Characterization of the Pro-Inflammatory and Pruritogenic Transcriptome in Skin Lesions of the Experimental Canine Atopic Acute IgE-Mediated Late Phase Reactions Model and Correlation to Acute Skin Lesions of Human Atopic Dermatitis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Inclusion
2.2. Intradermal Injections and Skin Biopsy Collection of Late Phase Reactions (LPRs)
2.3. Histopathology
2.4. RNA-Sequencing Analysis
2.5. Quantitative Reverse-Transcription PCR Analysis
2.6. Correlation Analysis to Previously Published Transcriptome Data Using RNA Sequencing for Acute Skin Lesions in Human and Canine Atopic Dermatitis
2.7. Statistical and Bioinformatics Analyses
3. Results
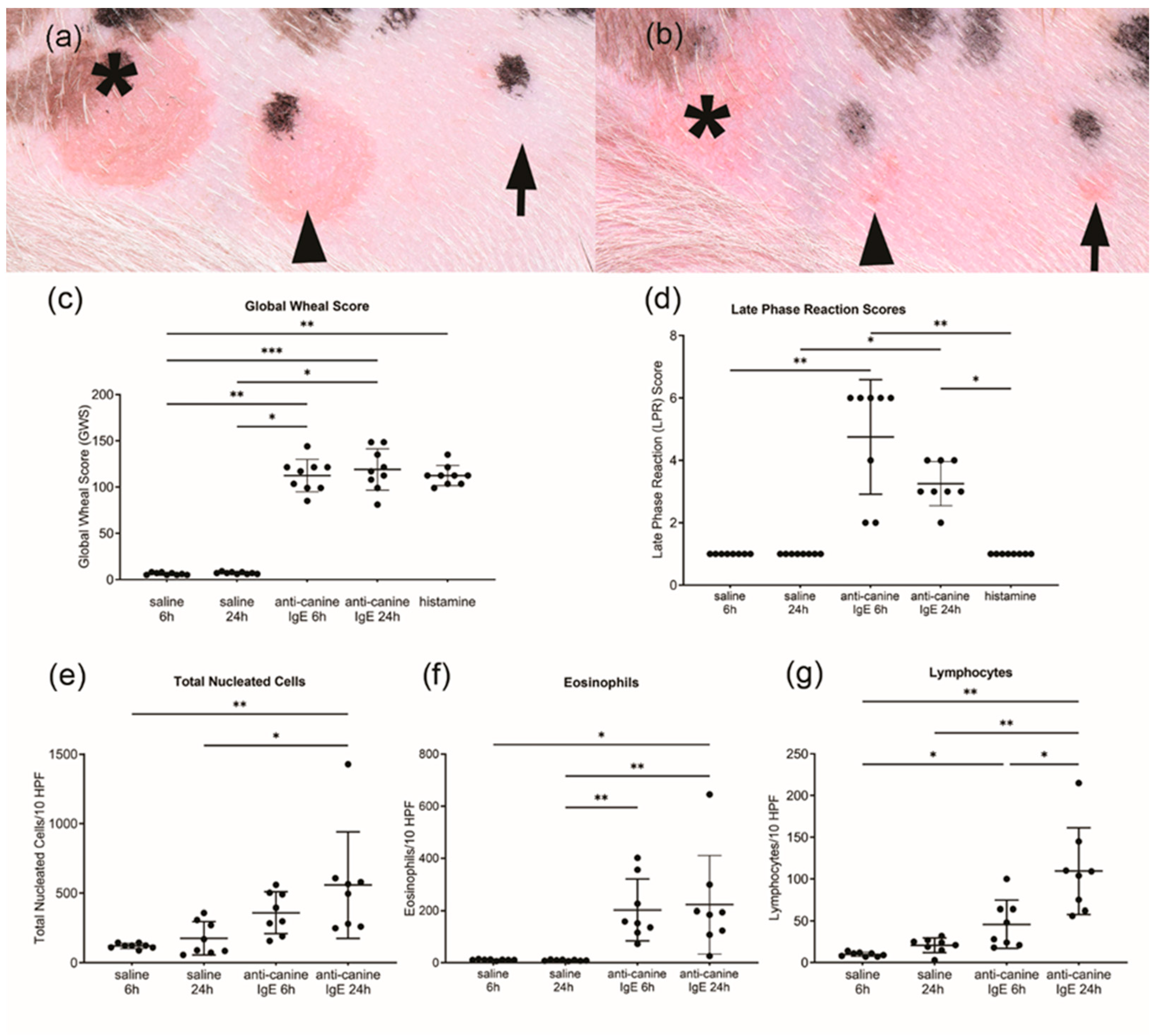
3.1. Global Wheal Score, Late Phase Reaction Scores and Histopathological Examination
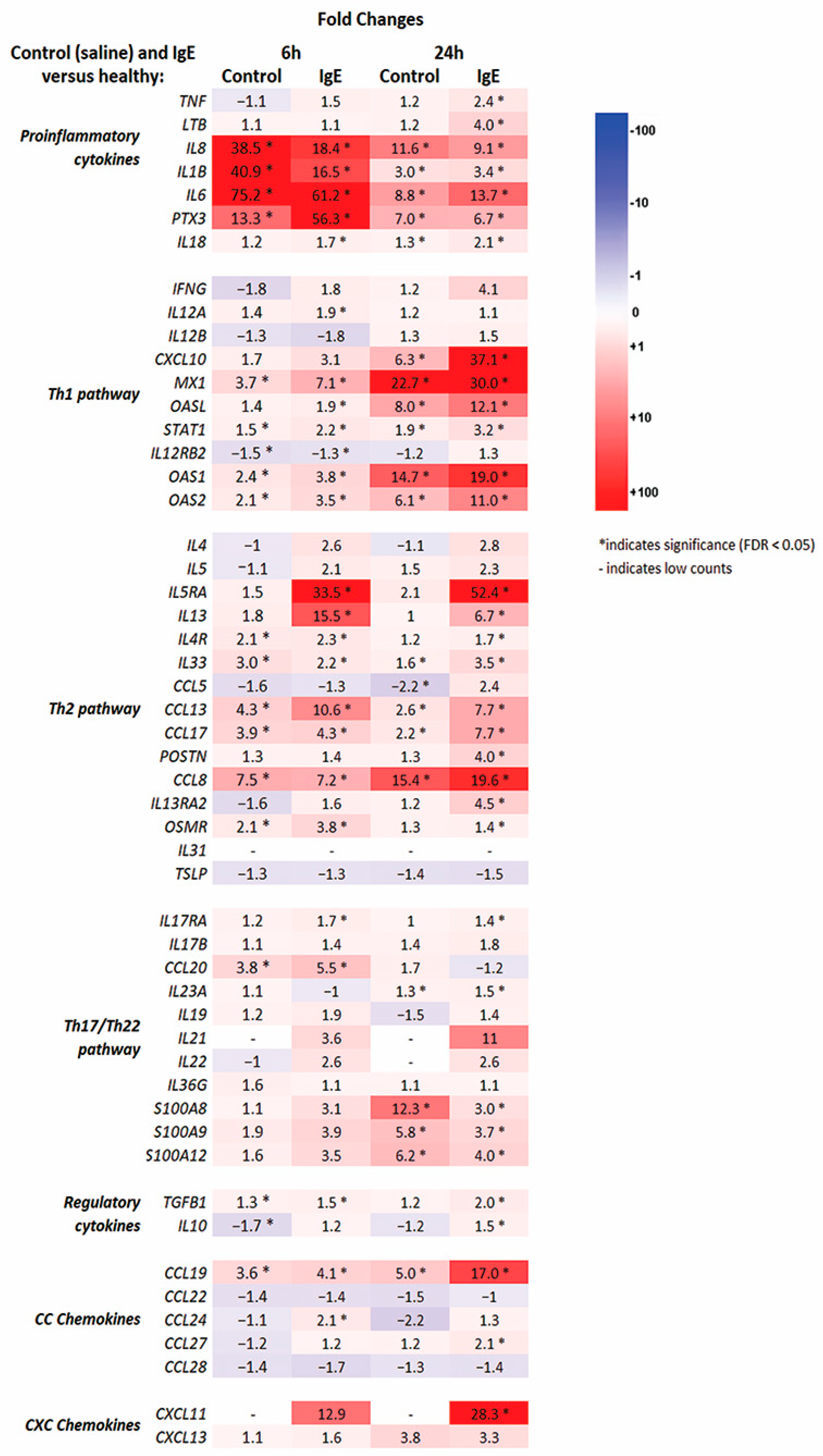
3.2. RNA-seq Molecular Profiling of Anti-Canine-IgE- and Saline-Mediated Late Phase Reactions (LPRs)
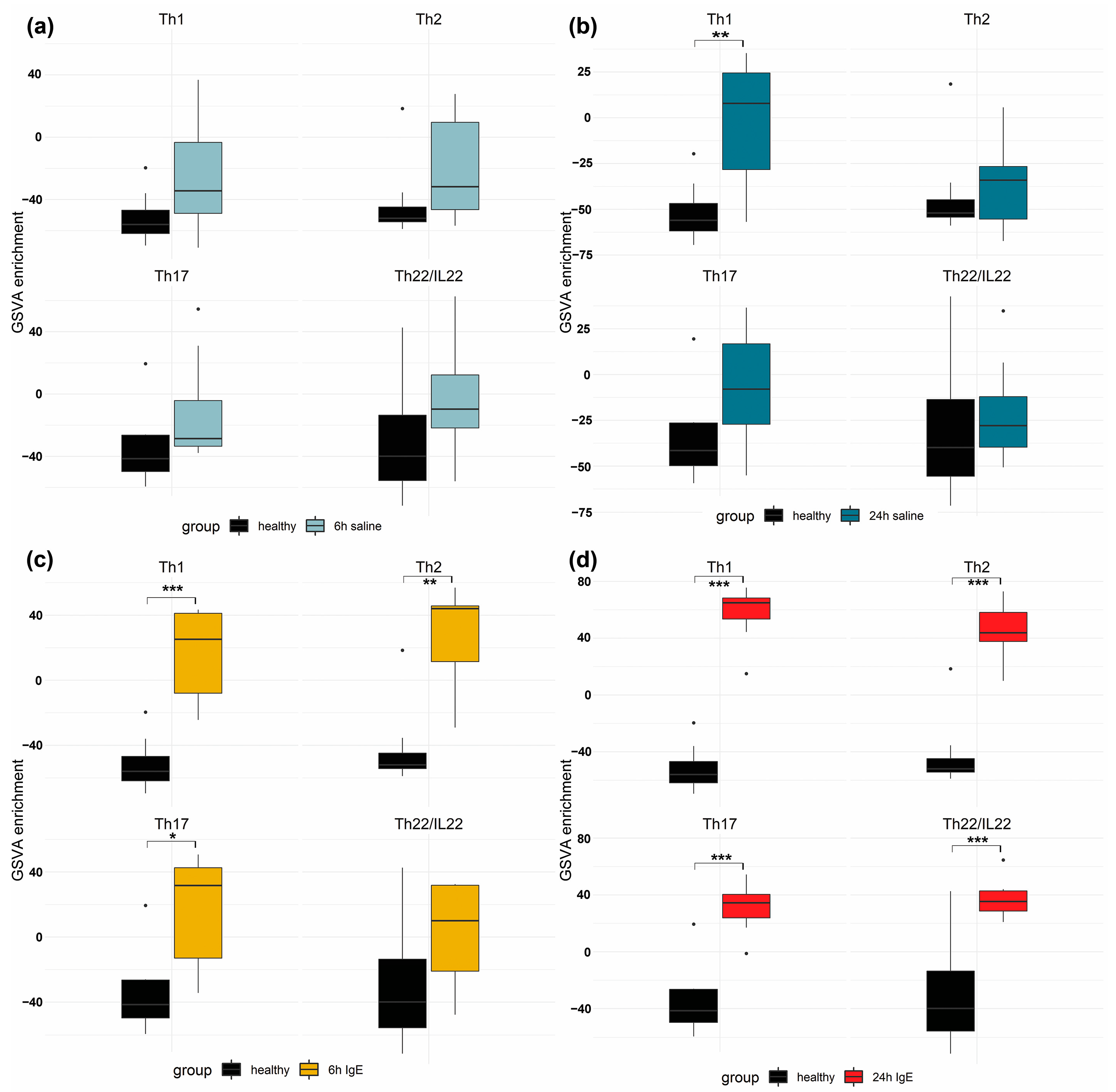
3.3. Pathway and Enrichment Analysis of of Anti-Canine-IgE- and Saline-Mediated Late Phase Reactions (LPRs)
3.4. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
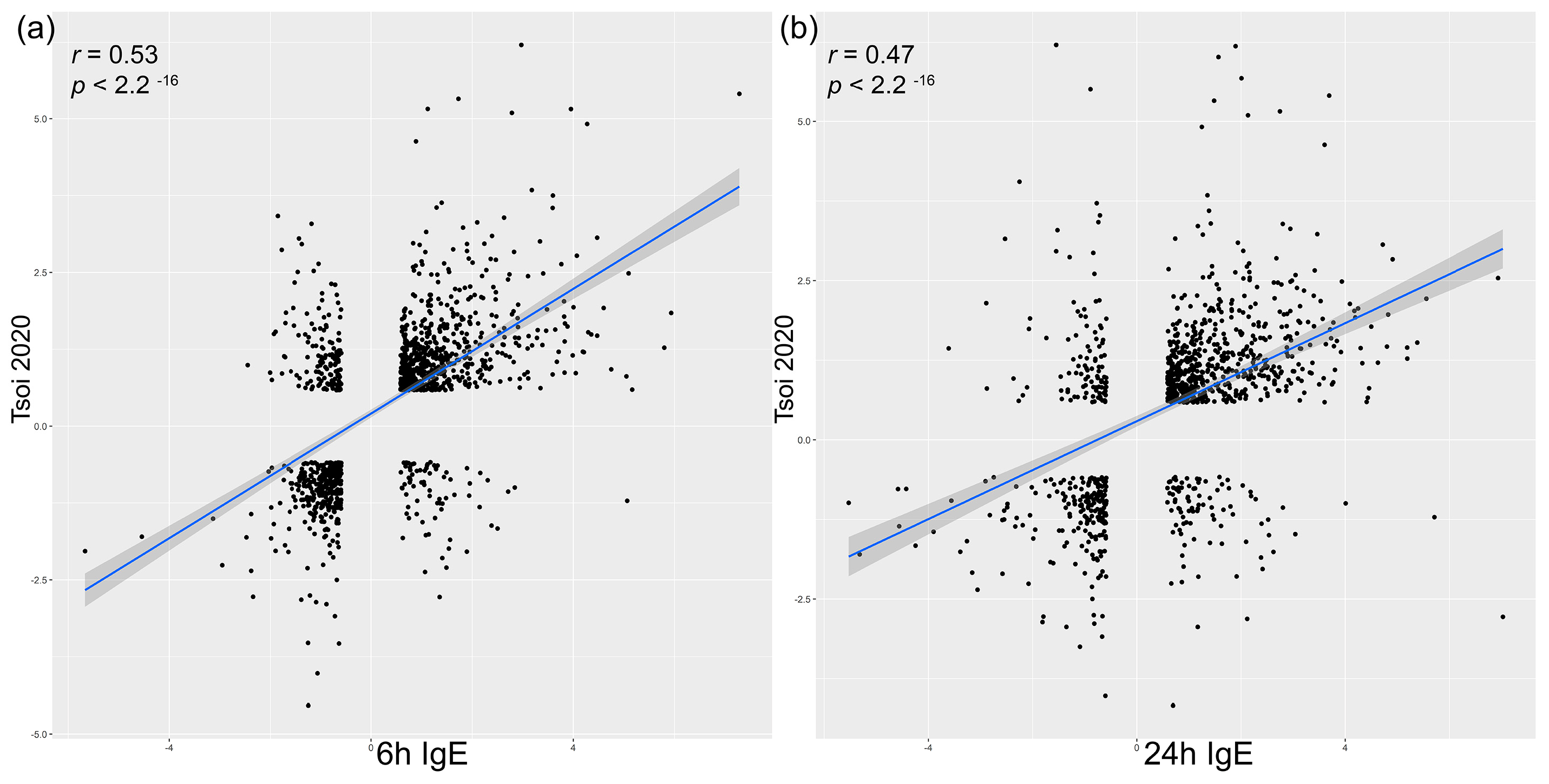
3.5. Correlation Analysis to Acute Skin Lesions in Human and Canine Spontaneous AD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marsella, R. Advances in our understanding of canine atopic dermatitis. Vet. Dermatol. 2021, 32, 547-e151. [Google Scholar] [CrossRef]
- Clark, R.A. Cell-mediated and IgE-mediated immune responses in atopic dermatitis. Arch. Dermatol. 1989, 125, 413–416. [Google Scholar] [CrossRef] [PubMed]
- Wollenberg, A.; Thomsen, S.F.; Lacour, J.P.; Jaumont, X.; Lazarewicz, S. Targeting immunoglobulin E in atopic dermatitis: A review of the existing evidence. World Allergy Organ J. 2021, 14, 100519. [Google Scholar] [CrossRef] [PubMed]
- Olivry, T.; Dunston, S.M.; Murphy, K.M.; Moore, P.F. Characterization of the inflammatory infiltrate during IgE-mediated late phase reactions in the skin of normal and atopic dogs. Vet. Dermatol. 2001, 12, 49–58. [Google Scholar] [CrossRef]
- Pucheu-Haston, C.M.; Shuster, D.; Olivry, T.; Brianceau, P.; Lockwood, P.; McClanahan, T.; de Waal Malefyt, R.; Mattson, J.D.; Hammerberg, B. A canine model of cutaneous late-phase reactions: Prednisolone inhibition of cellular and cytokine responses. Immunology 2006, 117, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Bizikova, P.; Linder, K.E.; Paps, J.; Olivry, T. Effect of a novel topical diester glucocorticoid spray on immediate- and late-phase cutaneous allergic reactions in Maltese-beagle atopic dogs: A placebo-controlled study. Vet. Dermatol. 2010, 21, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Pucheu-Haston, C.M.; Kasparek, K.A.; Stout, R.W.; Kearney, M.T.; Hammerberg, B. Effects of pentoxifylline on immediate and late-phase cutaneous reactions in response to anti-immunoglobulin E antibodies in clinically normal dogs. Am. J. Vet. Res. 2014, 75, 152–160. [Google Scholar] [CrossRef]
- Baumer, W.; Rossbach, K.; Schmidt, B.H. The selective glucocorticoid receptor agonist mapracorat displays a favourable safety-efficacy ratio for the topical treatment of inflammatory skin diseases in dogs. Vet. Dermatol. 2017, 28, 46-e11. [Google Scholar] [CrossRef] [PubMed]
- Blubaugh, A.; Rissi, D.; Elder, D.; Denley, T.; Eguiluz-Hernandez, S.; Banovic, F. The anti-inflammatory effect of topical tofacitinib on immediate and late-phase cutaneous allergic reactions in dogs: A placebo-controlled pilot study. Vet. Dermatol. 2018, 29, 250-e93. [Google Scholar] [CrossRef]
- Bieber, T. Insights into immunoglobulin E-mediated late-phase reactions in dogs. Vet. Dermatol. 2014, 25, 305–306. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2015, 14, 1471–2105. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Metacore. Metacore, a Cortellis Solution, © 2022 Clarivate. Available online: https://clarivate.com/cortellis/solutions/early-research-intelligence-solutions/ (accessed on 8 February 2023).
- Freudenberg, J.M.; Olivry, T.; Mayhew, D.N.; Rubenstein, D.S.; Rajpal, D.K. The Comparison of Skin Transcriptomes Confirms Canine Atopic Dermatitis Is a Natural Homologue to the Human Disease. J. Investig. Dermatol. 2019, 139, 968–971. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, R.D.; Pavel, A.B.; Glickman, J.; Chan, T.C.; Zheng, X.; Zhang, N.; Cueto, I.; Peng, X.; Estrada, Y.; Fuentes-Duculan, J.; et al. Atopic dermatitis in African American patients is T(H)2/T(H)22-skewed with T(H)1/T(H)17 attenuation. Ann. Allergy Asthma Immunol. 2019, 122, 99–110.e116. [Google Scholar] [CrossRef]
- Guttman-Yassky, E.; Diaz, A.; Pavel, A.B.; Fernandes, M.; Lefferdink, R.; Erickson, T.; Canter, T.; Rangel, S.; Peng, X.; Li, R.; et al. Use of Tape Strips to Detect Immune and Barrier Abnormalities in the Skin of Children with Early-Onset Atopic Dermatitis. JAMA Dermatol. 2019, 155, 1358–1370. [Google Scholar] [CrossRef]
- Eyerich, K.; Novak, N. Immunology of atopic eczema: Overcoming the Th1/Th2 paradigm. Allergy 2013, 68, 974–982. [Google Scholar] [CrossRef] [PubMed]
- Pucheu-Haston, C.M.; Bizikova, P.; Marsella, R.; Santoro, D.; Nuttall, T.; Eisenschenk, M.N. Review: Lymphocytes, cytokines, chemokines and the T-helper 1-T-helper 2 balance in canine atopic dermatitis. Vet. Dermatol. 2015, 26, 124-e32. [Google Scholar] [CrossRef]
- Tsoi, L.C.; Rodriguez, E.; Stolzl, D.; Wehkamp, U.; Sun, J.; Gerdes, S.; Sarkar, M.K.; Hubenthal, M.; Zeng, C.; Uppala, R.; et al. Progression of acute-to-chronic atopic dermatitis is associated with quantitative rather than qualitative changes in cytokine responses. J. Allergy Clin. Immunol. 2020, 145, 1406–1415. [Google Scholar] [CrossRef] [PubMed]
- Ewald, D.A.; Noda, S.; Oliva, M.; Litman, T.; Nakajima, S.; Li, X.; Xu, H.; Workman, C.T.; Scheipers, P.; Svitacheva, N.; et al. Major differences between human atopic dermatitis and murine models, as determined by using global transcriptomic profiling. J. Allergy Clin. Immunol. 2017, 139, 562–571. [Google Scholar] [CrossRef]
- Leung, D.Y.; Bhan, A.K.; Schneeberger, E.E.; Geha, R.S. Characterization of the mononuclear cell infiltrate in atopic dermatitis using monoclonal antibodies. J. Allergy Clin. Immunol. 1983, 71, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Olivry, T.; Hill, P.B. The ACVD task force on canine atopic dermatitis (XVIII): Histopathology of skin lesions. Vet. Immunol. Immunopathol. 2001, 81, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Bhagya, B.K.; Kamran, C.A.; Veeregowda, S.R.B. Histopathology of skin lesions of canine atopic dermatitis in pugs. J. Entomol. Zool. Stud. 2021, 9, 603–606. [Google Scholar] [CrossRef]
- Matucci, A.; Maggi, E.; Vultaggio, A. Eosinophils, the IL-5/IL-5Ralpha axis, and the biologic effects of benralizumab in severe asthma. Respir. Med. 2019, 160, 105819. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, L.C.; Rodriguez, E.; Degenhardt, F.; Baurecht, H.; Wehkamp, U.; Volks, N.; Szymczak, S.; Swindell, W.R.; Sarkar, M.K.; Raja, K.; et al. Atopic Dermatitis Is an IL-13-Dominant Disease with Greater Molecular Heterogeneity Compared to Psoriasis. J Investig. Dermatol. 2019, 139, 1480–1489. [Google Scholar] [CrossRef]
- Jassies-van der Lee, A.; Rutten, V.P.; Bruijn, J.; Willemse, T.; Broere, F. CD4+ and CD8+ skin-associated T lymphocytes in canine atopic dermatitis produce interleukin-13, interleukin-22 and interferon-gamma and contain a CD25+ FoxP3+ subset. Vet. Dermatol. 2014, 25, 456-e72. [Google Scholar] [CrossRef]
- Kunimura, K.; Fukui, Y. The molecular basis for IL-31 production and IL-31-mediated itch transmission: From biology to drug development. Int. Immunol. 2021, 33, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Pearson, J.; Leon, R.; Starr, H.; Kim, S.J.; Fogle, J.E.; Banovic, F. Establishment of an Intradermal Canine IL-31-Induced Pruritus Model to Evaluate Therapeutic Candidates in Atopic Dermatitis. Vet. Sci. 2023, 10, 329. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, A.J.; Humphrey, W.R.; Messamore, J.E.; Fleck, T.J.; Fici, G.J.; Shelly, J.A.; Teel, J.F.; Bammert, G.F.; Dunham, S.A.; Fuller, T.E.; et al. Interleukin-31: Its role in canine pruritus and naturally occurring canine atopic dermatitis. Vet. Dermatol. 2013, 24, 48-e12. [Google Scholar] [CrossRef]
- Tamamoto-Mochizuki, C.; Olivry, T. IL-31 and IL-31 receptor expression in acute experimental canine atopic dermatitis skin lesions. Vet. Dermatol. 2021, 32, 631-e169. [Google Scholar] [CrossRef]
- Arai, I.; Tsuji, M.; Miyagawa, K.; Takeda, H.; Akiyama, N.; Saito, S. Increased itching sensation depend on an increase in the dorsal root ganglia IL-31 receptor A (IL-31RA) expression in mice with atopic-dermatitis. Itch Pain 2014, 1, e467. [Google Scholar] [CrossRef]
- Arai, I.; Saito, S. Interleukin-31 Receptor A Expression in the Dorsal Root Ganglion of Mice with Atopic Dermatitis. Int. J. Mol. Sci. 2023, 24, 1047. [Google Scholar] [CrossRef]
- Kubo, T.; Sato, S.; Hida, T.; Minowa, T.; Hirohashi, Y.; Tsukahara, T.; Kanaseki, T.; Murata, K.; Uhara, H.; Torigoe, T. IL-13 modulates ∆Np63 levels causing altered expression of barrier- and inflammation-related molecules in human keratinocytes: A possible explanation for chronicity of atopic dermatitis. Immun Inflamm Dis 2021, 9, 734–745. [Google Scholar] [CrossRef] [PubMed]
- Mikhaylov, D.; Del Duca, E.; Guttman-Yassky, E. Proteomic signatures of inflammatory skin diseases: A focus on atopic dermatitis. Expert Rev. Proteom. 2021, 18, 345–361. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Parmigiani, G.; Johnson, W.E. ComBat-seq: Batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2020, 2, lqaa078. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blubaugh, A.; Hoover, K.; Kim, S.J.; Fogle, J.E.; Sow, F.B.; Banovic, F. Characterization of the Pro-Inflammatory and Pruritogenic Transcriptome in Skin Lesions of the Experimental Canine Atopic Acute IgE-Mediated Late Phase Reactions Model and Correlation to Acute Skin Lesions of Human Atopic Dermatitis. Vet. Sci. 2024, 11, 109. https://doi.org/10.3390/vetsci11030109
Blubaugh A, Hoover K, Kim SJ, Fogle JE, Sow FB, Banovic F. Characterization of the Pro-Inflammatory and Pruritogenic Transcriptome in Skin Lesions of the Experimental Canine Atopic Acute IgE-Mediated Late Phase Reactions Model and Correlation to Acute Skin Lesions of Human Atopic Dermatitis. Veterinary Sciences. 2024; 11(3):109. https://doi.org/10.3390/vetsci11030109
Chicago/Turabian StyleBlubaugh, Amanda, Kathleen Hoover, Sujung Jun Kim, Jonathan E. Fogle, Fatoumata B. Sow, and Frane Banovic. 2024. "Characterization of the Pro-Inflammatory and Pruritogenic Transcriptome in Skin Lesions of the Experimental Canine Atopic Acute IgE-Mediated Late Phase Reactions Model and Correlation to Acute Skin Lesions of Human Atopic Dermatitis" Veterinary Sciences 11, no. 3: 109. https://doi.org/10.3390/vetsci11030109
APA StyleBlubaugh, A., Hoover, K., Kim, S. J., Fogle, J. E., Sow, F. B., & Banovic, F. (2024). Characterization of the Pro-Inflammatory and Pruritogenic Transcriptome in Skin Lesions of the Experimental Canine Atopic Acute IgE-Mediated Late Phase Reactions Model and Correlation to Acute Skin Lesions of Human Atopic Dermatitis. Veterinary Sciences, 11(3), 109. https://doi.org/10.3390/vetsci11030109





