Integration Analysis of Transcriptome Sequencing and Whole-Genome Resequencing Reveal Wool Quality-Associated Key Genes in Zhexi Angora Rabbits
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Sequencing
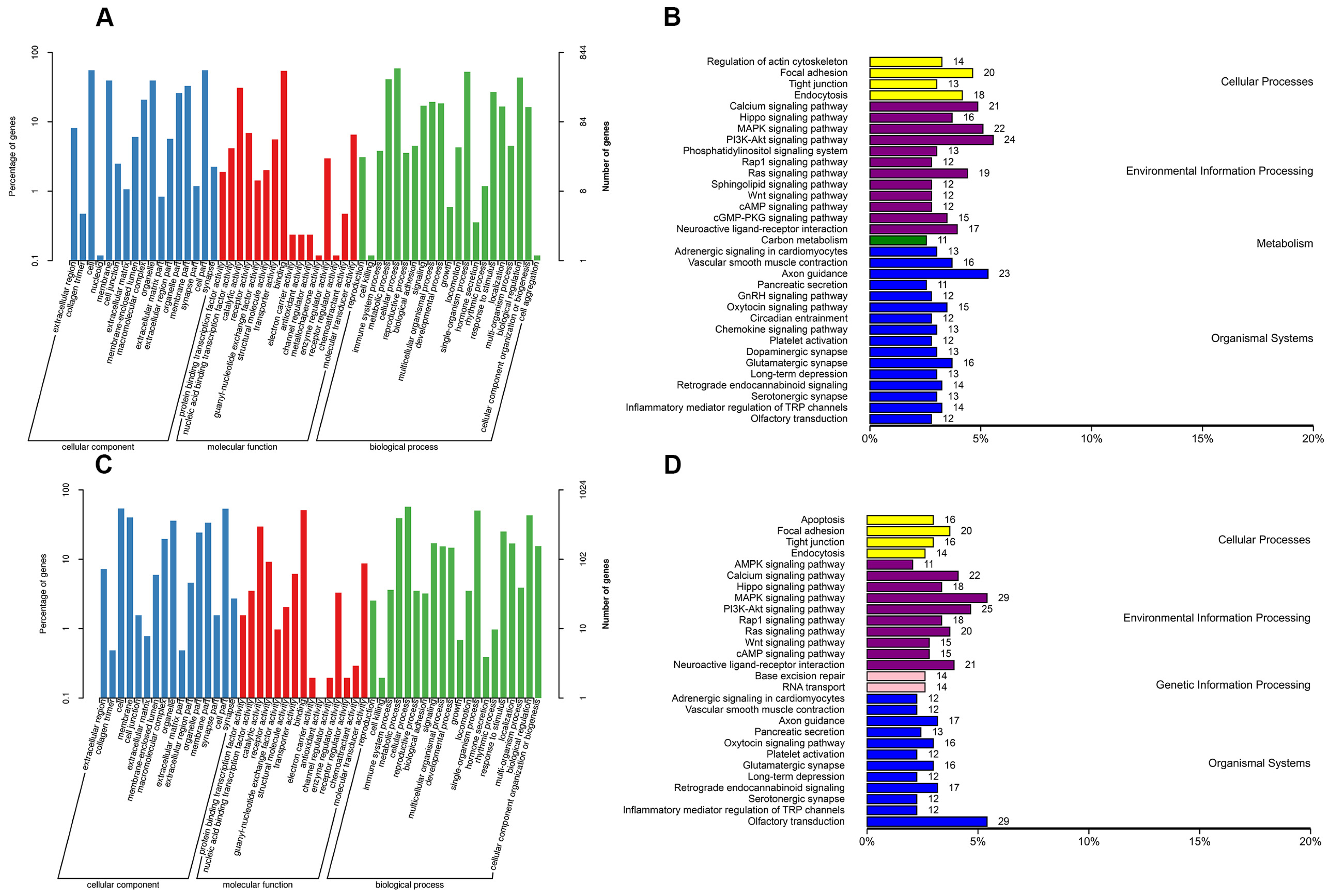
2.3. GO Enrichment and KEGG Pathway Analyses
2.4. Whole-Genome Resequencing
2.5. qPCR with Reverse Transcription
2.6. Joint Analysis of RNA-Seq and WGRS
2.7. Statistical Analysis
3. Results
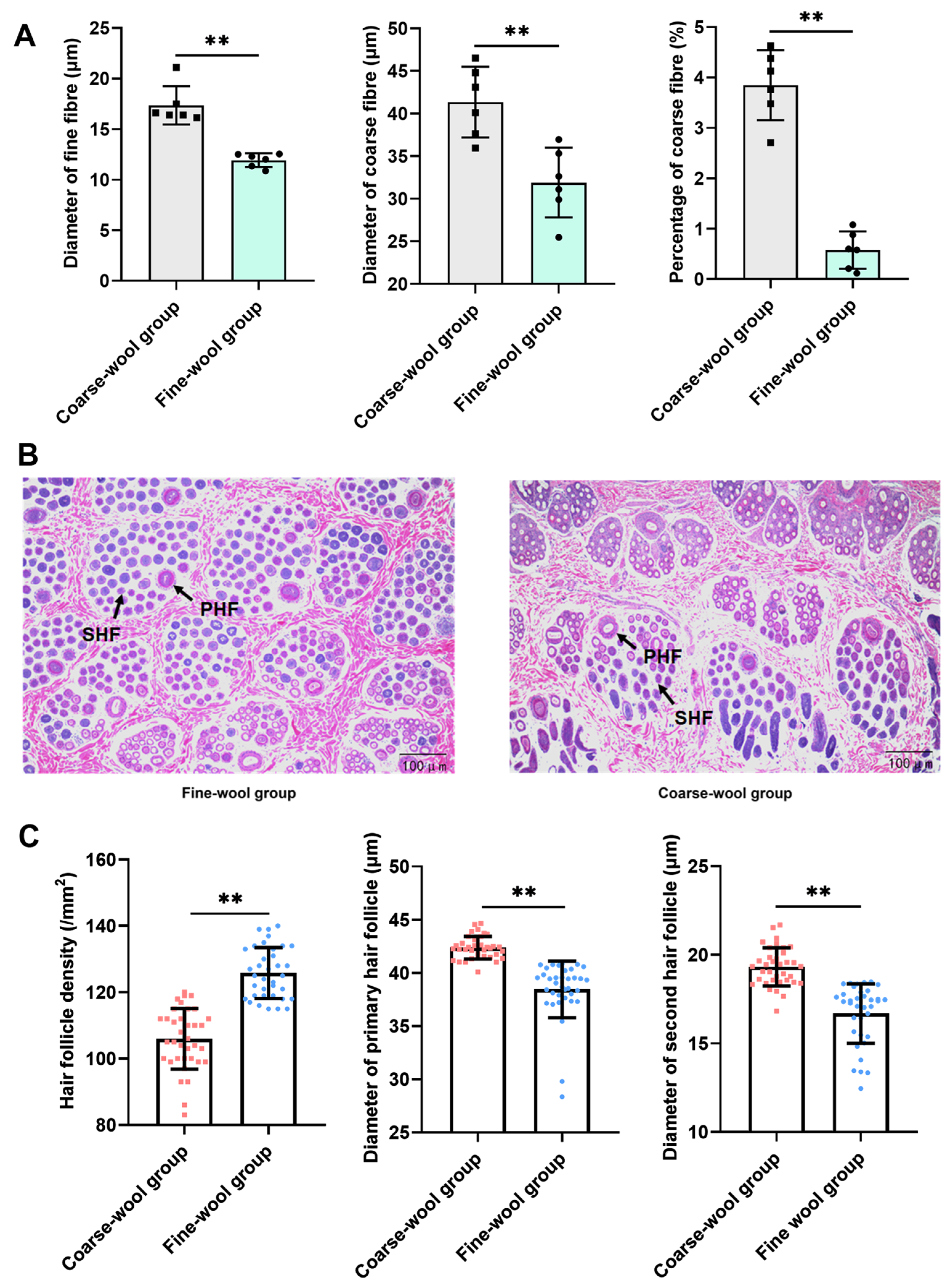
3.1. Morphological Characteristics of HFs in Zhexi Angora Rabbits
3.2. Screening of Fiber Formation-Associated DEGs Through RNA-Seq
3.3. Identification of Group-Specific Genetic Variation and Associated Genes Through WGRS
3.4. Joint Analysis of RNA-Seq and WGRS
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schneider, M.R.; Schmidt-Ullrich, R.; Paus, R. The hair follicle as a dynamic miniorgan. Curr. Biol. 2009, 19, R132–R142. [Google Scholar] [CrossRef] [PubMed]
- Galbraith, H. Fundamental hair follicle biology and fine fibre production in animals. Animal 2010, 4, 1490–1509. [Google Scholar] [CrossRef] [PubMed]
- Allafi, F.; Hossain, M.S.; Lalung, J.; Shaah, M.; Salehabadi, A.; Ahmad, M.I.; Shadi, A. Advancements in applications of natural wool fiber. J. Nat. Fibers 2022, 19, 497–512. [Google Scholar] [CrossRef]
- Laitala, K.; Klepp, I.G.; Henry, B. Does use matter? Comparison of environmental impacts of clothing based on fiber type. Sustainability 2018, 10, 2524. [Google Scholar] [CrossRef]
- Niranjan, S.; Sharma, S.; Gowane, G. Estimation of genetic parameters for wool traits in angora rabbit. Asian-Australas. J. Anim. Sci. 2011, 24, 1335–1340. [Google Scholar] [CrossRef]
- Doyle, E.K.; Preston, J.W.; McGregor, B.A.; Hynd, P.I. The science behind the wool industry. The importance and value of wool production from sheep. Anim. Front. 2021, 11, 15–23. [Google Scholar] [CrossRef]
- De Rochambeau, H.; Thébault, R.; Grun, J. Angora rabbit wool production: Non-genetic factors affecting quantity and quality of wool. Anim. Sci. 1991, 52, 383–393. [Google Scholar] [CrossRef]
- Murmu, S.B.; Debnath, S.; Bhutia, C.N. Evaluation of the german angora rabbit fiber produced in the northeast region of india. J. Nat. Fibers 2023, 20, 2210323. [Google Scholar] [CrossRef]
- Qian, Q.; Ma, J.; Zhang, G.; Xie, C.; Ren, L.; Qian, B. Breeding and Application of Zhexi Angora Rabbits. In Proceedings of the 11th World Rabbit Congress, Qingdao, China, 15–18 June 2016; pp. 861–864. [Google Scholar]
- Wang, D.; Xie, K.; Wang, Y.; Hu, J.; Li, W.; Yang, A.; Zhang, Q.; Ning, C.; Fan, X. Cost-effectively dissecting the genetic architecture of complex wool traits in rabbits by low-coverage sequencing. Genet. Sel. Evol. 2022, 54, 75. [Google Scholar] [CrossRef]
- Ning, C.; Xie, K.; Huang, J.; Di, Y.; Wang, Y.; Yang, A.; Hu, J.; Zhang, Q.; Wang, D.; Fan, X. Marker density and statistical model designs to increase accuracy of genomic selection for wool traits in angora rabbits. Front. Genet. 2022, 13, 968712. [Google Scholar] [CrossRef]
- Fatima, N.; Jia, L.; Liu, B.; Li, L.; Bai, L.; Wang, W.; Zhao, S.; Wang, R.; Liu, E. A homozygous missense mutation in the fibroblast growth factor 5 gene is associated with the long-hair trait in angora rabbits. BMC Genom. 2023, 24, 298. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Chen, Y.; Hu, S.; Yang, N.; Wang, M.; Liu, M.; Li, J.; Xiao, Y.; Wu, X. Systematic analysis of non-coding rnas involved in the angora rabbit (Oryctolagus cuniculus) hair follicle cycle by rna sequencing. Front. Genet. 2019, 10, 407. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Ding, H.; Wang, Y.; Cheng, G.; Wang, X.; Leng, T.; Zhao, H. Hair follicle transcriptome analysis reveals differentially expressed genes that regulate wool fiber diameter in angora rabbits. Biology 2023, 12, 445. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Chen, Y.; Hao, Y.; Yang, N.; Wang, M.; Mei, M.; Wang, J.; Qiu, X.; Wu, X. Transcriptomic analysis reveals differentially expressed genes associated with wool length in rabbit. Anim. Genet. 2018, 49, 428–437. [Google Scholar] [CrossRef]
- Zhang, W.; Jin, M.; Li, T.; Lu, Z.; Wang, H.; Yuan, Z.; Wei, C. Whole-genome resequencing reveals selection signal related to sheep wool fineness. Animals 2023, 13, 2944. [Google Scholar] [CrossRef]
- Qi, Y.; Fu, S.; He, X.; Wang, B.; Da, L.; Te, R.; Yuejun, M.; Suzhen, S.; Zhang, W.; Liu, Y. Preliminary comparison of skin transcriptome from sheep with different wool fibre diameters. Anim. Prod. Sci. 2021, 61, 708–714. [Google Scholar] [CrossRef]
- Yue, L.; Lu, Z.; Guo, T.; Liu, J.; Yang, B.; Yuan, C. Key genes and metabolites that regulate wool fibre diameter identified by combined transcriptome and metabolome analysis. Genomics 2024, 116, 110886. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. Hisat: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve years of samtools and bcftools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, snpeff: Snps in the genome of drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Källberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C. Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2016, 32, 1220–1222. [Google Scholar] [CrossRef] [PubMed]
- Boeva, V.; Popova, T.; Bleakley, K.; Chiche, P.; Cappo, J.; Schleiermacher, G.; Janoueix-Lerosey, I.; Delattre, O.; Barillot, E. Control-freec: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 2012, 28, 423–425. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K. Analyzing real-time pcr data by the comparative ct method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Li, S.; Chen, W.; Zheng, X.; Liu, Z.; Yang, G.; Hu, X.; Mou, C. Comparative investigation of coarse and fine wool sheep skin indicates the early regulators for skin and wool diversity. Genes 2020, 758, 144968. [Google Scholar] [CrossRef]
- Ding, H.; Zhao, H.; Cheng, G.; Yang, Y.; Wang, X.; Zhao, X.; Qi, Y.; Huang, D. Analyses of histological and transcriptome differences in the skin of short-hair and long-hair rabbits. BMC Genom. 2019, 20, 140. [Google Scholar] [CrossRef]
- Cui, C.-Y.; Kunisada, M.; Piao, Y.; Childress, V.; Ko, M.S.; Schlessinger, D. Dkk4 and eda regulate distinctive developmental mechanisms for subtypes of mouse hair. PLoS ONE 2010, 5, e10009. [Google Scholar] [CrossRef]
- Rendl, M.; Lewis, L.; Fuchs, E. Molecular dissection of mesenchymal–epithelial interactions in the hair follicle. PLoS Biol. 2005, 3, e331. [Google Scholar] [CrossRef]
- Chu, Q.; Cai, L.; Fu, Y.; Chen, X.; Yan, Z.; Lin, X.; Zhou, G.; Han, H.; Widelitz, R.B.; Chuong, C.-m. Dkk2/frzb in the dermal papillae regulates feather regeneration. Dev. Biol. 2014, 387, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Maglakelidze, N.; Gao, T.; Feehan, R.P.; Hobbs, R.P. Aire deficiency leads to the development of alopecia areata‒like lesions in mice. J. Investig. Dermatol. 2023, 143, 578–587.e573. [Google Scholar] [CrossRef] [PubMed]
- Niiyama, S.; Ishimatsu-Tsuji, Y.; Kishimoto, J. Niche formed by bone morphogenetic protein antagonists gremlin 1 and gremlin 2 in human hair follicles. Health Sci. Rep. 2022, 5, e486. [Google Scholar] [CrossRef]
- Wu, P.; Zhang, Y.; Xing, Y.; Xu, W.; Guo, H.; Deng, F.; Ma, X.; Li, Y. Signaling, The balance of bmp6 and wnt10b regulates the telogen-anagen transition of hair follicles. Cell Commun. 2019, 17, 1–10. [Google Scholar]
- Xue, X.; Mi, Z.; Wang, Z.; Pang, Z.; Liu, H.; Zhang, F. High expression of ace2 on keratinocytes reveals skin as a potential target for SARS-CoV-2. J. Investig. Dermatol. 2021, 141, 206. [Google Scholar] [CrossRef]
- Selleri, S.; Arnaboldi, F.; Palazzo, M.; Gariboldi, S.; Zanobbio, L.; Opizzi, E.; Shirai, Y.; Balsari, A.; Rumio, C. Toll-like receptor agonists regulate β-defensin 2 release in hair follicle. Br. J. Dermatol. 2007, 156, 1172–1177. [Google Scholar] [CrossRef]
- Xiong, L.; Zhevlakova, I.; West, X.Z.; Gao, D.; Murtazina, R.; Horak, A.; Brown, J.M.; Molokotina, I.; Podrez, E.A.; Byzova, T.V. Tlr2 regulates hair follicle cycle and regeneration via bmp signaling. eLife 2024, 12, RP89335. [Google Scholar] [CrossRef]
- Xie, K.; Ning, C.; Yang, A.; Zhang, Q.; Wang, D.; Fan, X. Resequencing analyses revealed genetic diversity and selection signatures during rabbit breeding and improvement. Genes 2024, 15, 433. [Google Scholar] [CrossRef]
- Zhao, B.; Chen, Y.; Yang, N.; Chen, Q.; Bao, Z.; Liu, M.; Hu, S.; Li, J.; Wu, X. Mir-218-5p regulates skin and hair follicle development through wnt/β-catenin signaling pathway by targeting sfrp2. J. Cell. Physiol. 2019, 234, 20329–20341. [Google Scholar] [CrossRef]
- Kim, B.-K.; Yoon, S.K. Expression of sfrp2 is increased in catagen of hair follicles and inhibits keratinocyte proliferation. Ann. Dermatol. 2014, 26, 79–87. [Google Scholar] [CrossRef]
- Bai, R.; Guo, Y.; Liu, W.; Song, Y.; Yu, Z.; Ma, X. The roles of wnt signaling pathways in skin development and mechanical-stretch-induced skin regeneration. Biomolecules 2023, 13, 1702. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Chen, X.; Zhao, Q.; Stanic, V.; Lin, Z.; Yang, S.; Chen, T.; Chen, J.; Yang, Y. Hair loss caused by gain-of-function mutant trpv3 is associated with premature differentiation of follicular keratinocytes. J. Investig. Dermatol. 2021, 141, 1964–1974. [Google Scholar] [CrossRef]
- Shah, K.; Mehmood, S.; Jan, A.; Abbe, I.; Hussain Ali, R.; Khan, A.; Chishti, M.S.; Lee, K.; Ahmad, F.; Ansar, M. Sequence variants in nine different genes underlying rare skin disorders in 10 consanguineous families. Int. J. Dermatol. 2017, 56, 1406–1413. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Li, A.G.; Liang, Y.-Y.; Owens, P.; He, W.; Lu, S.; Yoshimatsu, Y.; Wang, D.; Ten Dijke, P.; Lin, X. Smad7-induced β-catenin degradation alters epidermal appendage development. Dev. Cell 2006, 11, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Tervaniemi, M.H.; Katayama, S.; Skoog, T.; Siitonen, H.A.; Vuola, J.; Nuutila, K.; Sormunen, R.; Johnsson, A.; Linnarsson, S.; Suomela, S. Nod-like receptor signaling and inflammasome-related pathways are highlighted in psoriatic epidermis. Sci. Rep. 2016, 6, 22745. [Google Scholar] [CrossRef] [PubMed]
- Zhai, B.; Zhang, L.; Wang, C.; Zhao, Z.; Zhang, M.; Li, X. Identification of microrna-21 target genes associated with hair follicle development in sheep. PeerJ 2019, 7, e7167. [Google Scholar] [CrossRef]
- Tian, D.; Pei, Q.; Jiang, H.; Guo, J.; Ma, X.; Han, B.; Li, X.; Zhao, K. Comprehensive analysis of the expression profiles of mrna, lncrna, circrna, and mirna in primary hair follicles of coarse sheep fetal skin. BMC Genom. 2024, 25, 574. [Google Scholar] [CrossRef]
- Tomcik, M.; Palumbo-Zerr, K.; Zerr, P.; Sumova, B.; Avouac, J.; Dees, C.; Distler, A.; Becvar, R.; Distler, O.; Schett, G. Tribbles homologue 3 stimulates canonical tgf-β signalling to regulate fibroblast activation and tissue fibrosis. Ann. Rheum. Dis. 2016, 75, 609–616. [Google Scholar] [CrossRef]
- Grover, J.; Lee, E.; Mounkes, L.; Stewart, C.; Roughley, P. The consequence of prelp overexpression on skin. Matrix Biol. 2007, 26, 140–143. [Google Scholar] [CrossRef]
- Andl, T.; Reddy, S.T.; Gaddapara, T.; Millar, S.E. Wnt signals are required for the initiation of hair follicle development. Dev. Cell 2002, 2, 643–653. [Google Scholar] [CrossRef]
- Harel, S.; Higgins, C.A.; Cerise, J.E.; Dai, Z.; Chen, J.C.; Clynes, R.; Christiano, A.M. Pharmacologic inhibition of jak-stat signaling promotes hair growth. Sci. Adv. 2015, 1, e1500973. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Gao, Y.; Fan, Z.; Xiao, X.; Chen, Y.; Si, Y.; Kong, D.; Wang, S.; Liao, M.; Chen, X. Amphiregulin promotes hair regeneration of skin-derived precursors via the pi3k and mapk pathways. Cell Prolif. 2021, 54, e13106. [Google Scholar] [CrossRef] [PubMed]
- Oshimori, N.; Fuchs, E. Paracrine tgf-β signaling counterbalances bmp-mediated repression in hair follicle stem cell activation. Cell Stem Cell 2012, 10, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fan, Z.; Wang, X.; Mo, M.; Zeng, S.B.; Xu, R.-H.; Wang, X.; Wu, Y. Pi3k/akt signaling pathway is essential for de novo hair follicle regeneration. Stem Cell Res. Ther. 2020, 11, 144. [Google Scholar] [CrossRef]
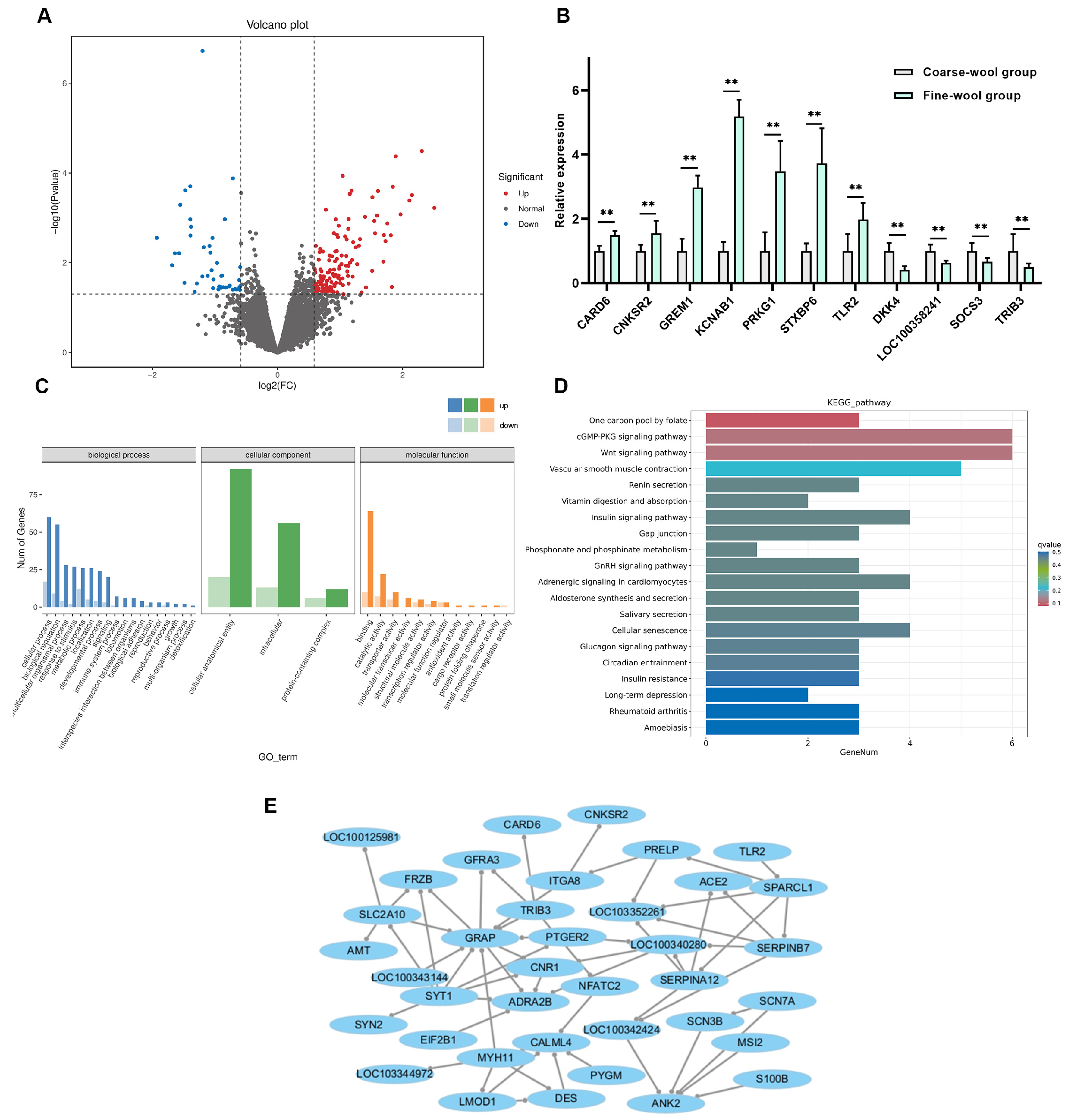
| Gene Name | Gene ID | p-Value | log2 Fold Change | Chromosome | SNP Number | Indel Number |
|---|---|---|---|---|---|---|
| KCNAB1 | ENSOCUG00000007885 | 0.0001 | 1.0415 | 14 | 2 | |
| TRIB3 | ENSOCUG00000007636 | 0.0002 | −1.3960 | 4 | 7 | 1 |
| TLR2 | ENSOCUG00000003367 | 0.0010 | 1.3993 | 15 | 2 | |
| STXBP6 | ENSOCUG00000005144 | 0.0015 | 0.9784 | 17 | 1 | |
| PRKG1 | ENSOCUG00000011473 | 0.0058 | 0.6725 | 18 | 1238 | 184 |
| ALDH1L2 | ENSOCUG00000006962 | 0.0060 | −1.0734 | 4 | 1 | |
| CNKSR2 | ENSOCUG00000014660 | 0.0089 | 0.8258 | X | 1 | |
| PRELP | ENSOCUG00000009707 | 0.0141 | 0.6867 | 16 | 2 | |
| CARD6 | ENSOCUG00000005374 | 0.0232 | 1.0014 | 11 | 1 | 2 |
| ANK2 | ENSOCUG00000010618 | 0.0265 | 0.6598 | 15 | 1 | |
| CDH19 | ENSOCUG00000017601 | 0.0461 | 1.3454 | 9 | 1 | |
| CA10 | ENSOCUG00000002345 | 0.0495 | 0.6092 | 19 | 1 |
| Overlapping Signaling Pathways | RNA-Seq | WGRS |
|---|---|---|
| Wnt signaling pathway | DKK4, FMNL1, FRZB, NFATC2, PLCB4, RSPO1 | INVS, CSNK1A1, RSPO2, DAAM2, PRKCA, TCF7L2 |
| JAK-STAT signaling pathway | FHL1, SOCS3 | STAT4, BCL2L1, STAT5B, SOCS2, IL13RA1, SOCS6, PTPN11, SOCS7, AKT3 |
| MAPK signaling pathway | GRAP, LOC100358241 | PLA2G4E, ANGPT1, PDGFRL, PPM1B, PRKCA, WDR7 |
| TGF-β signaling pathway | GREM1 | TGIF2, BMP6, SMAD5, RBL1, LTBP1 |
| PI3K-Akt signaling pathway | ITGA8, LOC100353654, TLR2 | PPP2R3A, COL6A6, BCL2L1, CREB5, COL4A3, TLR2, ANGPT1, PRKAA2 |
| VEGF signaling pathway | NFATC2 | PLA2G4E, PRKCA, PLA2G4F, PPP3CA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, B.; Yu, Y.; Sun, S.; Cai, J.; Bao, Z.; Chen, Y.; Wu, X. Integration Analysis of Transcriptome Sequencing and Whole-Genome Resequencing Reveal Wool Quality-Associated Key Genes in Zhexi Angora Rabbits. Vet. Sci. 2024, 11, 651. https://doi.org/10.3390/vetsci11120651
Zhao B, Yu Y, Sun S, Cai J, Bao Z, Chen Y, Wu X. Integration Analysis of Transcriptome Sequencing and Whole-Genome Resequencing Reveal Wool Quality-Associated Key Genes in Zhexi Angora Rabbits. Veterinary Sciences. 2024; 11(12):651. https://doi.org/10.3390/vetsci11120651
Chicago/Turabian StyleZhao, Bohao, Yongqi Yu, Shaoning Sun, Jiawei Cai, Zhiyuan Bao, Yang Chen, and Xinsheng Wu. 2024. "Integration Analysis of Transcriptome Sequencing and Whole-Genome Resequencing Reveal Wool Quality-Associated Key Genes in Zhexi Angora Rabbits" Veterinary Sciences 11, no. 12: 651. https://doi.org/10.3390/vetsci11120651
APA StyleZhao, B., Yu, Y., Sun, S., Cai, J., Bao, Z., Chen, Y., & Wu, X. (2024). Integration Analysis of Transcriptome Sequencing and Whole-Genome Resequencing Reveal Wool Quality-Associated Key Genes in Zhexi Angora Rabbits. Veterinary Sciences, 11(12), 651. https://doi.org/10.3390/vetsci11120651





