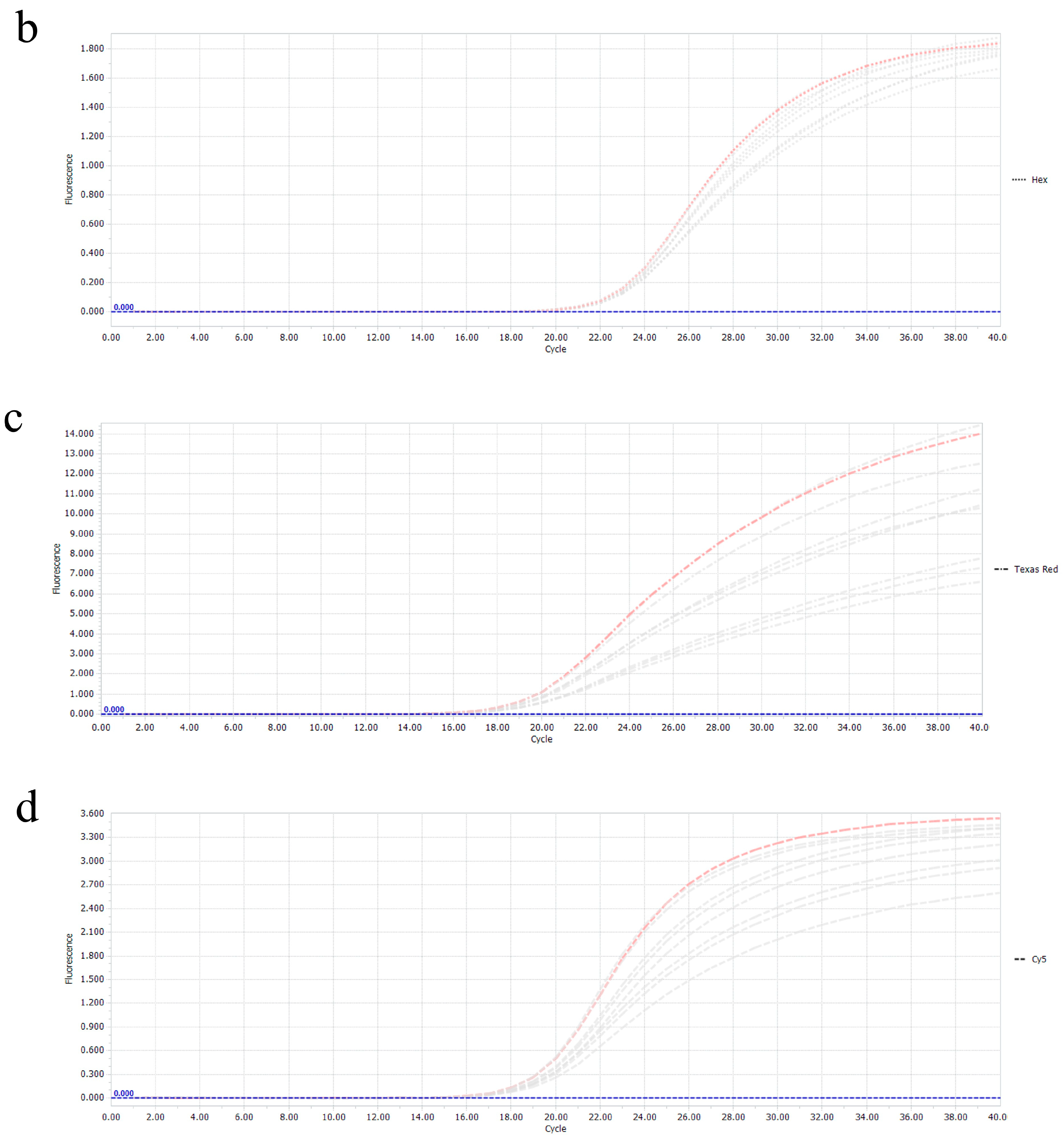
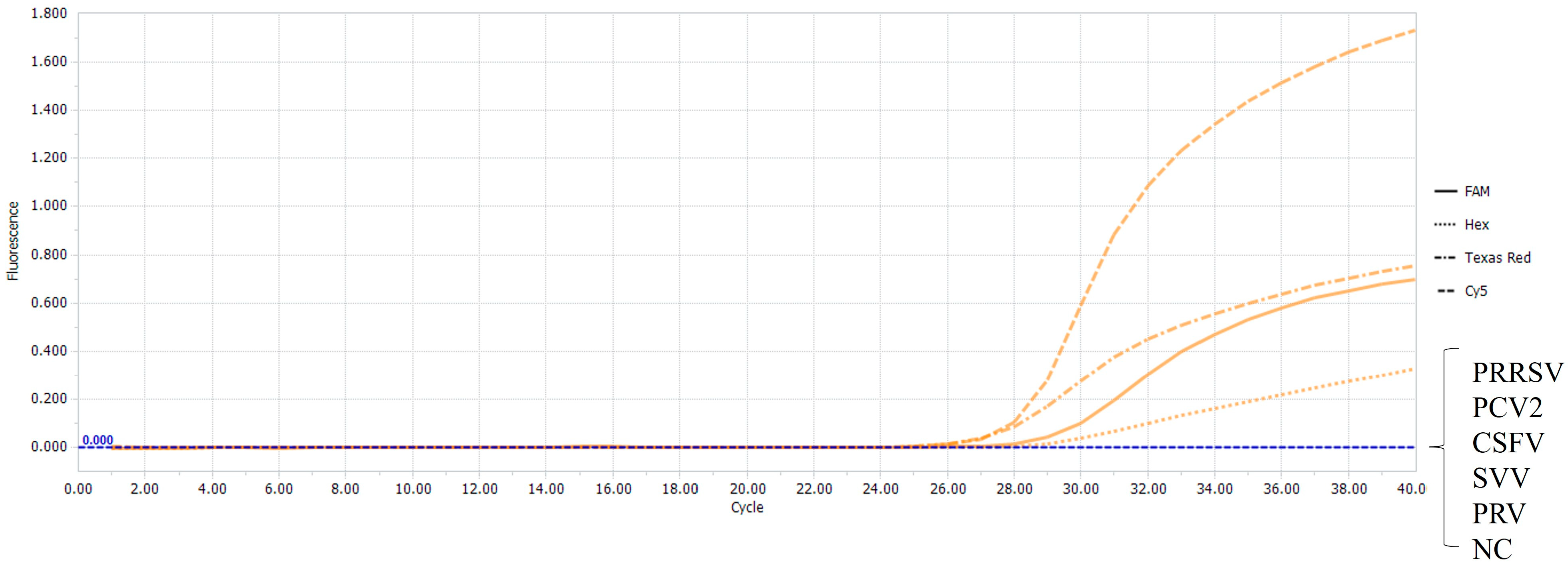
3.3. Specificity of the Multiplex Real-Time RT-PCR
3.4. Sensitivity of the Multiplex Real-Time PCR
3.5. Repeatability Test of the Multiplex Real-Time PCR
3.6. Detection of the Clinical Samples
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, J.; Liu, R.; Liu, H.; Chen, J.; Li, X.; Zhang, J.; Zhou, B. Development of a Multiplex Quantitative PCR for Detecting Porcine Epidemic Diarrhea Virus, Transmissible Gastroenteritis Virus, and Porcine Deltacoronavirus Simultaneously in China. Vet. Sci. 2023, 10, 402. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Shi, K.; Long, F.; Zhao, K.; Feng, S.; Yin, Y.; Xiong, C.; Qu, S.; Lu, W.; Li, Z. A Quadruplex qRT-PCR for Differential Detection of Four Porcine Enteric Coronaviruses. Vet. Sci. 2022, 9, 634. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef]
- Jang, G.; Lee, D.; Shin, S.; Lim, J.; Won, H.; Eo, Y.; Kim, C.-H.; Lee, C. Porcine epidemic diarrhea virus: An update overview of virus epidemiology, vaccines, and control strategies in South Korea. J. Vet. Sci. 2023, 24, e58. [Google Scholar] [CrossRef]
- Lee, C. Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus. Virol. J. 2015, 12, 193. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y.; Wang, X.; Zhou, J.; Ma, L.; Li, J.; Yang, L.; Ouyang, H.; Yuan, H.; Pang, D. Transmissible Gastroenteritis Virus: An Update Review and Perspective. Viruses 2023, 15, 359. [Google Scholar] [CrossRef] [PubMed]
- Niu, Z.; Zhang, S.; Xu, S.; Wang, J.; Wang, S.; Hu, X.; Zhang, L.; Ren, L.; Zhang, J.; Liu, X.; et al. Porcine Epidemic Diarrhea Virus Replication in Human Intestinal Cells Reveals Potential Susceptibility to Cross-Species Infection. Viruses 2023, 15, 956. [Google Scholar] [CrossRef] [PubMed]
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Elbadry, M.A.; Alam, M.; Stephenson, C.J.; Bonny, T.S.; Loeb, J.C.; Telisma, T.; Chavannes, S.; et al. Independent infections of porcine deltacoronavirus among Haitian children. Nature 2021, 600, 133–137. [Google Scholar] [CrossRef]
- Li, M.; Guo, L.; Feng, L. Interplay between swine enteric coronaviruses and host innate immune. Front. Vet. Sci. 2022, 9, 1083605. [Google Scholar] [CrossRef]
- Vlasova, A.N.; Amimo, J.O.; Saif, L.J. Porcine Rotaviruses: Epidemiology, Immune Responses and Control Strategies. Viruses 2017, 9, 48. [Google Scholar] [CrossRef]
- Raev, S.A.; Amimo, J.O.; Saif, L.J.; Vlasova, A.N. Intestinal mucin-type O-glycans: The major players in the host-bacteria-rotavirus interactions. Gut Microbes 2023, 15, 2197833. [Google Scholar] [CrossRef]
- Malik, Y.S.; Ansari, M.I.; Karikalan, M.; Sircar, S.; Selvaraj, I.; Ghosh, S.; Singh, K. Molecular Characterization of Rotavirus C from Rescued Sloth Bears, India: Evidence of Zooanthroponotic Transmission. Pathogens 2023, 12, 934. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.B.; Flores, P.S.; Amorim, A.R.; Mendes, G.d.S.; Santos, N. Porcine rotavirus C strains carrying human-like NSP4 and NSP5. Zoonoses Public Health 2020, 67, 849–861. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.-J.; Matthijnssens, J.; Kim, D.-S.; Kim, J.-Y.; Alfajaro, M.M.; Park, J.-G.; Hosmillo, M.; Son, K.-Y.; Soliman, M.; Baek, Y.-B.; et al. Genetic diversity of the VP7, VP4 and VP6 genes of Korean porcine group C rotaviruses. Vet. Microbiol. 2015, 176, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.S.; Jare, V.M.; Gopalkrishna, V. Group C rotavirus infection in patients with acute gastroenteritis in outbreaks in western India between 2006 and 2014. Epidemiol. Infect. 2017, 145, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, D.; Ghosh, S.; Kuzuya, M.; Wang, Y.-H.; Zhou, X.; Chawla-Sarkar, M.; Paul, S.K.; Ishino, M.; Kobayashi, N. Whole-genome characterization of human group C rotaviruses: Identification of two lineages in the VP3 gene. J. Gen. Virol. 2011, 92 Pt 2, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.-J.; Park, S.-I.; Hosmillo, M.; Shin, D.-J.; Chun, Y.-H.; Kim, H.-J.; Kwon, H.-J.; Kang, S.-Y.; Woo, S.-K.; Park, S.-J.; et al. Detection and molecular characterization of porcine group C rotaviruses in South Korea. Vet. Microbiol. 2009, 138, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Fan, M.; Zhu, Z.; Li, X. Establishment and Application of a Triplex Real-Time RT-PCR Assay for Differentiation of PEDV, PoRV, and PDCoV. Viruses 2023, 15, 1238. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Han, F.; Hu, Y.; Hao, C.; Li, Z.; Wei, Z.; Zhang, H. Co-infection of porcine deltacoronavirus and porcine epidemic diarrhoea virus alters gut microbiota diversity and composition in the colon of piglets. Virus Res. 2022, 322, 198954. [Google Scholar] [CrossRef]
- Mai, K.; Feng, J.; Chen, G.; Li, D.; Zhou, L.; Bai, Y.; Wu, Q.; Ma, J. The detection and phylogenetic analysis of porcine deltacoronavirus from Guangdong Province in Southern China. Transbound. Emerg. Dis. 2018, 65, 166–173. [Google Scholar] [CrossRef]
- Zhang, F.; Luo, S.; Gu, J.; Li, Z.; Li, K.; Yuan, W.; Ye, Y.; Li, H.; Ding, Z.; Song, D.; et al. Prevalence and phylogenetic analysis of porcine diarrhea associated viruses in southern China from 2012 to 2018. BMC Vet. Res. 2019, 15, 470. [Google Scholar] [CrossRef] [PubMed]
- Thakor, J.C.; Dinesh, M.; Manikandan, R.; Bindu, S.; Sahoo, M.; Sahoo, D.; Dhawan, M.; Pandey, M.K.; Tiwari, R.; Bin Emran, T.; et al. Swine coronaviruses (SCoVs) and their emerging threats to swine population, inter-species transmission, exploring the susceptibility of pigs for SARS-CoV-2 and zoonotic concerns. Vet. Q. 2022, 42, 125–147. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Lu, J.; Wang, N.; He, W.-T.; Zhang, L.; Zhao, W.; Su, S. Development of a Taq Man-probe-based multiplex real-time PCR for the simultaneous detection of emerging and reemerging swine coronaviruses. Virulence 2020, 11, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Chen, J.; Yao, G.; Guo, Q.; Wang, J.; Liu, G. A TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses. Appl. Microbiol. Biotechnol. 2019, 103, 4943–4952. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, G.; D’anza, E.; Rossi, A.; Improda, E.; Iovane, V.; Pagnini, U.; Iovane, G.; Montagnaro, S. A Serological Investigation of Porcine Reproductive and Respiratory Syndrome and Three Coronaviruses in the Campania Region, Southern Italy. Viruses 2023, 15, 300. [Google Scholar] [CrossRef] [PubMed]
- Okda, F.; Liu, X.; Singrey, A.; Clement, T.; Nelson, J.; Christopher-Hennings, J.; Nelson, E.A.; Lawson, S. Development of an indirect ELISA, blocking ELISA, fluorescent microsphere immunoassay and fluorescent focus neutralization assay for serologic evaluation of exposure to North American strains of Porcine Epidemic Diarrhea Virus. BMC Vet. Res. 2015, 11, 180. [Google Scholar] [CrossRef] [PubMed]
- Malbec, R.; Kimpston-Burkgren, K.; Vandenkoornhuyse, E.; Olivier, C.; Souplet, V.; Audebert, C.; Carrillo-Ávila, J.A.; Baum, D.; Giménez-Lirola, L. Agrodiag PorCoV: A multiplex immunoassay for the differential diagnosis of porcine enteric coronaviruses. J. Immunol. Methods 2020, 483, 112808. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, T.; Friendship, R.; Carman, S.; Pearl, D.L.; McEwen, B.; Dewey, C. Seroprevalence of bovine viral diarrhea virus neutralizing antibodies in finisher hogs in Ontario swine herds and targeted diagnostic testing of 2 suspect herds. Can. Vet. J. 2011, 52, 1342–1344. [Google Scholar]
- Gatto, I.R.H.; Linhares, D.C.L.; de Souza Almeida, H.M.; Mathias, L.A.; de Medeiros, A.S.R.; Poljak, Z.; Samara, S.I.; de Oliveira, L.G. Description of risk factors associated with the detection of BVDV antibodies in Brazilian pig herds. Trop. Anim. Health Prod. 2018, 50, 773–778. [Google Scholar] [CrossRef]
- Tao, J.; Liao, J.; Wang, Y.; Zhang, X.; Wang, J.; Zhu, G. Bovine viral diarrhea virus (BVDV) infections in pigs. Vet. Microbiol. 2013, 165, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lu, H.; Geng, C.; Yang, K.; Liu, W.; Liu, Z.; Yuan, F.; Gao, T.; Wang, S.; Wen, P.; et al. Epidemic and Evolutionary Characteristics of Swine Enteric Viruses in South-Central China from 2018 to 2021. Viruses 2022, 14, 1420. [Google Scholar] [CrossRef] [PubMed]
- Si, G.; Niu, J.; Zhou, X.; Xie, Y.; Chen, Z.; Li, G.; Chen, R.; He, D. Use of dual priming oligonucleotide system-based multiplex RT-PCR assay to detect five diarrhea viruses in pig herds in South China. AMB Express 2021, 11, 99. [Google Scholar] [CrossRef]
- Zhu, J.-H.; Rawal, G.; Aljets, E.; Yim-Im, W.; Yang, Y.-L.; Huang, Y.-W.; Krueger, K.; Gauger, P.; Main, R.; Zhang, J. Development and Clinical Applications of a 5-Plex Real-Time RT-PCR for Swine Enteric Coronaviruses. Viruses 2022, 14, 1536. [Google Scholar] [CrossRef]
- Saeng-Chuto, K.; Jermsutjarit, P.; Stott, C.J.; Vui, D.T.; Tantituvanont, A.; Nilubol, D. Retrospective study, full-length genome characterization and evaluation of viral infectivity and pathogenicity of chimeric porcine deltacoronavirus detected in Vietnam. Transbound. Emerg. Dis. 2020, 67, 183–198. [Google Scholar] [CrossRef] [PubMed]
| Pathogens | Primers and Probe | Sequences (5′ to 3′) | Size (bp) | Gene |
|---|---|---|---|---|
| PEDV | Forward | TACTCTGCGTTCTTGTATGG | 92 | M |
| Reverse | CTAGCCCATGCATCAAAAAG | |||
| Probe | TAMRA-AATAGCCATCTTGACACCATACA-FAM | |||
| TGEV | Forward | GTGACAAGATTTTATGGAAC | 157 | N |
| Reverse | CTTCCTTTGAAGTCCAATAG | |||
| Probe | HEX-CTAGACACAGATGGAACACATTCA-BHQ1 | |||
| PDCoV | Forward | ACTCCCTCCTAATGATAC | 87 | N |
| Reverse | GCAACATGAGGTTTAATAG | |||
| Probe | Texas Red-CAGCAACCACTCGTGTTACTT-BHQ2 | |||
| PoRV-A | Forward | CGAACACATGTACTATAAGA | 99 | VP7 |
| Reverse | CAGCTGTTATGTCAAGTATA | |||
| Probe | Cy5-TTGGACCTCCTACCTGAATTAC-BHQ3 |
| PEDV | TGEV | ||||||
| Probe concentration (nM) | Primer concentration (nM) | Probe concentration (nM) | Primer concentration (nM) | ||||
| 300 | 400 | 500 | 300 | 400 | 500 | ||
| 100 | 18.80 | 18.76 | 18.77 | 100 | 21.07 | 21.35 | 21.67 |
| 200 | 18.90 | 18.85 | 18.92 | 200 | 21.66 | 21.84 | 21.65 |
| 300 | 19.32 | 19.37 | 19.63 | 300 | 21.61 | 22.16 | 22.2 |
| PDCoV | PoRVA | ||||||
| Probe concentration (nM) | Primer concentration (nM) | Probe concentration (nM) | Primer concentration (nM) | ||||
| 300 | 400 | 500 | 300 | 400 | 500 | ||
| 100 | 15.84 | 15.33 | 15.6 | 100 | 16.85 | 16.9 | 16.79 |
| 200 | 16.02 | 15.92 | 16.18 | 200 | 17.24 | 17.05 | 16.99 |
| 300 | 16.29 | 17.00 | 16.46 | 300 | 17.48 | 17.27 | 17.19 |
| Pathogens | Concentration | Total | Positive | Detection Rate | 90% Detection Rate |
|---|---|---|---|---|---|
| PEDV | 1000 copies/μL | 15 | 15 | 100% | >90% |
| 100 copies/μL | 15 | 15 | 100% | >90% | |
| 10 copies/μL | 15 | 9 | 60% | <90% | |
| TGEV | 1000 copies/μL | 15 | 15 | 100% | >90% |
| 100 copies/μL | 15 | 15 | 100% | >90% | |
| 10 copies/μL | 15 | 0 | 0% | <90% | |
| PDCoV | 1000 copies/μL | 15 | 15 | 100% | >90% |
| 100 copies/μL | 15 | 15 | 100% | >90% | |
| 10 copies/μL | 15 | 3 | 20% | <90% | |
| PoRVA | 1000 copies/μL | 15 | 15 | 100% | >90% |
| 100 copies/μL | 15 | 15 | 100% | >90% | |
| 10 copies/μL | 15 | 6 | 40% | <90% |
| Plasmid | Dilutions | Intra-Assay | Inter-Assay | ||
|---|---|---|---|---|---|
| Mean ± SD | CV% | Mean ± SD | CV% | ||
| PoRVA | 107 | 14.92 ± 0.05 | 0.37 | 15.00 ± 0.09 | 0.60 |
| 106 | 17.35 ± 0.01 | 0.10 | 17.38 ± 0.04 | 0.25 | |
| 105 | 22.25 ± 0.13 | 0.59 | 20.27 ± 0.64 | 3.18 | |
| 104 | 24.63 ± 0.08 | 0.34 | 24.70 ± 0.07 | 0.28 | |
| 103 | 29.35 ± 0.34 | 1.17 | 29.31 ± 0.25 | 0.86 | |
| 102 | 32.51 ± 0.53 | 1.64 | 32.95 ± 0.35 | 1.08 | |
| NTC | ND | ND | ND | ND | |
| TGEV | 107 | 15.27 ± 0.04 | 0.29 | 15.32 ± 0.07 | 0.46 |
| 106 | 19.11 ± 0.08 | 0.40 | 19.04 ± 0.13 | 0.66 | |
| 105 | 21.11 ± 0.04 | 0.19 | 21.19 ± 0.05 | 0.23 | |
| 104 | 24.14 ± 0.03 | 0.13 | 24.16 ± 0.03 | 0.12 | |
| 103 | 27.58 ± 0.08 | 0.28 | 27.63 ± 0.06 | 0.21 | |
| 102 | 31.67 ± 0.15 | 0.47 | 31.55 ± 0.36 | 1.15 | |
| NTC | ND | ND | ND | ND | |
| PEDV | 107 | 15.03 ± 0.06 | 0.40 | 15.04 ± 0.14 | 0.91 |
| 106 | 18.36 ± 0.02 | 0.08 | 18.40 ± 0.02 | 0.11 | |
| 105 | 22.40 ± 0.11 | 0.50 | 23.34 ± 0.45 | 1.91 | |
| 104 | 26.57 ± 0.07 | 0.27 | 26.74 ± 0.25 | 0.92 | |
| 103 | 28.99 ± 0.25 | 0.87 | 28.88 ± 0.05 | 1.74 | |
| 102 | 32.42 ± 0.33 | 1.03 | 32.55 ± 0.46 | 1.37 | |
| NTC | ND | ND | ND | ND | |
| PDCoV | 107 | 11.82 ± 0.15 | 1.27 | 12.44 ± 0.32 | 2.54 |
| 106 | 15.68 ± 0.08 | 0.48 | 15.74 ± 0.13 | 0.85 | |
| 105 | 19.27 ± 0.08 | 0.42 | 19.13 ± 0.23 | 1.18 | |
| 104 | 22.54 ± 0.08 | 0.35 | 22.61 ± 0.26 | 1.14 | |
| 103 | 25.23 ± 0.12 | 0.46 | 25.44 ± 0.40 | 1.58 | |
| 102 | 28.02 ± 0.14 | 0.5 | 28.50 ± 0.47 | 1.65 | |
| NTC | ND | ND | ND | ND | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xin, Z.; Li, S.; Lu, X.; Liu, L.; Gao, Y.; Hu, F.; Yu, K.; Ma, X.; Li, Y.; Huang, B.; et al. Development and Clinical Application of a Molecular Assay for Four Common Porcine Enteroviruses. Vet. Sci. 2024, 11, 305. https://doi.org/10.3390/vetsci11070305
Xin Z, Li S, Lu X, Liu L, Gao Y, Hu F, Yu K, Ma X, Li Y, Huang B, et al. Development and Clinical Application of a Molecular Assay for Four Common Porcine Enteroviruses. Veterinary Sciences. 2024; 11(7):305. https://doi.org/10.3390/vetsci11070305
Chicago/Turabian StyleXin, Zhonghao, Shiheng Li, Xiao Lu, Liping Liu, Yuehua Gao, Feng Hu, Kexiang Yu, Xiuli Ma, Yufeng Li, Bing Huang, and et al. 2024. "Development and Clinical Application of a Molecular Assay for Four Common Porcine Enteroviruses" Veterinary Sciences 11, no. 7: 305. https://doi.org/10.3390/vetsci11070305
APA StyleXin, Z., Li, S., Lu, X., Liu, L., Gao, Y., Hu, F., Yu, K., Ma, X., Li, Y., Huang, B., Wu, J., & Guo, X. (2024). Development and Clinical Application of a Molecular Assay for Four Common Porcine Enteroviruses. Veterinary Sciences, 11(7), 305. https://doi.org/10.3390/vetsci11070305






