Investigation of the Seroprevalence of Brucella Antibodies and Characterization of Field Strains in Immunized Dairy Cows by B. abortus A19
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Serological Tests
2.3. The Seroprevalence Investigation of Brucella Antibody in Vaccinated Cattle in Lingwu City
2.4. Investigation of the Positive Rate of Brucella Antibodies in Dairy Herds of Three Brucellosis-Free Dairy Farms
2.5. Identification of the DNA Sequence That Distinguishes Brucella A19 from Other Strains
2.6. PCR-Based Differential Diagnostic Approach
2.7. Processing of Swab Samples
2.8. Investigation of Brucella Field Strains among Cattle in 10 Dairy Farms
2.9. PCR Detection of Brucellosis Utilizing the Mathematical Expectation Method
3. Results
3.1. Seroprevalence of Brucella Antibody in Vaccinated Cattle
3.2. Seroprevalence of Brucella Antibodies in Three Brucellosis-Free Dairy Farms
3.3. Distinctive Differential Sequences of Brucella abortus Strain A19
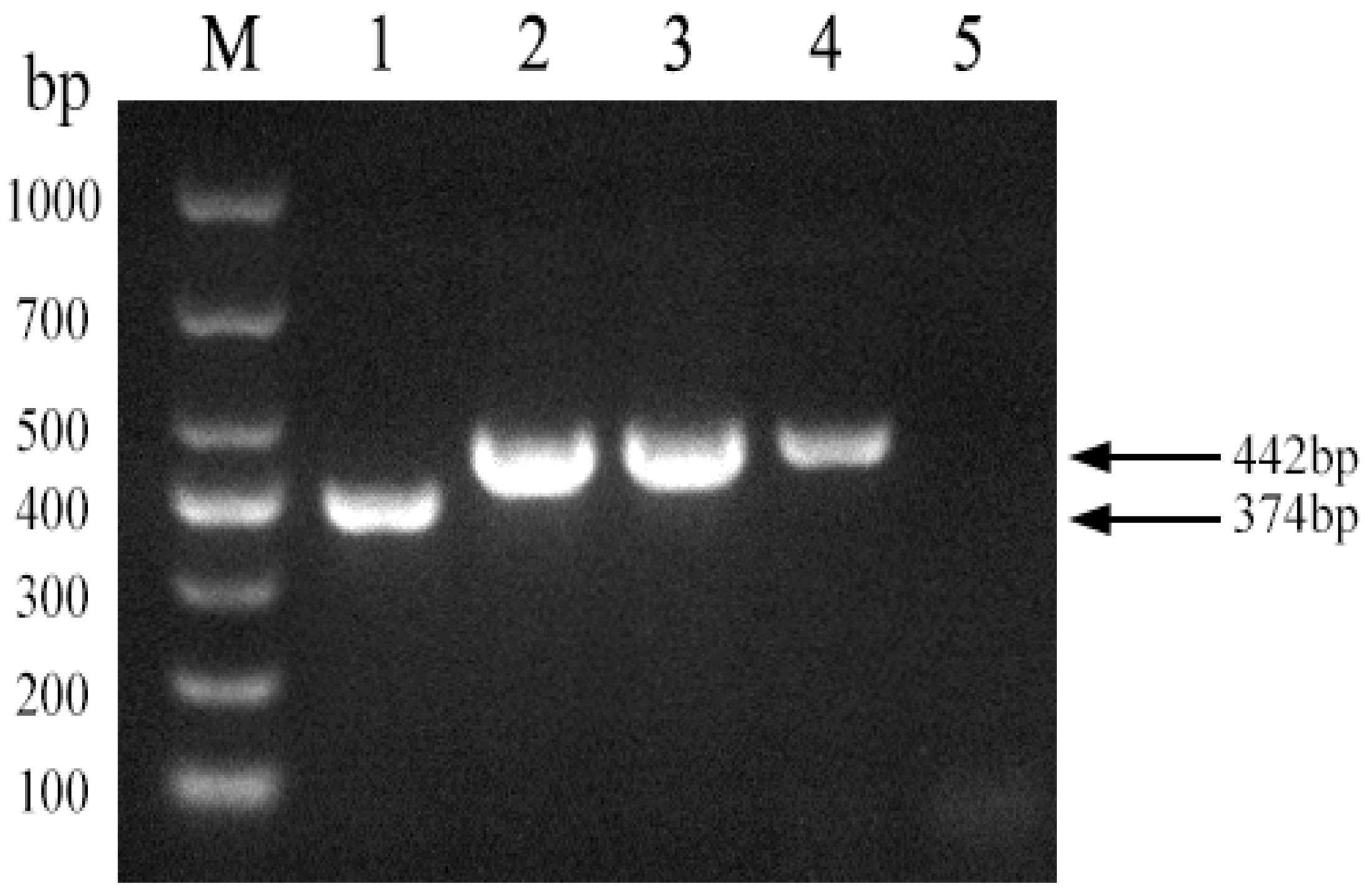
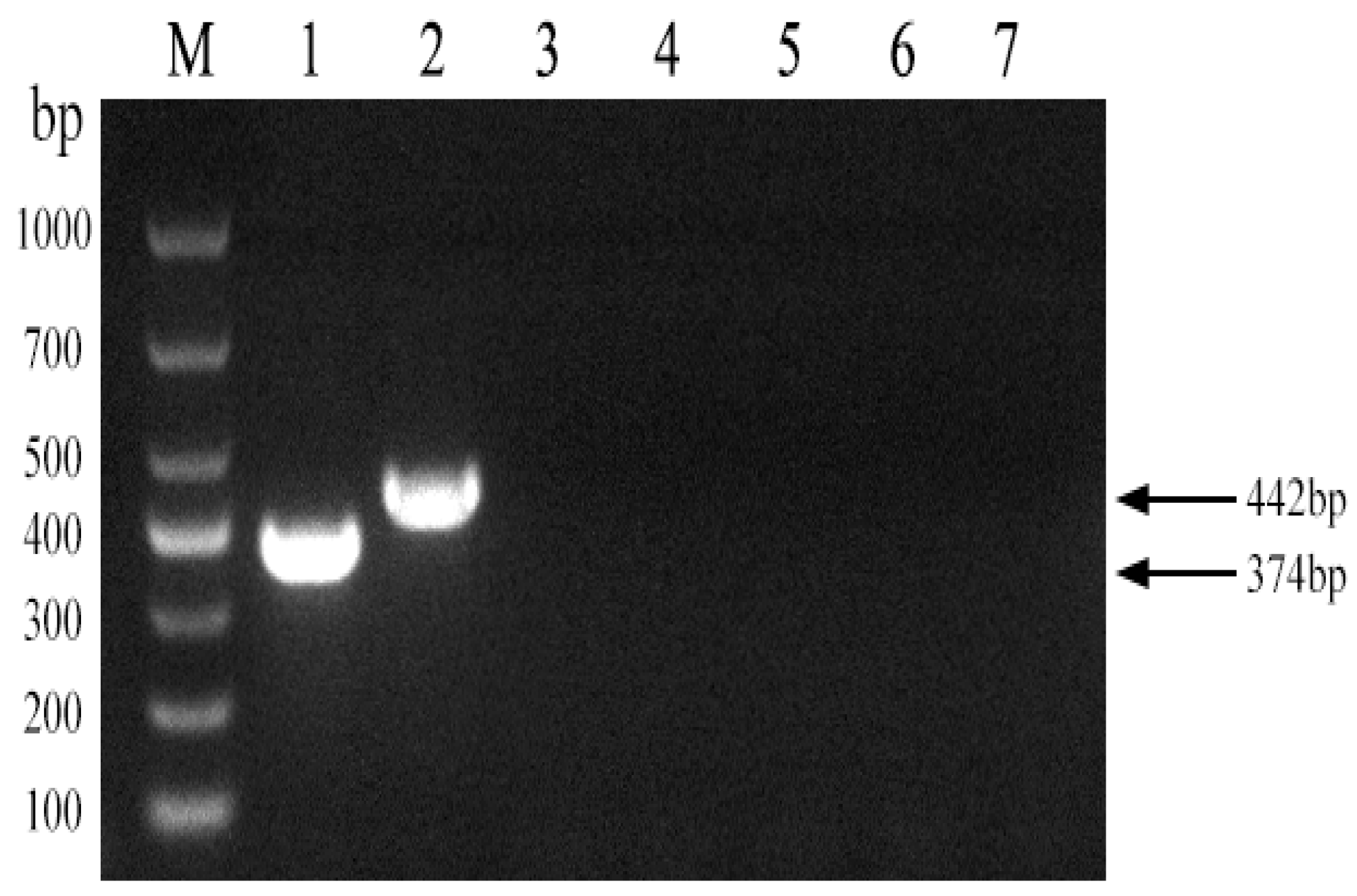
3.4. PCR-Based Identification of Brucella abortus Strain A19
3.5. PCR Assay Specificity and Sensitivity
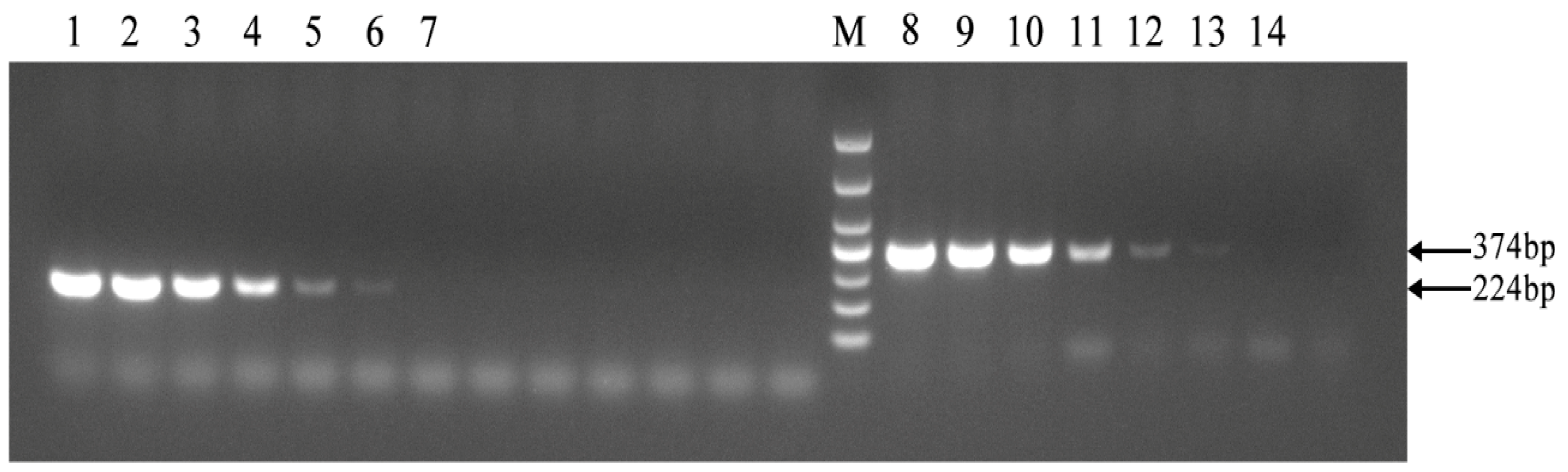
3.6. PCR Detection of Brucellosis Utilizing Mathematical Expectation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moreno, E. Retrospective and prospective perspectives on zoonotic brucellosis. Front. Microbiol. 2014, 5, 213. [Google Scholar] [CrossRef] [PubMed]
- de Macedo, G.C.; Herrera, H.M.; de Oliveira Porfirio, G.E.; Santos, F.M.; de Assis, W.O.; de Andrade, G.B.; Nantes, W.A.G.; de Mendoza, J.H.; Fernandez-Llario, P.; de Oliveira, C.E. Brucellosis in the Brazilian Pantanal wetland: Threat to animal production and wildlife conservation. Braz. J. Microbiol. 2022, 53, 2287–2297. [Google Scholar] [CrossRef] [PubMed]
- Godfroid, J. Brucellosis in livestock and wildlife: Zoonotic diseases without pandemic potential in need of innovative one health approaches. Arch. Public Health 2017, 75, 34. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.T.; Wang, C.L.; Liu, L.N.; Wang, D.; Li, D.; Li, Z.J.; Liu, Z.G. Epidemiologically characteristics of human brucellosis and antimicrobial susceptibility pattern of Brucella melitensis in Hinggan League of the Inner Mongolia Autonomous Region, China. Infect. Dis. Poverty 2020, 9, 79. [Google Scholar] [CrossRef] [PubMed]
- Dean, A.S.; Crump, L.; Greter, H.; Schelling, E.; Zinsstag, J. Global burden of human brucellosis: A systematic review of disease frequency. PLoS Negl. Trop. Dis. 2012, 6, e1865. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.B.; Khatkar, M.S.; Aulakh, R.S.; Gill, J.P.S.; Dhand, N.K. Estimation of the health and economic burden of human brucellosis in India. Prev. Vet. Med. 2018, 154, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Pappas, G.; Papadimitriou, P.; Akritidis, N.; Christou, L.; Tsianos, E.V. The new global map of human brucellosis. Lancet Infect. Dis. 2006, 6, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Seleem, M.N.; Boyle, S.M.; Sriranganathan, N. Brucellosis: A re-emerging zoonosis. Vet. Microbiol. 2010, 140, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Di Bari, C.; Venkateswaran, N.; Bruce, M.; Fastl, C.; Huntington, B.; Patterson, G.T.; Rushton, J.; Torgerson, P.; Pigott, D.M.; Devleesschauwer, B. Methodological choices in brucellosis burden of disease assessments: A systematic review. PLoS Negl. Trop. Dis. 2022, 16, e0010468. [Google Scholar]
- Zhang, N.; Huang, D.; Wu, W.; Liu, J.; Liang, F.; Zhou, B.; Guan, P. Animal brucellosis control or eradication programs worldwide: A systematic review of experiences and lessons learned. Prev. Vet. Med. 2018, 160, 105–115. [Google Scholar] [CrossRef]
- Barbier, T.; Collard, F.; Zuniga-Ripa, A.; Moriyon, I.; Godard, T.; Becker, J.; Wittmann, C.; Van Schaftingen, E.; Letesson, J.J. Erythritol feeds the pentose phosphate pathway via three new isomerases leading to D-erythrose-4-phosphate in Brucella. Proc. Natl. Acad. Sci. USA 2014, 111, 17815–17820. [Google Scholar] [CrossRef] [PubMed]
- Bowman, D.D. Introduction to the alpha-proteobacteria: Wolbachia and Bartonella, Rickettsia, Brucella, Ehrlichia, and Anaplasma. Top. Companion Anim. Med. 2011, 26, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Vergnaud, G.; Hauck, Y.; Christiany, D.; Daoud, B.; Pourcel, C.; Jacques, I.; Cloeckaert, A.; Zygmunt, M.S. Genotypic Expansion Within the Population Structure of Classical Brucella Species Revealed by MLVA16 Typing of 1404 Brucella Isolates From Different Animal and Geographic Origins, 1974–2006. Front. Microbiol. 2018, 9, 1545. [Google Scholar] [CrossRef] [PubMed]
- Tuon, F.F.; Gondolfo, R.B.; Cerchiari, N. Human-to-human transmission of Brucella—A systematic review. Trop. Med. Int. Health 2017, 22, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Kamal, I.H.; Al Gashgari, B.; Moselhy, S.S.; Kumosani, T.A.; Abulnaja, K.O. Two-stage PCR assay for detection of human brucellosis in endemic areas. BMC Infect. Dis. 2013, 13, 145. [Google Scholar] [CrossRef] [PubMed]
- Casalinuovo, F.; Ciambrone, L.; Cacia, A.; Rippa, P. Contamination of Bovine, Sheep and Goat Meat with Brucella spp. Ital. J. Food Saf. 2016, 5, 5913. [Google Scholar] [CrossRef]
- Ye, H.Y.; Xing, F.F.; Yang, J.; Lo, S.K.; Lau, R.W.; Chen, J.H.; Chiu, K.H.; Yuen, K.Y. High index of suspicion for brucellosis in a highly cosmopolitan city in southern China. BMC Infect. Dis. 2020, 20, 22. [Google Scholar] [CrossRef]
- Ashby, B.; Best, A. Herd immunity. Curr. Biol. 2021, 31, R174–R177. [Google Scholar] [CrossRef]
- Fine, P.; Eames, K.; Heymann, D.L. “Herd immunity”: A rough guide. Clin. Infect. Dis. 2011, 52, 911–916. [Google Scholar] [CrossRef]
- Rubach, M.P.; Halliday, J.E.; Cleaveland, S.; Crump, J.A. Brucellosis in low-income and middle-income countries. Curr. Opin. Infect. Dis. 2013, 26, 404–412. [Google Scholar] [CrossRef]
- Schuchat, A.; Bell, B.P. Monitoring the impact of vaccines postlicensure: New challenges, new opportunities. Expert Rev. Vaccines 2008, 7, 437–456. [Google Scholar] [CrossRef]
- Moriyon, I.; Grillo, M.J.; Monreal, D.; Gonzalez, D.; Marin, C.; Lopez-Goni, I.; Mainar-Jaime, R.C.; Moreno, E.; Blasco, J.M. Rough vaccines in animal brucellosis: Structural and genetic basis and present status. Vet. Res. 2004, 35, 1–38. [Google Scholar] [CrossRef]
- Avila-Calderon, E.D.; Lopez-Merino, A.; Sriranganathan, N.; Boyle, S.M.; Contreras-Rodriguez, A. A history of the development of Brucella vaccines. Biomed. Res. Int. 2013, 2013, 743509. [Google Scholar] [CrossRef]
- Zamri-Saad, M.; Kamarudin, M.I. Control of animal brucellosis: The Malaysian experience. Asian Pac. J. Trop. Med. 2016, 9, 1136–1140. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Liu, X.; Peng, Q. The advances in brucellosis vaccines. Vaccine 2019, 37, 3981–3988. [Google Scholar] [CrossRef]
- Jiang, H.; Dong, H.; Peng, X.; Feng, Y.; Zhu, L.; Niu, K.; Peng, Y.; Fan, H.; Ding, J. Transcriptome analysis of gene expression profiling of infected macrophages between Brucella suis 1330 and live attenuated vaccine strain S2 displays mechanistic implication for regulation of virulence. Microb. Pathog. 2018, 119, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Qiao, Z.; Hu, S.; Liu, W.; Zheng, H.; Liu, S.; Zhao, X.; Bu, Z. Comparison of genomes of Brucella melitensis M28 and the B. melitensis M5-90 derivative vaccine strain highlights the translation elongation factor Tu gene tuf2 as an attenuation-related gene. Infect. Immun. 2013, 81, 2812–2818. [Google Scholar] [CrossRef] [PubMed]
- He, C.Y.; Zhang, Y.Z.; Liu, M.Z.; Zhao, H.L.; Ren, L.S.; Liu, B.S.; He, S.; Chen, Z.L. Combined immunization with inactivated vaccine reduces the dose of live B. abortus A19 vaccine. BMC Vet. Res. 2022, 18, 128. [Google Scholar] [CrossRef]
- Wang, S.; Wang, W.; Sun, K.; Bateer, H.; Zhao, X. Comparative genomic analysis between newly sequenced Brucella abortus vaccine strain A19 and another Brucella abortus vaccine S19. Genomics 2020, 112, 1444–1453. [Google Scholar] [CrossRef]
- Olsen, S.C.; Stoffregen, W.S. Essential role of vaccines in brucellosis control and eradication programs for livestock. Expert Rev. Vaccines 2005, 4, 915–928. [Google Scholar] [CrossRef]
- Perkins, S.D.; Smither, S.J.; Atkins, H.S. Towards a Brucella vaccine for humans. FEMS Microbiol. Rev. 2010, 34, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Manthei, C.A. Brucellosis. Application of research to bovine brucellosis control and eradication programs. J. Dairy Sci. 1968, 51, 1115–1120. [Google Scholar] [CrossRef] [PubMed]
- Ragan, V.E. The Animal and Plant Health Inspection Service (APHIS) brucellosis eradication program in the United States. Vet. Microbiol. 2002, 90, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Ran, X.; Cheng, J.; Wang, M.; Chen, X.; Wang, H.; Ge, Y.; Ni, H.; Zhang, X.X.; Wen, X. Brucellosis seroprevalence in dairy cattle in China during 2008–2018: A systematic review and meta-analysis. Acta Trop. 2019, 189, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Mamani, M.; Majzoobi, M.M.; Keramat, F.; Varmaghani, N.; Moghimbeigi, A. Seroprevalence of Brucellosis in Butchers, Veterinarians and Slaughterhouse Workers in Hamadan, Western Iran. J. Res. Health Sci. 2018, 18, e00406. [Google Scholar] [PubMed]
- Liu, Z.G.; Wang, M.; Ta, N.; Fang, M.G.; Mi, J.C.; Yu, R.P.; Luo, Y.; Cao, X.; Li, Z.J. Seroprevalence of human brucellosis and molecular characteristics of Brucella strains in Inner Mongolia Autonomous region of China, from 2012 to 2016. Emerg. Microbes Infect. 2020, 9, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Omer, M.K.; Skjerve, E.; Woldehiwet, Z.; Holstad, G. Risk factors for Brucella spp. infection In dairy cattle farms in Asmara, State of Eritrea. Prev. Vet. Med. 2000, 46, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Diao, Z. Serological Investigation of Brucella in a Largescale Dairy Farm and Establishment of Quadruple PCR Detection Method; Northwest A&F University: Xianyang, China, 2023. [Google Scholar]
- Dadar, M.; Tiwari, R.; Sharun, K.; Dhama, K. Importance of brucellosis control programs of livestock on the improvement of one health. Vet. Q. 2021, 41, 137–151. [Google Scholar] [CrossRef]
- Liu, B.; Ye, Y.; Zhai, J.; Zhang, Y.; Yang, J.; Cheng, D.; Ma, C.; Yu, D.; Yang, B.; Zhu, R.; et al. Combined nucleic acid assays for diagnosis of A19 vaccine-caused human brucellosis. Transbound. Emerg. Dis. 2021, 68, 368–374. [Google Scholar]
- Wu, X.; Qi, R.; Yan, F.; Ma, T.; Duan, H.; Gao, J. Analysis of epidemic characteristics and prediction of incidence trend of human brucellosis in Ningxia from 2012 to 2021. Ningxia Med. J. 2023, 45, 107–109. [Google Scholar]
- Yang, Q.; Zhang, S.; Liu, L.; Cao, X.; Lei, C.; Qi, X.; Lin, F.; Qu, W.; Qi, X.; Liu, J.; et al. Application of mathematical expectation (ME) strategy for detecting low frequency mutations: An example for evaluating 14-bp insertion/deletion (indel) within the bovine PRNP gene. Prion 2016, 10, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhu, X.; Ma, L.; Xu, H.; Cao, X.; Luo, R.; Chen, H.; Sun, X.; Cai, Y.; Lan, X. Detection of a new 20-bp insertion/deletion (indel) within sheep PRND gene using mathematical expectation (ME) method. Prion 2017, 11, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Arif, S.; Heller, J.; Hernandez-Jover, M.; McGill, D.M.; Thomson, P.C. Evaluation of three serological tests for diagnosis of bovine brucellosis in smallholder farms in Pakistan by estimating sensitivity and specificity using Bayesian latent class analysis. Prev. Vet. Med. 2018, 149, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Alamian, S.; Amiry, K.; Bahreinipour, A.; Etemadi, A.; Yousefi, A.R.; Dadar, M. Evaluation of serological diagnostic tests for bovine brucellosis in dairy cattle herds in an endemic area: A multicenter study. Trop. Anim. Health Prod. 2023, 55, 104. [Google Scholar] [CrossRef] [PubMed]

| Year | Number | Number of Positives | Positive Rate |
|---|---|---|---|
| 2021 | 1726 | 404 | 23.4% |
| 2022 | 2146 | 574 | 26.7% |
| 2023 | 1563 | 469 | 30.0% |
| Year | Number | Number of Positives | Positive Rate |
|---|---|---|---|
| 2021 | 234 | 77 | 32.9% |
| 2022 | 265 | 96 | 36.2% |
| 2023 | 123 | 68 | 55.3% |
| Farm | Number | Serological Testing | |
|---|---|---|---|
| Positive Number | Positive Rate | ||
| A | 253 | 67 | 26.5% |
| B | 150 | 27 | 18.0% |
| C | 130 | 7 | 5.4% |
| Farm | Number | Serological Testing | PCR Testing | ||
|---|---|---|---|---|---|
| Positive Number | Positive Rate | Positive Number | Positive Rate | ||
| 1 | 185 | 32 | 17.3% | 0 | 0.0% |
| 2 | 153 | 23 | 15.0% | 0 | 0.0% |
| 3 | 143 | 15 | 10.5% | 0 | 0.0% |
| 4 | 106 | 16 | 15.1% | 0 | 0.0% |
| 5 | 69 | 23 | 33.3% | 2 | 2.9% |
| 6 | 97 | 40 | 41.2% | 2 | 2.1% |
| 7 | 75 | 27 | 36.0% | 1 | 1.3% |
| 8 | 40 | 12 | 30.0% | 0 | 0.0% |
| 9 | 56 | 23 | 41.1% | 2 | 3.6% |
| 10 | 80 | 31 | 38.8% | 3 | 3.8% |
| Breeds | Data |
|---|---|
| Sizes | 1537 |
| Assumed prevalence | 3.0% |
| Number of individuals in one reaction time (NR1) | 1 |
| Reaction times (RT1) | 1537 |
| Number of individuals in one mixed group (NG6) | 6 |
| Reaction times (RT6) | 306 |
| Reduction rate (RR) | 80.1% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Y.; Cui, Y.; Wudong, G.; Li, S.; Yuan, Y.; Zhao, D.; Yin, S.; Diao, Z.; Li, B.; Zhou, D.; et al. Investigation of the Seroprevalence of Brucella Antibodies and Characterization of Field Strains in Immunized Dairy Cows by B. abortus A19. Vet. Sci. 2024, 11, 288. https://doi.org/10.3390/vetsci11070288
Shi Y, Cui Y, Wudong G, Li S, Yuan Y, Zhao D, Yin S, Diao Z, Li B, Zhou D, et al. Investigation of the Seroprevalence of Brucella Antibodies and Characterization of Field Strains in Immunized Dairy Cows by B. abortus A19. Veterinary Sciences. 2024; 11(7):288. https://doi.org/10.3390/vetsci11070288
Chicago/Turabian StyleShi, Yong, Yimeng Cui, Gaowa Wudong, Shengnan Li, Ye Yuan, Danyu Zhao, Shurong Yin, Ziyang Diao, Bin Li, Dong Zhou, and et al. 2024. "Investigation of the Seroprevalence of Brucella Antibodies and Characterization of Field Strains in Immunized Dairy Cows by B. abortus A19" Veterinary Sciences 11, no. 7: 288. https://doi.org/10.3390/vetsci11070288
APA StyleShi, Y., Cui, Y., Wudong, G., Li, S., Yuan, Y., Zhao, D., Yin, S., Diao, Z., Li, B., Zhou, D., Li, X., Wang, Z., Zhang, F., Xie, M., Zhao, Z., Wang, A., & Jin, Y. (2024). Investigation of the Seroprevalence of Brucella Antibodies and Characterization of Field Strains in Immunized Dairy Cows by B. abortus A19. Veterinary Sciences, 11(7), 288. https://doi.org/10.3390/vetsci11070288




