Evolution of Population Structure, Reproductive Performance, Inbreeding, and Genetic Diversity in Ecuadorian Charolais Cattle
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Charolais Genealogical Database
2.3. Demographic Analysis
- Demographic distribution: Number of females per number of births, number of males and females, and average age of offspring per year were used as criteria to determine the population structure in subsequent generations.
- Pedigree completeness: It was analyzed through the known ancestors up to 5 generations of individuals born in the proposed periods according to MacCluer et al. [28]. In addition, the numbers of complete (GCom), maximum (GMax), and equivalent (GEqu) generations were considered. In the case of GEqu, the following formula was used:Here, “n” is the number of generations in which the individual is separated from each ancestor with a known record. The pedigree completeness was analyzed through the numbers of GEqu, GMax, and GCom [29] and the pedigree completeness index [28].
- Generation interval (GI): The GI was defined as the average age of the parents at the birth of their offspring. The GI was calculated for the 4 gametic pathways: Sire–Son, Sire–Daughter, Dam–Son, and Dam–Daughter [6].
2.4. Reproductive-Performance-Derived Parameters
2.5. Inbreeding, Relatedness, Coancestry, Non-Random Mating, and Genetic Conservation Parameters
2.6. Gene Origin and Genetic-Diversity-Derived Parameters
2.7. Data Analysis and Software
3. Results
3.1. Evolution of Population Structure and Reproductive Performance in Ecuadorian Charolais Cattle
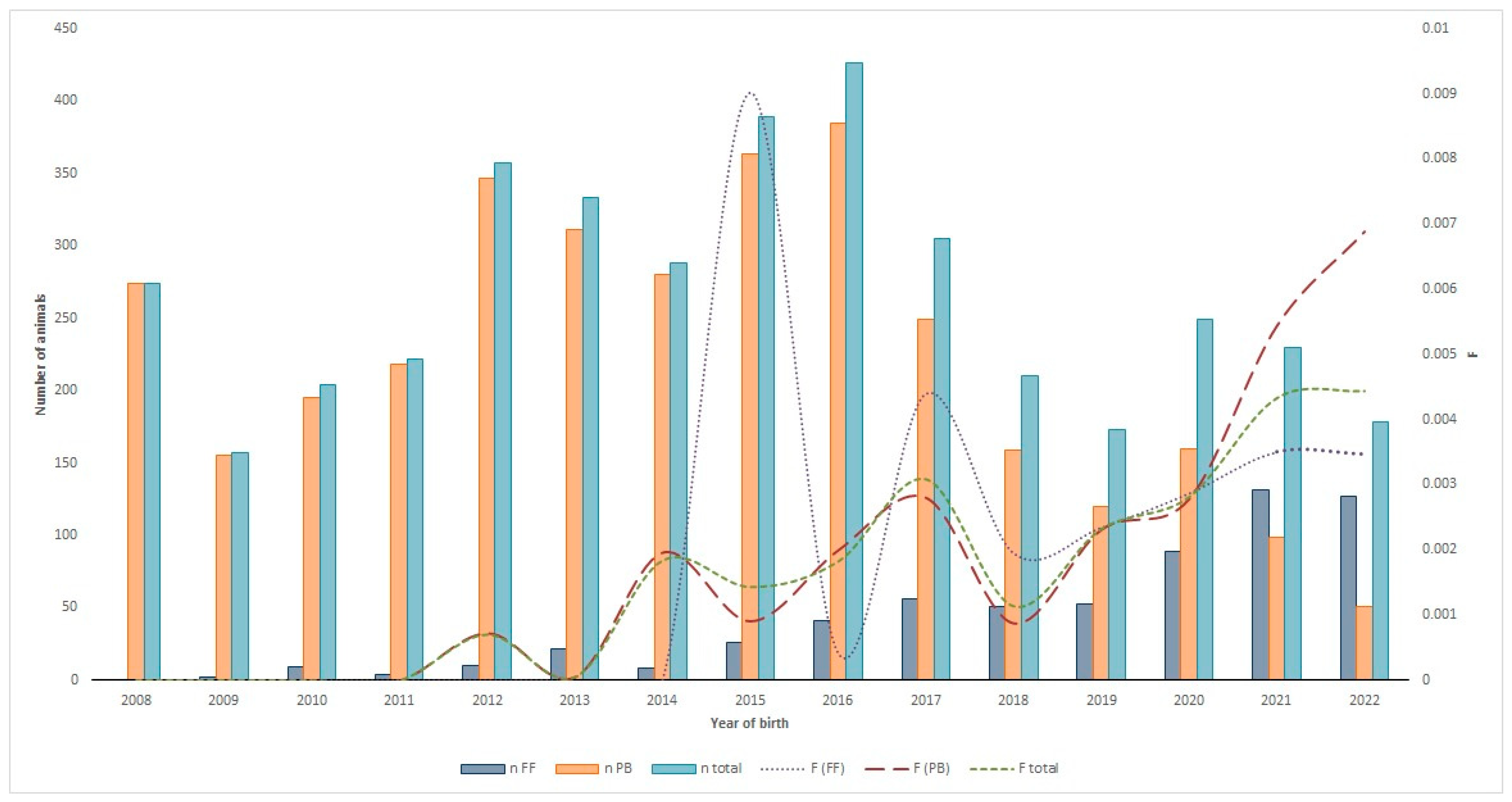
3.1.1. Demographic Structure and Reproductive Performance (Calving Rates)
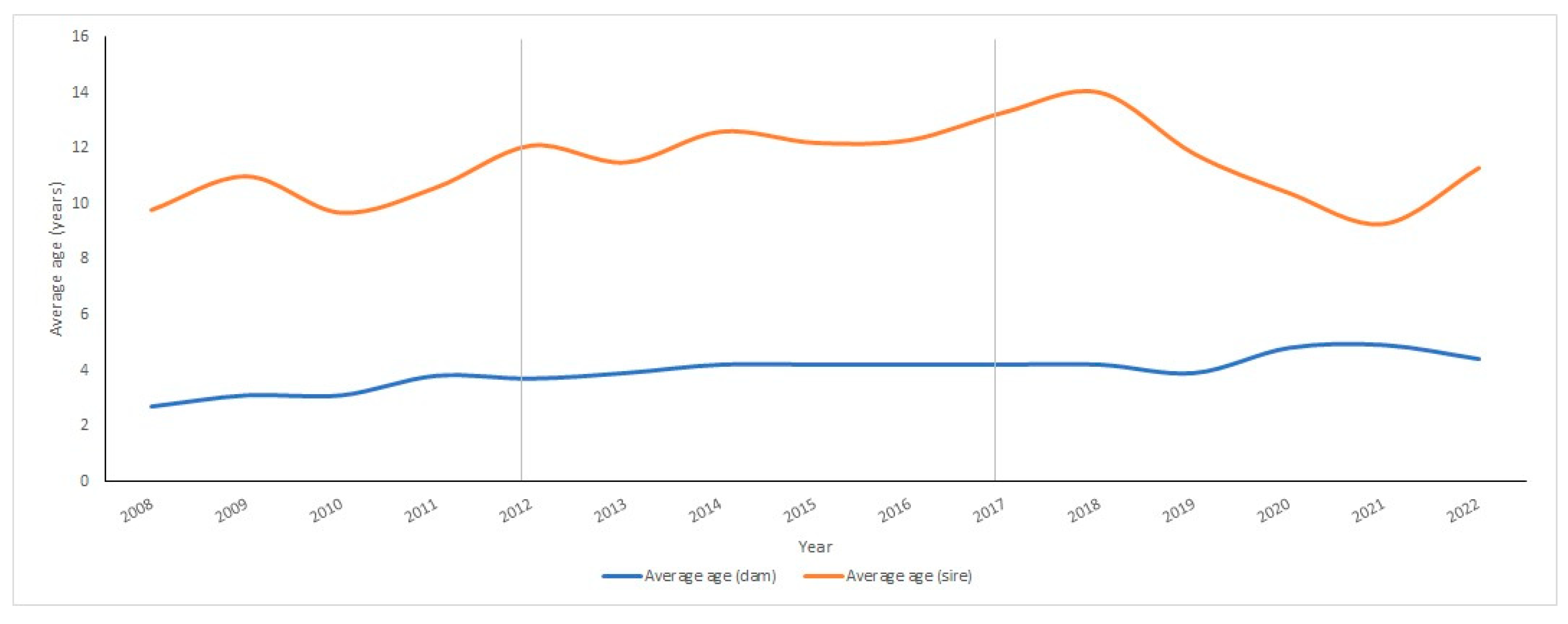
3.1.2. Breeding Sires’ and Dams’ Average Ages
3.1.3. Evolution of the Use of ARTs in the Ecuadorian Charolais Cattle Population from 2008 to 2022
3.1.4. Number of Calves Obtained per Cow from 2008 to 2022
3.2. Evolution of Pedigree Completeness and Generation Intervals in Ecuadorian Charolais Cattle
3.2.1. Pedigree Completeness in the Ecuadorian Charolais Cattle Population from 2008 to 2022
3.2.2. Generation Intervals in Ecuadorian Charolais Cattle Population from 2008 to 2022
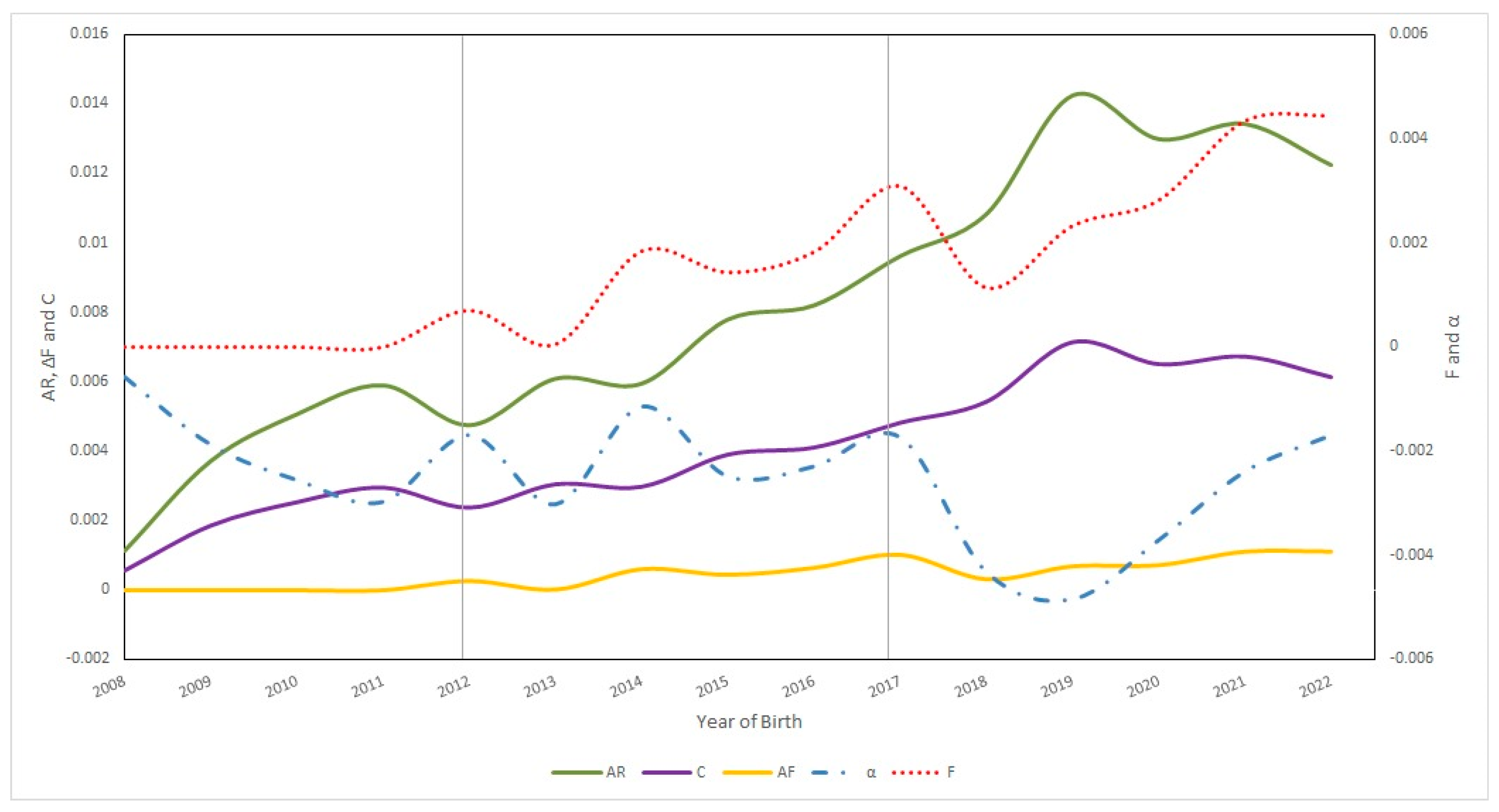
3.2.3. Inbreeding, Average Relatedness, Coancestry, and Non-Random Mating in Ecuadorian Charolais Cattle Population from 2008 to 2022
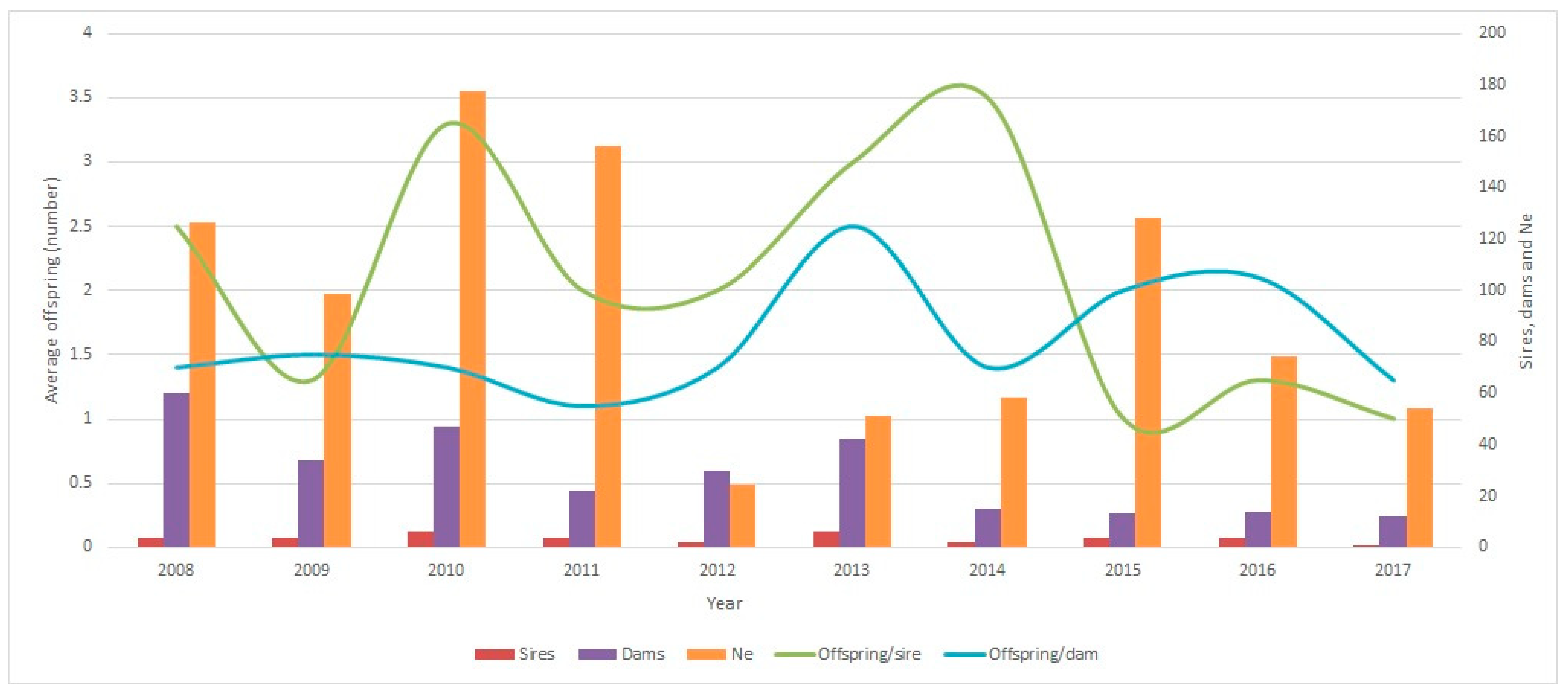
3.2.4. Effective Size (Ne) in Ecuadorian Charolais Cattle Population from 2008 to 2022
3.3. Evolution of the Gene Origin Probability-Derived Parameters in Ecuadorian Charolais Cattle
3.3.1. Gene Origin Probability in Ecuadorian Charolais Cattle Population from 2008 to 2022
3.3.2. Genetic Diversity in Ecuadorian Charolais Cattle Population from 2008 to 2022
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lozada, E.; Chacón, E.; Sambache, E.; Revelo, M.; Gutiérrez, M.; Delgado, J.V.; Cartuche, L.F.; Navas, F.J. Diversidad Genética y Estructura de La Población de La Raza Charolais En Ecuador a Través Del Pedigrí. Arch. Zootec. 2023, 72, 32–42. [Google Scholar] [CrossRef]
- Gutiérrez-Reinoso, M.A.; Aponte, P.M.; García-Herreros, M. A Review of Inbreeding Depression in Dairy Cattle: Current Status, Emerging Control Strategies, and Future Prospects. J. Dairy Res. 2022, 89, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Forneris, N.S.; Garcia-Baccino, C.A.; Cantet, R.J.C.; Vitezica, Z.G. Estimating Inbreeding Depression for Growth and Reproductive Traits Using Pedigree and Genomic Methods in Argentinean Brangus Cattle. J. Anim. Sci. 2021, 99, skab289. [Google Scholar] [CrossRef]
- Gutiérrez-Reinoso, M.A.; Aponte, P.M.; Cabezas, J.; Rodriguez-Alvarez, L.; Garcia-Herreros, M. Genomic Evaluation of Primiparous High-Producing Dairy Cows: Inbreeding Effects on Genotypic and Phenotypic Production–Reproductive Traits. Animals 2020, 10, 1704. [Google Scholar] [CrossRef]
- Howard, J.T.; Pryce, J.E.; Baes, C.; Maltecca, C. Invited Review: Inbreeding in the Genomics Era: Inbreeding, Inbreeding Depression, and Management of Genomic Variability. J. Dairy Sci. 2017, 100, 6009–6024. [Google Scholar] [CrossRef]
- James, J.W. A Note on Selection Differential and Generation Length When Generations Overlap. Anim. Sci. 1977, 24, 109–112. [Google Scholar] [CrossRef]
- Gutiérrez-Reinoso, M.A.; Aponte, P.M.; García-Herreros, M. Genomic and Phenotypic Udder Evaluation for Dairy Cattle Selection: A Review. Animals 2023, 13, 1588. [Google Scholar] [CrossRef]
- Baumung, R.; Simianer, H.; Hoffmann, I. Genetic Diversity Studies in Farm Animals—A SurveyStudien Zur Genetischen Diversität Landwirtschaftlicher Nutztiere—Ein Globaler Überblick. J. Anim. Breed. Genet. 2004, 121, 361–373. [Google Scholar] [CrossRef]
- Hill, W.G.; Mackay, T.F.C. Introduction to Quantitative Genetics. Genetics 2004, 167, 1529–1536. [Google Scholar] [CrossRef]
- Gutiérrez, J.P.; Cervantes, I.; Goyache, F. Improving the Estimation of Realized Effective Population Sizes in Farm Animals. J. Anim. Breed. Genet. 2009, 126, 327–332. [Google Scholar] [CrossRef]
- Leroy, G.; Mary-Huard, T.; Verrier, E.; Danvy, S.; Charvolin, E.; Danchin-Burge, C. Methods to Estimate Effective Population Size Using Pedigree Data: Examples in Dog, Sheep, Cattle and Horse. Genet. Sel. Evol. 2013, 45, 1. [Google Scholar] [CrossRef]
- James, J.W. Computation of Genetic Contributions from Pedigrees. TAG Theor. Appl. Genet. Theor. Und Angew. Genet. 1972, 42, 272–273. [Google Scholar] [CrossRef] [PubMed]
- Lacy, R.C. Analysis of Founder Representation in Pedigrees: Founder Equivalents and Founder Genome Equivalents. Zoo Biol. 1989, 8, 111–123. [Google Scholar] [CrossRef]
- Boichard, D.; Maignel, L.; Verrier, É. The Value of Using Probabilities of Gene Origin to Measure Genetic Variability in a Population. Genet. Sel. Evol. 1997, 29, 5–23. [Google Scholar] [CrossRef]
- Colleau, J.J.; Sargolzaei, M. A Proximal Decomposition of Inbreeding, Coancestry and Contributions. Genet. Res. 2008, 90, 191–198. [Google Scholar] [CrossRef]
- Gutierrez-Reinoso, M.A.; Aponte, P.M.; Garcia-Herreros, M. Genomic Analysis, Progress and Future Perspectives in Dairy Cattle Selection: A Review. Animals 2021, 11, 599. [Google Scholar] [CrossRef]
- Amaya, A.; Martínez, R.; Cerón-Muñoz, M. Population Structure and Genetic Diversity in Colombian Simmental Cattle. Trop. Anim. Health Prod. 2020, 52, 1133–1139. [Google Scholar] [CrossRef]
- Barbosa, A.C.B.; Malhado, C.H.M.; Carneiro, P.L.S.; Muniz, L.M.S.; Ambrosini, D.P.; Carrillo, J.A.; Filho, R.M. Population Structure of Nellore Cattle in Northeastern Brazil. Rev. Bras. Zootec. 2013, 42, 639–644. [Google Scholar] [CrossRef]
- Bernardes, P.A.; Grossi, D.A.; Savegnago, R.P.; Buzanskas, M.E.; Ramos, S.B.; Romanzini, E.P.; Guidolin, D.G.F.; Bezerra, L.A.F.; Lôbo, R.B.; Munari, D.P. Population Structure of Tabapuã Beef Cattle Using Pedigree Analysis. Livest. Sci. 2016, 187, 96–101. [Google Scholar] [CrossRef]
- Fabbri, M.C.; de Rezende, M.P.G.; Dadousis, C.; Biffani, S.; Negrini, R.; Carneiro, P.L.S.; Bozzi, R. Population Structure and Genetic Diversity of Italian Beef Breeds as a Tool for Planning Conservation and Selection Strategies. Animals 2019, 9, 880. [Google Scholar] [CrossRef]
- García-Ruiz, A.; Cole, J.B.; VanRaden, P.M.; Wiggans, G.R.; Ruiz-López, F.J.; Van Tassell, C.P. Changes in Genetic Selection Differentials and Generation Intervals in US Holstein Dairy Cattle as a Result of Genomic Selection. Proc. Natl. Acad. Sci. USA 2016, 113, E3995–E4004. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.P.; Goyache, F. A Note on ENDOG: A Computer Program for Analysing Pedigree Information. J. Anim. Breed. Genet. 2005, 122, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Sargolzaei, M.; Iwaisaki, H.; Colleau, J.J. CFC: A Tool for Monitoring Genetic Diversity. In Proceedings of the 8th World Congress on Genetics Applied to Livestock Production, Belo Horizonte, Minas Gerais, Brazil, 13–18 August 2006; WCGALP: Rotterdam, The Netherlands, 2006. [Google Scholar]
- Groeneveld, E.; Westhuizen, B.D.; Maiwashe, A.; Voordewind, F.; Ferraz, J.B. POPREP: A Generic Report for Population Management. Genet. Mol. Res. GMR 2009, 8, 1158–1178. [Google Scholar] [CrossRef] [PubMed]
- Berg, P.; Nielsen, J.; Sørensen, M.K. EVA: Realized and Predicted Optimal Genetic Contributions. In Proceedings of the 8th World Congress on Genetics Applied to Livestock Production, Belo Horizonte, Minas Gerais, Brazil, 13–18 August 2006; WCGALP: Rotterdam, The Netherlands, 2006. [Google Scholar]
- Boichard, D. Pedig: A Fortran Package for Pedigree Analysis Suited for Large Populations. Didier Boichard to Cite This Version: HAL Id: Hal-02833573. In Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, 19–23 August 2002; Volume 32, pp. 525–528. [Google Scholar]
- Navas, F.J.; Jordana, J.; León, J.M.; Barba, C.; Delgado, J.V. A Model to Infer the Demographic Structure Evolution of Endangered Donkey Populations. Animal 2017, 11, 2129–2138. [Google Scholar] [CrossRef]
- Maccluer, J.W.; Boyce, A.J.; Dyke, B.; Weitkamp, L.R.; Pfenning, D.W.; Parsons, C.J. Inbreeding and Pedigree Structure in Standardbred Horses. J. Hered. 1983, 74, 394–399. [Google Scholar] [CrossRef]
- Maignell, L.; Boichatt, D.; Vetiel, E. Genetic Variability of French Dairy Breeds Estimated from Pedigree Information. Interbull Bull. 1996, 14, 49. [Google Scholar]
- Meuwissen, T.H.E.; Luo, Z. Computing Inbreeding Coefficients in Large Populations. Genet. Sel. Evol. 1992, 24, 305–313. [Google Scholar] [CrossRef]
- Colleau, J.-J. An Indirect Approach to the Extensive Calculation of Relationship Coefficients. Genet. Sel. Evol. GSE 2002, 34, 409–421. [Google Scholar] [CrossRef]
- Caballero, A.; Toro, M.A. Interrelations between Effective Population Size and Other Pedigree Tools for the Management of Conserved Populations. Genet. Res. 2000, 75, 331–343. [Google Scholar] [CrossRef]
- Sheppard, P.M.; Wright, S. Evolution and the Genetics of Populations. Vol. 2. The Theory of Gene Frequencies. J. Anim. Ecol. 1969, 40, 266. [Google Scholar] [CrossRef]
- Gutiérrez, J.P.; Cervantes, I.; Molina, A.; Valera, M.; Goyache, F. Individual Increase in Inbreeding Allows Estimating Effective Sizes from Pedigrees. Genet. Sel. Evol. 2008, 40, 359–378. [Google Scholar] [CrossRef] [PubMed]
- Hill, W.G. A Note on Effective Population Size with Overlapping Generations. Genetics 1979, 92, 317. [Google Scholar] [CrossRef] [PubMed]
- Alderson, G.L. A System to Maximize the Maintenance of Genetic Variability in Small Populations. In Genetic Conservation of Domestic Livestock; CAB International: Wallingford, UK, 1992; pp. 18–29. ISBN 0851988091. [Google Scholar]
- de Rezende, M.P.G.; Malhado, C.H.M.; Biffani, S.; Souza Carneiro, P.L.; Bozzi, R. Genetic Diversity Derived from Pedigree Information and Estimation of Genetic Parameters for Reproductive Traits of Limousine and Charolais Cattle Raised in Italy. Ital. J. Anim. Sci. 2020, 19, 762–771. [Google Scholar] [CrossRef]
- Macor, L. Evaluación de La Variabilidad Genética Mediante el Número Efectivo en Braford Argentino. Master’s Thesis, Universidad Politecnica de Valencia, Valencia, Spain, 2013. [Google Scholar]
- Jarnecka, O.; Bauer, E.A.; Jagusiak, W. Pedigree Analysis in the Polish Red Cattle Population. Animal 2021, 15, 100238. [Google Scholar] [CrossRef] [PubMed]
- Toušová, R.; Ducháček, J.; Stádník, L.; Ptáček, M.; Beran, J. The Effect of Selected Factors on the Growth Ability of Charolais Cattle. Acta Univ. Agric. Silvic. Mendel. Brun. 2014, 62, 255–260. [Google Scholar] [CrossRef]
- Bouquet, A.; Venot, E.; Laloë, D.; Forabosco, F.; Fogh, A.; Pabiou, T.; Moore, K.; Eriksson, J.Å.; Renand, G.; Phocas, F. Genetic Structure of the European Charolais and Limousin Cattle Metapopulations Using Pedigree Analyses. J. Anim. Sci. 2011, 89, 1719–1730. [Google Scholar] [CrossRef]
- Ríos-Utrera, Á.; Montaño-Bermúdez, M.; Vega-Murillo, V.E.; Martínez-Velázquez, G.; Baeza-Rodríguez, J.J.; Román-Ponce, S.I. Genetic Diversity Evolution in the Mexican Charolais Cattle Population. Anim. Biosci. 2021, 34, 1116–1122. [Google Scholar] [CrossRef]
- Bhardwaj, S.; Togla, O.; Mumtaz, S.; Yadav, N.; Tiwari, J.; Muansangi, L.; Illa, S.K.; Wani, Y.M.; Mukherjee, S.; Mukherjee, A. Comparative Assessment of the Effective Population Size and Linkage Disequilibrium of Karan Fries Cattle Revealed Viable Population Dynamics. Anim. Biosci. 2024, 37, 795–806. [Google Scholar] [CrossRef]
- Boichard, D.; Maignel, L.; Verrier, E. Analyse Généalogique Des Races Bovines Laitières Françaises. INRAE Prod. Anim. 1996, 9, 323–335. [Google Scholar] [CrossRef]
- Čepon, M.; Simčič, M.; Malovrh, Š. Stima Dei Parametri Genetici Relativi Al Peso Corporeo Su Vitelli Charolais Allevati in Slovenia. Ital. J. Anim. Sci. 2009, 8, 735–742. [Google Scholar] [CrossRef]
- Doublet, A.C.; Restoux, G.; Fritz, S.; Balberini, L.; Fayolle, G.; Hozé, C.; Laloë, D.; Croiseau, P. Intensified Use of Reproductive Technologies and Reduced Dimensions of Breeding Schemes Put Genetic Diversity at Risk in Dairy Cattle Breeds. Animals 2020, 10, 1903. [Google Scholar] [CrossRef] [PubMed]
- Doublet, A.C.; Croiseau, P.; Fritz, S.; Michenet, A.; Hozé, C.; Danchin-Burge, C.; Laloë, D.; Restoux, G. The Impact of Genomic Selection on Genetic Diversity and Genetic Gain in Three French Dairy Cattle Breeds. Genet. Sel. Evol. 2019, 51, 52. [Google Scholar] [CrossRef] [PubMed]
- IDELE Les Races Bovines Allaitantes Affichent Un Faible Niveau de Consanguinité|Réussir Bovins Viande. Available online: https://www.reussir.fr/bovins-viande/les-races-bovines-allaitantes-affichent-un-faible-niveau-de-consanguinite (accessed on 26 August 2024).



| Demographic Parameter | Historical Period * | 2008–2012 Period | 2013–2017 Period | 2018–2022 Period |
| Number of animals with pedigree | 4961 | 1266 | 1781 | 1067 |
| Number of animals (reference population) | 3098 | 546 | 1254 | 1000 |
| Sires (total) | 470 | 105 | 157 | 133 |
| Dams (total) | 1593 | 356 | 717 | 485 |
| Individuals with progeny (offspring) | 2063 | 604 | 554 | 114 |
| Individuals without progeny (offspring) | 2898 | 662 | 1237 | 953 |
| Number of animals with both known parents | 3098 | 546 | 1254 | 1000 |
| Number of animals only with known sire | 258 | 88 | 98 | 6 |
| Number of animals only with known dam | 26 | 15 | 8 | 2 |
| Number of animals with no known parents | 1579 | 617 | 421 | 59 |
| Reproductive Parameter | Historical Period | 2008–2012 Period | 2013–2017 Period | 2018–2022 Period |
| Average number of calves per sire | 7.14 | 7.54 | 7.87 | 5.80 |
| Maximum number of calves per sire | 113 | 40 | 35 | 24 |
| Average number of calves per dam | 1.96 | 2.13 | 2.30 | 1.86 |
| Maximum number of calves per dam | 30 | 13 | 30 | 8 |
| Pedigree Completeness | Historical Period * (n = 4961) | 2008–2012 Period (n = 1266) | 2013–2017 Period (n = 1781) | 2018–2022 Period (n = 1067) |
|---|---|---|---|---|
| First generation (%) | 65.35 | 47.20 | 73.39 | 94.10 |
| Second generation (%) | 47.84 | 28.46 | 53.54 | 84.70 |
| Third generation (%) | 33.23 | 16.24 | 36.34 | 68.38 |
| Fourth generation (%) | 18.77 | 5.96 | 18.90 | 46.45 |
| Fifth generation (%) | 9.25 | 2.20 | 7.75 | 26.85 |
| Average GMax-FF | 7.83 | 6.00 | 6.86 | 8.36 |
| Average GCom-FF | 2.05 | 1.97 | 2.02 | 2.07 |
| Average GEqu-FF | 3.76 | 3.01 | 3.49 | 3.93 |
| Average GMax-PB | 3.57 | 2.23 | 4.05 | 6.38 |
| Average GCom-PB | 0.75 | 0.46 | 0.84 | 1.39 |
| Average GEqu-PB | 1.49 | 0.92 | 1.68 | 2.75 |
| Number of generations | 11 | 9 | 10 | 11 |
| Gene-Origin/Genetic Diversity Parameters | Reference Population (n = 3068) | 2008–2012 Period (n = 549) | 2013–2017 Period (n = 1253) | 2018–2022 Period (n = 970) |
|---|---|---|---|---|
| Historical population * (n) | 4937 | 1271 | 1777 | 1041 |
| Number of ancestors contributing to the reference population (n) | 1063 | 372 | 646 | 361 |
| Number of founders contributing to the reference population (n) | 1146 | 539 | 847 | 544 |
| Effective number of founders (fe) | 207.72 | 168.92 | 189.65 | 131.81 |
| Effective number of non-founders (Nenf) | 91.94 | 95.96 | 90.64 | 37.23 |
| Founder genome equivalents (fg) | 63.73 | 61.19 | 61.33 | 29.03 |
| Effective number of ancestors (fa) | 97 | 78 | 90 | 43 |
| fa/fe ratio | 0.47 | 0.46 | 0.47 | 0.33 |
| fg/fa ratio | 0.66 | 0.78 | 0.68 | 0.68 |
| fg/fe ratio | 0.31 | 0.36 | 0.32 | 0.22 |
| Number of ancestors to explain: | ||||
| 25% of gene pool | 10 | 8 | 10 | 5 |
| 50% of gene pool | 41 | 40 | 37 | 18 |
| 75% of gene pool | 203 | 146 | 161 | 58 |
| GD | 99.22 | 99.18 | 99.18 | 98.28 |
| 1-GD (GD loss) | 0.78 | 0.82 | 0.82 | 1.72 |
| DG* | 99.76 | 99.70 | 99.74 | 99.62 |
| Proportion of unequal contributions of the founders in GD loss (%) | 0.24 | 0.30 | 0.26 | 0.38 |
| Proportion of random genetic drift and bottle necks in GD loss (%) | 0.78 | 0.82 | 0.82 | 1.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cartuche-Macas, L.F.; Lozada, E.F.; Gutiérrez-Reinoso, M.A.; Chacón, E.; Navas, F.J.; García-Herreros, M. Evolution of Population Structure, Reproductive Performance, Inbreeding, and Genetic Diversity in Ecuadorian Charolais Cattle. Vet. Sci. 2024, 11, 566. https://doi.org/10.3390/vetsci11110566
Cartuche-Macas LF, Lozada EF, Gutiérrez-Reinoso MA, Chacón E, Navas FJ, García-Herreros M. Evolution of Population Structure, Reproductive Performance, Inbreeding, and Genetic Diversity in Ecuadorian Charolais Cattle. Veterinary Sciences. 2024; 11(11):566. https://doi.org/10.3390/vetsci11110566
Chicago/Turabian StyleCartuche-Macas, Luis F., Edwin F. Lozada, Miguel A. Gutiérrez-Reinoso, Edilberto Chacón, Francisco J. Navas, and Manuel García-Herreros. 2024. "Evolution of Population Structure, Reproductive Performance, Inbreeding, and Genetic Diversity in Ecuadorian Charolais Cattle" Veterinary Sciences 11, no. 11: 566. https://doi.org/10.3390/vetsci11110566
APA StyleCartuche-Macas, L. F., Lozada, E. F., Gutiérrez-Reinoso, M. A., Chacón, E., Navas, F. J., & García-Herreros, M. (2024). Evolution of Population Structure, Reproductive Performance, Inbreeding, and Genetic Diversity in Ecuadorian Charolais Cattle. Veterinary Sciences, 11(11), 566. https://doi.org/10.3390/vetsci11110566









