Feline Infectious Peritonitis Effusion Index: A Novel Diagnostic Method and Validation of Flow Cytometry-Based Delta Total Nucleated Cells Analysis on the Sysmex XN-1000V®
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection, Sampling, and Diagnostic Procedures
- Only samples from cats with effusions reported by veterinary practitioners who were clinically suspected of FIP (e.g., lethargy, anorexia, weight loss, fever, jaundice, dyspnoea, abdominal lymphadenopathy, thickened intestinal walls, renomegaly, uveitis, vasculitis/phlebitis, retinal changes, multifocal neurologic signs) were included in this study.
- Availability of laboratory requisition with the relevant clinical information such as clinical suspicion/clinical signs and requested analyses.
- Availability of complete documentation of physicochemical analysis of the effusion including specific gravity (SG), protein content, absolute counts of white blood cells and total nucleated cells in body fluid, and total nucleated cells derived from the ratio between specific channel (TNCC-WDF and TNCC-WNR) counts on the Sysmex XN-1000V®.
- Absence of complete clinical documentation: Samples lacking a comprehensive laboratory requisition or clinical information including clinical suspicion/clinical signs and requested analyses were excluded.
- Lack of confirmed FCoV real-time RT-qPCR diagnosis: Cases without a confirmed diagnosis of FCoV through real-time RT-qPCR testing were excluded from the study.
- Incomplete physicochemical analysis data: Effusion samples without full documentation of physicochemical analysis such as SG, protein content, absolute counts of white blood cells and total nucleated cells in body fluid, and the total nucleated cells derived from the ratio between the TNCC-WDF and TNCC-WNR counts on the Sysmex XN-1000V® were not included.
- Non-compliance with sampling protocols: Samples collected in non-EDTA tubes or samples submitted beyond the 18–24 h window post-collection were excluded to maintain the integrity of the cell count analysis.
- Samples with cytological findings that were septic, unclear, or inconclusive were excluded from the study.
- Previous treatments influencing effusion composition: Samples from cats that had undergone treatments known to affect the composition of effusions, such as recent chemotherapy or immunosuppressive therapy, were excluded to avoid confounding results.
- Insufficient sample volume: Effusions with insufficient volume to perform all required analyses were excluded to ensure that all parameters could be adequately measured.
2.2. Biochemical Analysis
2.3. Rivalta Test
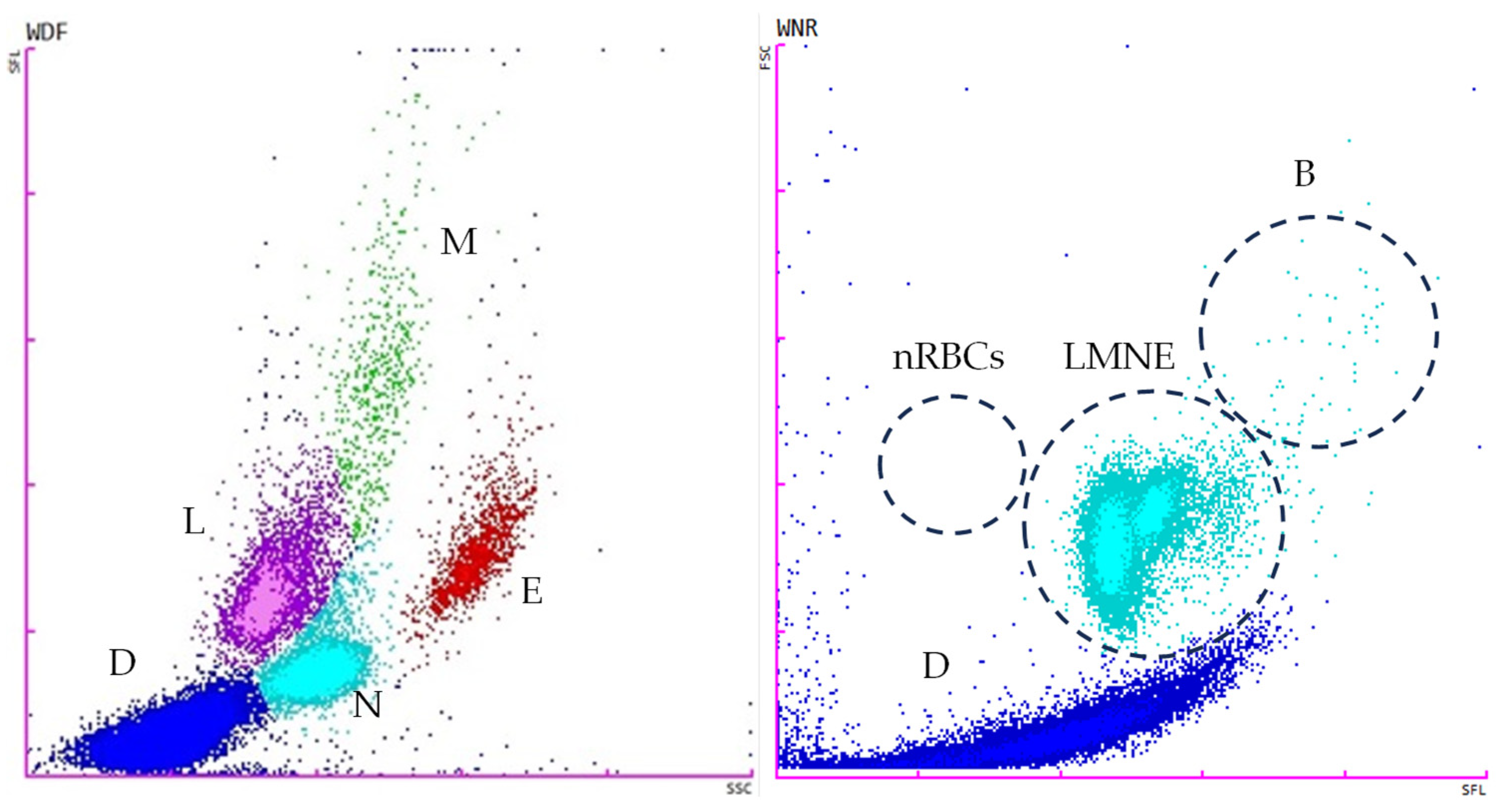
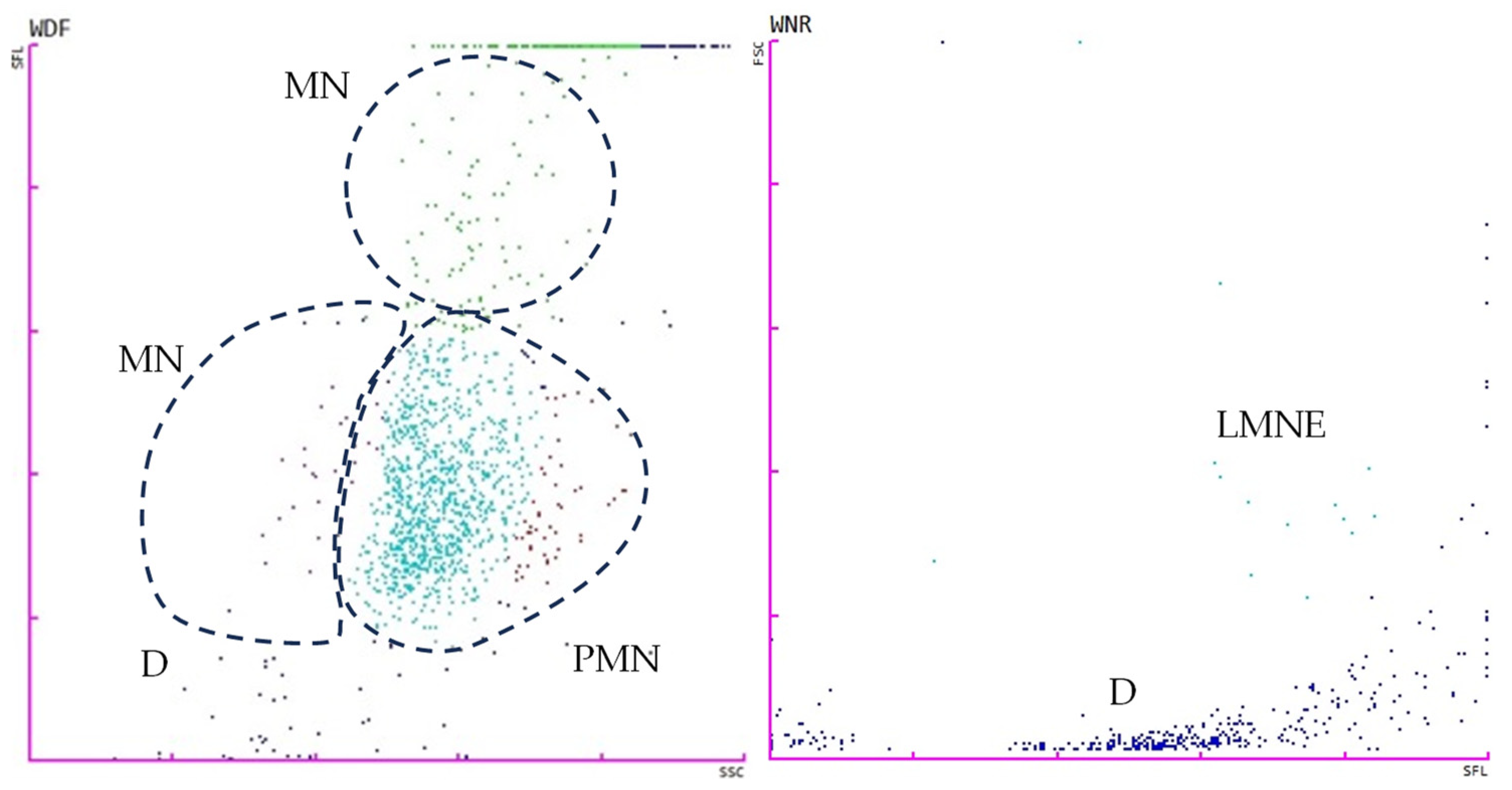
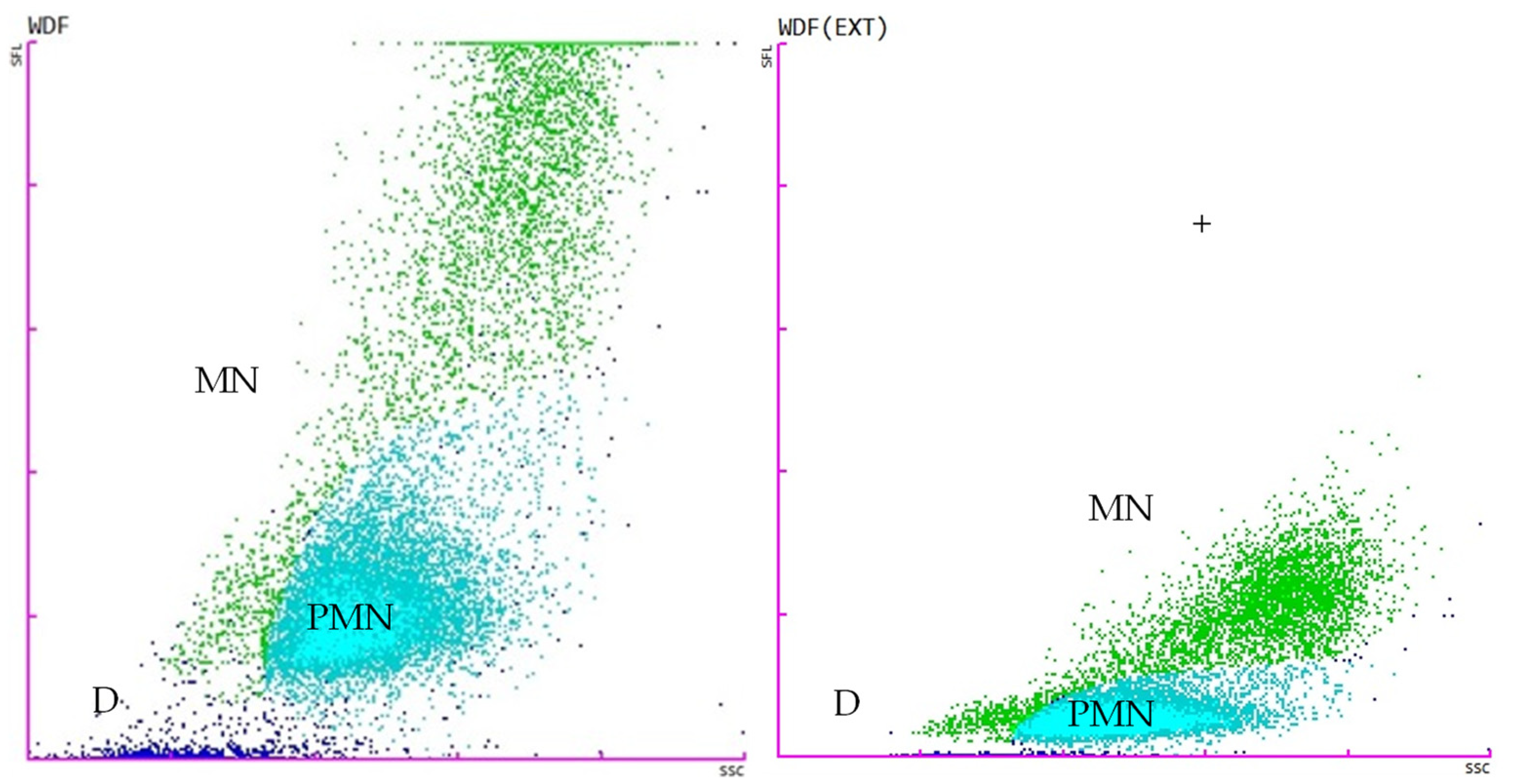
2.4. Flow Cytometry Analysis
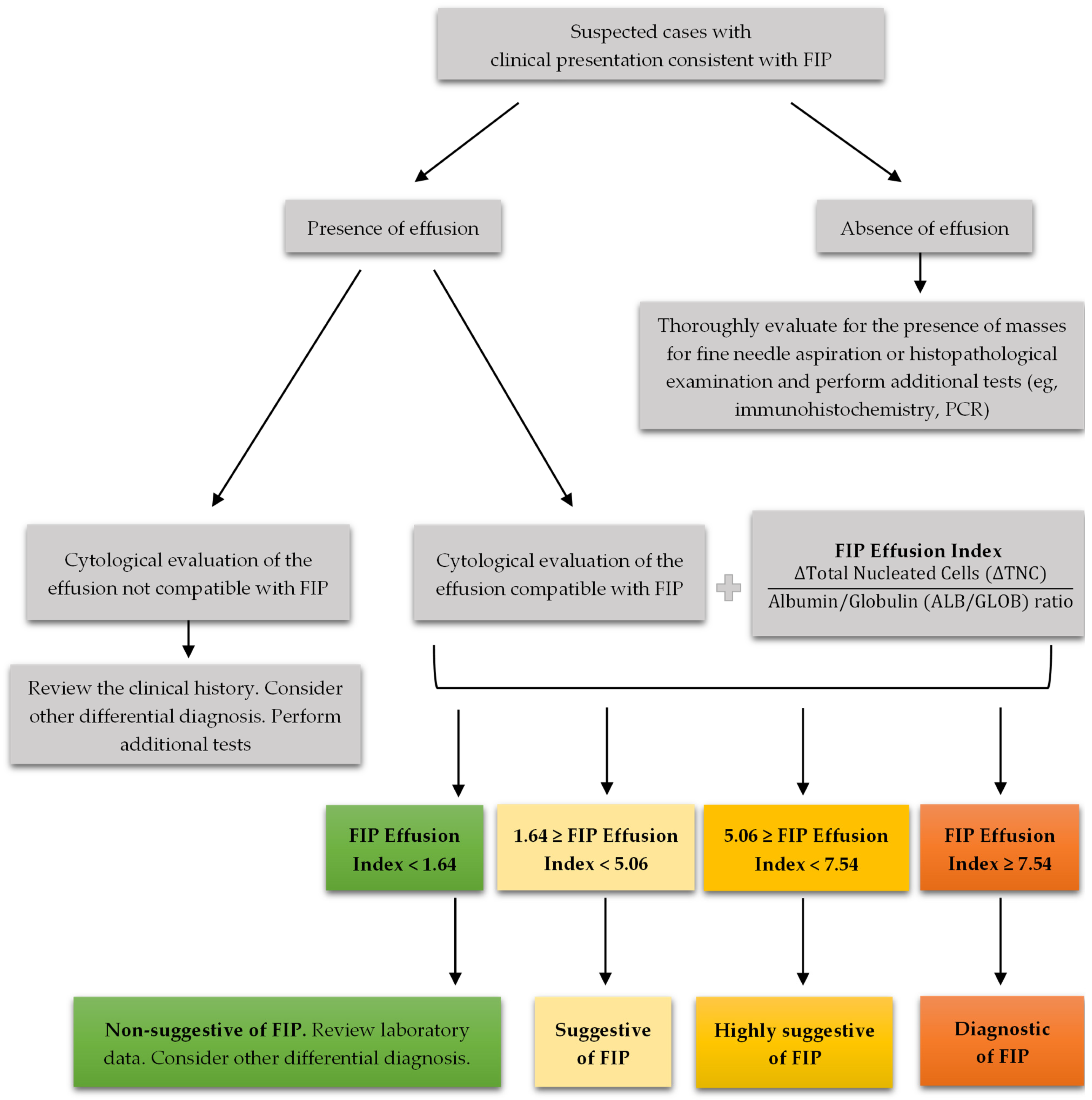
2.5. FIP Effusion Index
2.6. Effusion Classification
2.7. Molecular Analysis
- Amplification of the internal extraction control;
- No amplification of the negative control;
- Amplification of the positive control.
2.8. Statistical Analysis
3. Results
3.1. Qualitative Variables
3.1.1. Descriptive Data
3.1.2. Breed
3.1.3. Sex
3.1.4. Age
3.1.5. Rivalta Test Results
3.1.6. Effusion Classification
3.2. Quantitative Variables
3.2.1. Biochemical and Flow Cytometry Data
3.2.2. Diagnostic Accuracy of the ∆TNC
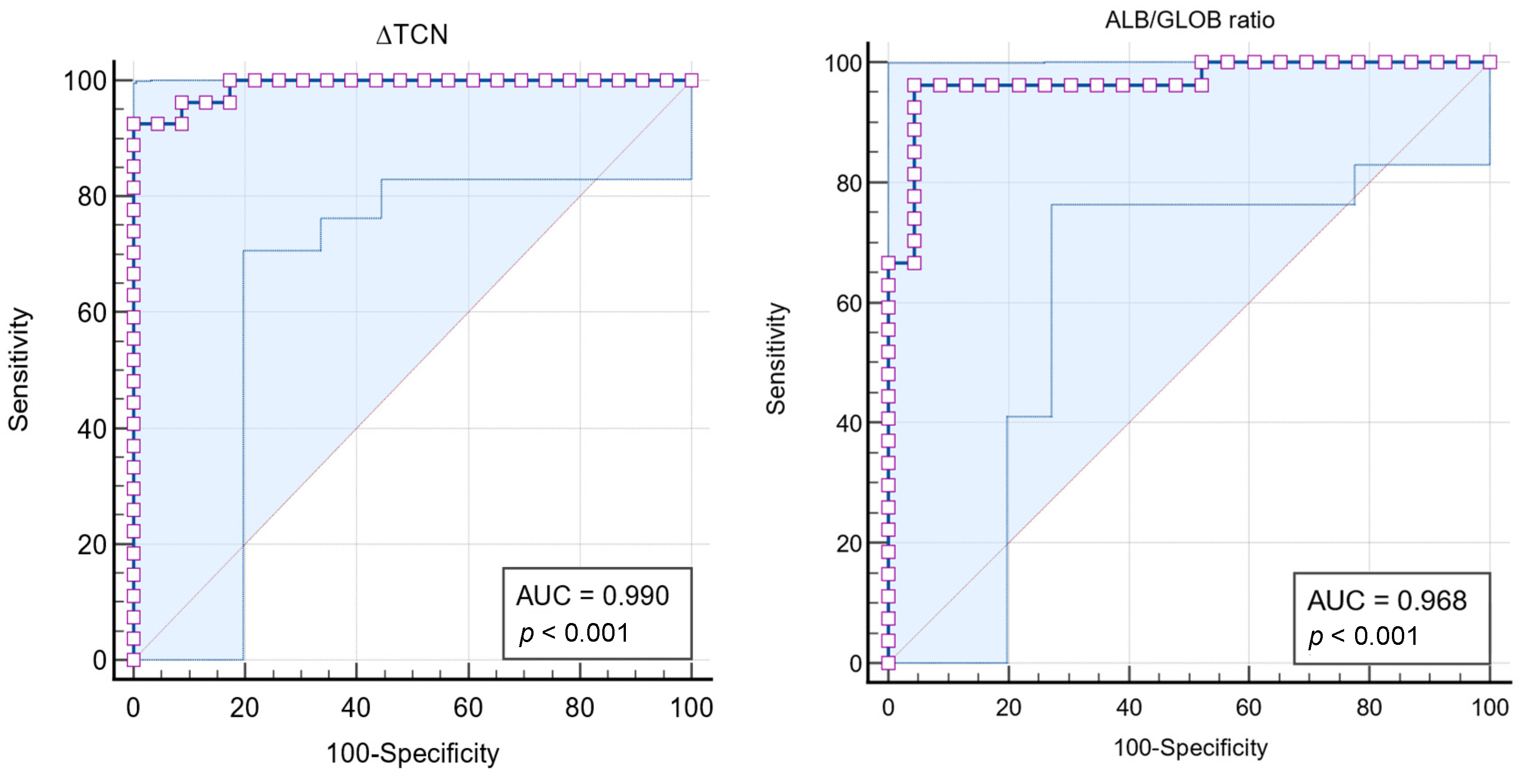
3.2.3. Receiver Operating Characteristic (ROC) Curve Analyses
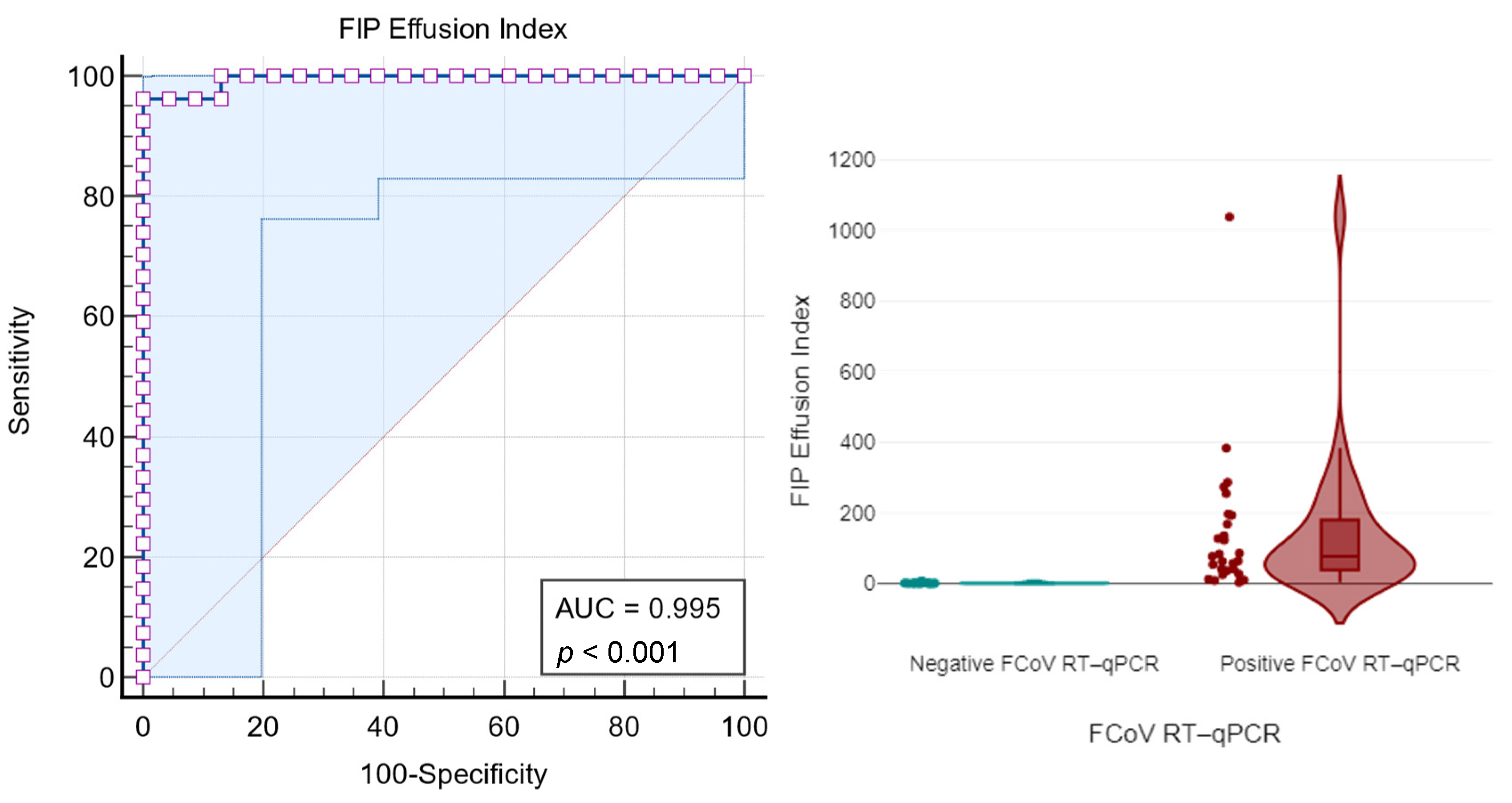
3.2.4. FIP Effusion Index
4. Discussion
4.1. Sex, Breed and Age
4.2. Effusion Classification
4.3. ∆TNC-XN, Rivalta Test, and ALB/GLOB Ratio in FIP Diagnostic Approaches
4.4. FIP Effusion Index: A Novel Diagnostic Method
4.5. Recommendations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Naseer, S.; Khalid, S.; Parveen, S.; Abbass, K.; Song, H.; Achim, M.V. COVID-19 outbreak: Impact on global economy. Front. Public Health 2023, 10, 1009393. [Google Scholar] [CrossRef] [PubMed]
- Clemente-Suárez, V.J.; Navarro-Jiménez, E.; Moreno-Luna, L.; Saavedra-Serrano, M.C.; Jimenez, M.; Simón, J.A.; Tornero-Aguilera, J.F. The impact of the COVID-19 pandemic on social, health, and economy. Sustainability 2021, 13, 6314. [Google Scholar] [CrossRef]
- Hiscott, J.; Alexandridi, M.; Muscolini, M.; Tassone, E.; Palermo, E.; Soultsioti, M.; Zevini, A. The global impact of the coronavirus pandemic. Cytokine Growth Factor Rev. 2020, 53, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Di Gennaro, F.; Pizzol, D.; Marotta, C.; Antunes, M.; Racalbuto, V.; Veronese, N.; Smith, L. Coronavirus diseases (COVID-19) Current status and future perspectives: A narrative review. Int. J. Environ. Res. Public Health 2020, 17, 2690. [Google Scholar] [CrossRef]
- Arshad Ali, S.; Baloch, M.; Ahmed, N.; Arshad Ali, A.; Iqbal, A. The outbreak of coronavirus disease 2019 (COVID-19)—An emerging global health threat. J. Infect. Public Health 2020, 13, 644–646. [Google Scholar] [CrossRef]
- Amoutzias, G.D.; Nikolaidis, M.; Tryfonopoulou, E.; Chlichlia, K.; Markoulatos, P.; Oliver, S.G. The remarkable evolutionary plasticity of coronaviruses by mutation and recombination: Insights for the COVID-19 pandemic and the future evolutionary paths of SARS-CoV-2. Viruses 2022, 14, 78. [Google Scholar] [CrossRef]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Krentz, D.; Zenger, K.; Alberer, M.; Felten, S.; Bergmann, M.; Dorsch, R.; Matiasek, K.; Kolberg, L.; Hofmann-Lehmann, R.; Meli, M.L.; et al. Curing cats with feline infectious peritonitis with an oral multi-component drug containing GS-441524. Viruses 2021, 13, 2228. [Google Scholar] [CrossRef]
- Guo, Y.-R.; Cao, Q.-D.; Hong, Z.-S.; Tan, Y.-Y.; Chen, S.-D.; Jin, H.-J.; Tan, K.-S.; Wang, D.-Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—An update on the status. Mil. Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef]
- Li, X.; Geng, M.; Peng, Y.; Meng, L.; Lu, S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020, 10, 102–108. [Google Scholar] [CrossRef]
- Prompetchara, E.; Ketloy, C.; Palaga, T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020, 38, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pavlin, D.; Grubelnik, G.; Kogoj, R.; Suljič, A.; Tozon, N.; Tatjana, A.Ž.; Korva, M. Investigation of possible recombination between feline coronavirus and SARS-CoV-2 in cats with feline infectious peritonitis. In Proceedings of the European Congress of Veterinary Internal Medicine—Companion Animals (ECVIM-CA), Lyon, France, 5–7 September 2024; p. 319. [Google Scholar]
- Wynants, L.; Van Calster, B.; Collins, G.S.; Riley, R.D.; Heinze, G.; Schuit, E.; Albu, E.; Arshi, B.; Bellou, V.; Bonten, M.M.J.; et al. Prediction models for diagnosis and prognosis of COVID-19: Systematic review and critical appraisal. BMJ 2020, 369, m1328. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Li, X.; Zhao, W.; Jia, S.; Zhang, X.; Irwin, D.M.; Zhang, S. Adaptive evolution of feline coronavirus genes based on selection analysis. BioMed Res. Int. 2020, 2020, 9089768. [Google Scholar] [CrossRef] [PubMed]
- Zuzzi-Krebitz, A.-M.; Buchta, K.; Bergmann, M.; Krentz, D.; Zwicklbauer, K.; Dorsch, R.; Wess, G.; Fischer, A.; Matiasek, K.; Hönl, A.; et al. Short treatment of 42 days with oral GS-441524 results in equal efficacy as the recommended 84-day treatment in cats suffering from feline infectious peritonitis with effusion—A prospective randomized controlled study. Viruses 2024, 16, 1144. [Google Scholar] [CrossRef]
- Sweet, A.N.; André, N.M.; Stout, A.E.; Licitra, B.N.; Whittaker, G.R. Clinical and molecular relationships between COVID-19 and feline infectious peritonitis (FIP). Viruses 2022, 14, 481. [Google Scholar] [CrossRef]
- Chu, H.; Chan, J.F.-W.; Wang, Y.; Yuen, T.T.-T.; Chai, Y.; Hou, Y.; Shuai, H.; Yang, D.; Hu, B.; Huang, X.; et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: An ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 2020, 71, 1400–1409. [Google Scholar] [CrossRef]
- Gao, Y.-Y.; Wang, Q.; Liang, X.-Y.; Zhang, S.; Bao, D.; Zhao, H.; Li, S.-B.; Wang, K.; Hu, G.-X.; Gao, F.-S. An updated review of feline coronavirus: Mind the two biotypes. Virus Res. 2023, 326, 199059. [Google Scholar] [CrossRef]
- Hosie, M.J.; Hofmann-Lehmann, R.; Hartmann, K.; Egberink, H.; Truyen, U.; Addie, D.D.; Belák, S.; Boucraut-Baralon, C.; Frymus, T.; Lloret, A.; et al. Anthropogenic infection of cats during the 2020 COVID-19 pandemic. Viruses 2021, 13, 185. [Google Scholar] [CrossRef]
- Michelitsch, A.; Hoffmann, D.; Wernike, K.; Beer, M. Occurrence of antibodies against SARS-CoV-2 in the domestic cat population of Germany. Vaccines 2020, 8, 772. [Google Scholar] [CrossRef]
- Bosco-Lauth, A.M.; Hartwig, A.E.; Porter, S.M.; Gordy, P.W.; Nehring, M.; Byas, A.D.; VandeWoude, S.; Ragan, I.K.; Maison, R.M.; Bowen, R.A. Experimental infection of domestic dogs and cats with SARS-CoV-2: Pathogenesis, transmission, and response to reexposure in cats. Proc. Natl. Acad. Sci. USA 2020, 117, 26382–26388. [Google Scholar] [CrossRef]
- Moyadee, W.; Sunpongsri, S.; Choowongkomon, K.; Roytrakul, S.; Rattanasrisomporn, A.; Tansakul, N.; Rattanasrisomporn, J. Feline infectious peritonitis: A comprehensive evaluation of clinical manifestations, laboratory diagnosis, and therapeutic approaches. J. Adv. Vet. Anim. Res. 2024, 11, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhu, Z.; Du, J.; Zhu, X.; Pan, C.; Yin, C.; Sun, W. Development of multiplex real-time PCR for simultaneous detection of SARS-CoV-2, CCoV, and FIPV. Front. Vet. Sci. 2024, 11, 1337690. [Google Scholar] [CrossRef] [PubMed]
- Deng, K.; Nemser, S.M.; Frost, K.; Goodman, L.B.; Ip, H.S.; Killian, M.L.; Ulaszek, J.; Kiener, S.; Kmet, M.; Uhlig, S.; et al. Successful detection of delta and omicron variants of SARS-CoV-2 by veterinary diagnostic laboratory participants in an interlaboratory comparison exercise. J. Appl. Lab. Med. 2023, 8, 726–741. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–Coronavirus 2. Science (1979) 2020, 368, 1016–1020. [Google Scholar] [CrossRef]
- Alexander, M.R.; Schoeder, C.T.; Brown, J.A.; Smart, C.D.; Moth, C.; Wikswo, J.P.; Capra, J.A.; Meiler, J.; Chen, W.; Madhur, M.S. Predicting susceptibility to SARS-CoV-2 infection based on structural differences in ACE2 across species. FASEB J. 2020, 34, 15946–15960. [Google Scholar] [CrossRef]
- Decaro, N.; Mari, V.; Lanave, G.; Lorusso, E.; Lucente, M.S.; Desario, C.; Colaianni, M.L.; Elia, G.; Ferringo, F.; Alfano, F.; et al. Mutation analysis of the spike protein in italian feline infectious peritonitis virus and feline enteric coronavirus sequences. Res. Vet. Sci. 2021, 135, 15–19. [Google Scholar] [CrossRef]
- Meli, M.; Helfer-Hungerbuehler, A.K.; Spiri, A.; Meili, T.; Riond, B.; Krentz, D.; Zwicklbauer, K.; Bucht, K.; Zuzzi-Krebitz, A.M.; Hartmann, K.; et al. Alpha-1-acid glycoprotein quantification via spatial proximity analyte reagent capture luminescence assay—Validation and application as diagnostic and prognostic marker in serum and effusions of cats with feline infectious peritonitis during GS-441524 treatment. In Proceedings of the European Congress of Veterinary Internal Medicine—Companion Animals (ECVIM-CA), Lyon, France, 5–7 September 2024; p. 305. [Google Scholar]
- Farsijani, F.; Safi, S.; Shirazi Beheshtiha, S.H. Comparison of the performance of bioresonance, electrophoresis and RT-PCR in the diagnosis of feline infectious peritonitis. Arch. Razi Inst. 2023, 78, 1077–1085. [Google Scholar]
- Thayer, V.; Gogolski, S.; Felten, S.; Hartmann, K.; Kennedy, M.; Olah, G.A. 2022 AAFP/EveryCat feline infectious peritonitis diagnosis guidelines. J. Feline Med. Surg. 2022, 24, 905–933, Erratum in J. Feline Med. Surg. 2022, 24, e676. [Google Scholar] [CrossRef]
- Jaimes, J.A.; Millet, J.K.; Stout, A.E.; André, N.M.; Whittaker, G.R. A tale of two viruses: The distinct spike glycoproteins of feline coronaviruses. Viruses 2020, 12, 83. [Google Scholar] [CrossRef]
- Cook, S.; Castillo, D.; Williams, S.; Haake, C.; Murphy, B. Serotype I and II feline coronavirus replication and gene expression patterns of feline cells—Building a better understanding of serotype I FIPV biology. Viruses 2022, 14, 1356. [Google Scholar] [CrossRef]
- Kennedy, M.A. Feline infectious peritonitis: Update on pathogenesis, diagnostics, and treatment. Vet. Clin. N. Am. Small Anim. Pract. 2020, 50, 1001–1011. [Google Scholar] [CrossRef] [PubMed]
- Felten, S.; Hartmann, K. Diagnosis of feline infectious peritonitis: A review of the current literature. Viruses 2019, 11, 1068. [Google Scholar] [CrossRef] [PubMed]
- Licitra, B.N.; Millet, J.K.; Regan, A.D.; Hamilton, B.S.; Rinaldi, V.D.; Duhamel, G.E.; Whittaker, G.R. Mutation in spike protein cleavage site and pathogenesis of feline coronavirus. Emerg. Infect. Dis. 2013, 19, 1066–1073. [Google Scholar] [CrossRef]
- Stranieri, A.; Giordano, A.; Paltrinieri, S.; Giudice, C.; Cannito, V.; Lauzi, S. Comparison of the performance of laboratory tests in the diagnosis of feline infectious peritonitis. J. Vet. Diagn. Investig. 2018, 30, 459–463. [Google Scholar] [CrossRef]
- Geks, A.K.; Walter, H.; Steffensen, N.; Volk, H.; Rieder, J.C.; Busse, C. Detection of S gene mutation (M 1058L and feline coronavirus ribonucleic acid in the aqueous humour of three living cats with uveitis and three negative controls. In Proceedings of the European Congress of Veterinary Internal Medicine—Companion Animals (ECVIM-CA), Lyon, France, 5–7 September 2024; p. 328. [Google Scholar]
- Tasker, S.; Addie, D.D.; Egberink, H.; Hofmann-Lehmann, R.; Hosie, M.J.; Truyen, U.; Belák, S.; Boucraut-Baralon, C.; Frymus, T.; Lloret, A.; et al. Feline infectious peritonitis: European advisory board on cat diseases guidelines. Viruses 2023, 15, 1847. [Google Scholar] [CrossRef]
- Addie, D.; Belák, S.; Boucraut-Baralon, C.; Egberink, H.; Frymus, T.; Gruffydd-Jones, T.; Hartmann, K.; Hosie, M.J.; Lloret, A.; Lutz, H.; et al. Feline infectious peritonitis: ABCD guidelines on prevention and management. J. Feline Med. Surg. 2009, 11, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Fonte-Oliveira, L.; Lopes, C.; Malhão, F.; Canadas-Sousa, A.; Gregório, H.; Marcos, R.; Santos, M. The utility of the cell tube block technique in the diagnosis of effusive feline infectious peritonitis. In Proceedings of the Congress of European Society of Veterinary Clinical Pathology (ESVCP/ECVCP), University of Veterinary Medicine, Budapest, Hungary, 28–31 August 2024; p. 148. [Google Scholar]
- Kipar, A.; Meli, M.L. Feline infectious peritonitis. Vet. Pathol. 2014, 51, 505–526. [Google Scholar] [CrossRef]
- Giordano, A.; Stranieri, A.; Rossi, G.; Paltrinieri, S. High diagnostic accuracy of the Sysmex XT-2000iV delta total nucleated cells on effusions for feline infectious peritonitis. Vet. Clin. Pathol. 2015, 44, 295–302. [Google Scholar] [CrossRef]
- Zanetti, M.; Paltrinieri, S.; Meazzi, S.; Novellini, S.; Martini, V.; Lauzi, S.; Rossi, S.; Mangiagalli, G.; Giordano, A. Delta total nucleated cells assessed via Sysmex XT-2000iV and Sysmex XN-1000V on feline effusions: Comparable diagnostic accuracy for feline infectious peritonitis. In Proceedings of the European Congress of Veterinary Pathology & Clinical Pathology (ESVP/ECVP/ESVCP/ECVCP), Lisbon, Portugal, 30 August–2 September 2023. [Google Scholar]
- Wenk, J.M.; Meli, M.L.; De Witt, C.C.; Bouzon, A.B.; Pineroli, B.; Felten, S.; Meunier, S.M.; Zwicklbauer, K.; Hartmann, K. Feline coronavirus loads and presence of co-infections in cats diagnosed with feline infectious peritonitis. In Proceedings of the European Congress of Veterinary Internal Medicine—Companion Animals (ECVIM-CA), Lyon, France, 5–7 September 2024; p. 306. [Google Scholar]
- Yu, B.; Xu, C.; Huang, S.; Ni, J.; Zhou, J.; Zhang, Y.; Wu, M.; Zhang, J.; Fang, L. Development of a universal real-time RT-PCR assay for detection of pan-SARS-coronaviruses with an RNA-based internal control. Front. Microbiol. 2023, 14, 1181097. [Google Scholar] [CrossRef]
- Phongroop, K.; Rattanasrisomporn, J.; Tangtrongsup, S.; Rungsipipat, A.; Piewbang, C.; Techangamsuwan, S. High-resolution melting analysis for simultaneous detection and discrimination between wild-type and vaccine strains of feline calicivirus. Vet. Q. 2023, 43, 1–12. [Google Scholar] [CrossRef]
- Jähne, S.; Felten, S.; Bergmann, M.; Erber, K.; Matiasek, K.; Meli, M.L.; Hofmann-Lehmann, R.; Hartmann, K. Detection of feline coronavirus variants in cats without feline infectious peritonitis. Viruses 2022, 14, 1671. [Google Scholar] [CrossRef] [PubMed]
- Felten, S.; Leutenegger, C.M.; Balzer, H.-J.; Pantchev, N.; Matiasek, K.; Wess, G.; Egberink, H.; Hartmann, K. Sensitivity and specificity of a real-time reverse transcriptase polymerase chain reaction detecting feline coronavirus mutations in effusion and serum/plasma of cats to diagnose feline infectious peritonitis. BMC Vet. Res. 2017, 13, 228. [Google Scholar] [CrossRef] [PubMed]
- Fischer, Y.; Sauter-Louis, C.; Hartmann, K. Diagnostic accuracy of the rivalta test for feline infectious peritonitis. Vet. Clin. Pathol. 2012, 41, 558–567. [Google Scholar] [CrossRef] [PubMed]
- Sakai, N.; Iijima, S.; Shiba, K. Reinvestigation of clinical value of rivalta reaction of puncture fluid. Rinsho Byori 2004, 52, 877–882. [Google Scholar]
- Bossuyt, P.M.; Reitsma, J.B.; Bruns, D.E.; Gatsonis, C.A.; Glasziou, P.P.; Irwig, L.; Lijmer, J.G.; Moher, D.; Rennie, D.; de Vet, H.C.W.; et al. STARD 2015: An updated list of essential items for reporting diagnostic accuracy studies. Clin. Chem. 2015, 61, 1446–1452. [Google Scholar] [CrossRef]
- Washabau, R.J.; Day, M.J. Canine and Feline Gastroenterology; Elsevier: Amsterdam, The Netherlands, 2013. [Google Scholar] [CrossRef]
- Stockham, S.L.; Scott, M.A. Fundamentals of Veterinary Clinical Pathology, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Gavazza, A.; Turinelli, V.; Lubas, G. Effusion in the cat: Classification of 396 fluids according to a problem-oriented scheme. Comp. Clin. Path. 2013, 22, 517–521. [Google Scholar] [CrossRef]
- Zuraidawati, Z.; Dewi, M.; Syah, N.Y.; Zamzami, R.S. Use of vinegar as a candidate substitute of acetic acid (CH3COOH) in the rivalta test method in diagnosis feline infectious peritonitis (FIP) domestic cat. Int. J. Trop. Vet. Biomed. Res. 2023, 7, 11–14. [Google Scholar] [CrossRef]
- Fischer, Y.; Weber, K.; Sauter-Louis, C.; Hartmann, K. The rivalta’s test as a diagnostic variable in feline effusions—Evaluation of optimum reaction and storage conditions. Tierärztl. Prax. Ausg. K Kleintiere/Heimtiere 2013, 41, 297–303. [Google Scholar] [CrossRef]
- Valenciano, A.C.; Rizzi, T.E. Abdominal, thoracic, and pericardial effusions. In Cowell and Tyler’s Diagnostic Cytology and Hematology of the Dog and Cat; Elsevier: Amsterdam, The Netherlands, 2020; pp. 229–246. [Google Scholar] [CrossRef]
- Hazuchova, K.; Held, S.; Klemm, I.; Bauer, N. Simplified Light’s criteria and acute phase proteins reflect aetiology of feline body cavity effusions better than the traditional classification scheme. Animals 2023, 13, 1918. [Google Scholar] [CrossRef]
- Lythgoe, K.A.; Hall, M.; Ferretti, L.; de Cesare, M.; MacIntyre-Cockett, G.; Trebes, A.; Andersson, M.; Otecko, N.; Wise, E.L.; Moore, N.; et al. SARS-CoV-2 within-host diversity and transmission. Science (1979) 2021, 372, eabg0821. [Google Scholar] [CrossRef]
- Tasker, S. Diagnosis of feline infectious peritonitis: Update on evidence supporting available tests. J. Feline Med. Surg. 2018, 20, 228–243. [Google Scholar] [CrossRef]
- Hartmann, K. New treatment options for cats with feline infectious peritonitis. In Proceedings of the European Congress of Veterinary Internal Medicine—Companion Animals (ECVIM-CA), Lyon, France, 5–7 September 2024; p. 340. [Google Scholar]
- Buchta, K.; Zuzzi-Krebitz, A.M.; Bergmann, M.; Krentz, D.; Zwicklbauer, K.; Dorsch, R.; Wess, G.; Fischer, A.; Hofmann-Lehmann, R.; Meli, M.L.; et al. Shorter treatment duration of 42 days with oral GS-441524 also leads to complete remission in cats with feline infectious peritonitis. In Proceedings of the European Congress of Veterinary Internal Medicine—Companion Animals (ECVIM-CA), Lyon, France, 5–7 September 2024; p. 309. [Google Scholar]
- Schlachet, A.; Schuller, S.; Carranza Valencia, A. Unlicensed antiviral treatment with GS-441524: How are clinicians approaching feline infectious peritonitis in primary care practices? In Proceedings of the European Congress of Veterinary Internal Medicine—Companion Animals (ECVIM-CA), Lyon, France, 5–7 September 2024; p. 308. [Google Scholar]
- Cerna, P.; Ross, E.; Willis, M.; Visser, L.; Hawley, J.; Lappin, M. Assessment of cardiac changes in cats with feline infectious peritonitis. In Proceedings of the European Congress of Veterinary Internal Medicine—Companion Animals (ECVIM-CA), Lyon, France, 5–7 September 2024; p. 307. [Google Scholar]
- Riemer, F.; Kuehner, K.A.; Ritz, S.; Sauter-Louis, C.; Hartmann, K. Clinical and laboratory features of cats with feline infectious peritonitis—A retrospective study of 231 confirmed cases (2000–2010). J. Feline Med. Surg. 2016, 18, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Li, T.; Wang, C.; Liu, X.; Ouyang, H.; Ji, W.; Liu, J.; Liao, X.; Li, J.; Hu, C. A retrospective study of clinical and laboratory features and treatment on cats highly suspected of feline infectious peritonitis in Wuhan, China. Sci. Rep. 2021, 11, 5208. [Google Scholar] [CrossRef] [PubMed]
- Pesteanu-Somogyi, L.D.; Radzai, C.; Pressler, B.M. Prevalence of feline infectious peritonitis in specific cat breeds. J. Feline Med. Surg. 2006, 8, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Norris, J.; Bosward, K.; White, J.; Baral, R.; Catt, M.; Malik, R. Clinicopathological findings associated with feline infectious peritonitis in Sydney, Australia: 42 cases (1990–2002). Aust. Vet. J. 2005, 83, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Worthing, K.A.; Wigney, D.I.; Dhand, N.K.; Fawcett, A.; McDonagh, P.; Malik, R.; Norris, J.M. Risk factors for feline infectious peritonitis in australian cats. J. Feline Med. Surg. 2012, 14, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.C.; Liu, H.; Gandolfi, B.; Lyons, L.A. The influence of age and genetics on natural resistance to experimentally induced feline infectious peritonitis. Vet. Immunol. Immunopathol. 2014, 162, 33–40. [Google Scholar] [CrossRef]
- Savary, K.; Sellon, R.; Law, J. Chylous abdominal effusion in a cat with feline infectious peritonitis. J. Am. Anim. Hosp. Assoc. 2001, 37, 35–40. [Google Scholar] [CrossRef]
- Zwicklbauer, K.; von La Roche, D.; Krentz, D.; Kolberg, L.; Alberer, M.; Hartmann, K.; von Both, U.U.; Härtle, S. Characterization of the lymphocyte response in cats with feline infectious peritonitis during antiviral treatment using the smart tube technology for flow cytometry in feline full blood samples. In Proceedings of the European Congress of Veterinary Internal Medicine—Companion Animals (ECVIM-CA), Lyon, France, 5–7 September 2024; p. 304. [Google Scholar]
- Herrewegh, A.A.; de Groot, R.J.; Cepica, A.; Egberink, H.F.; Horzinek, M.C.; Rottier, P.J. Detection of feline coronavirus RNA in feces, tissues, and body fluids of naturally infected cats by reverse transcriptase PCR. J. Clin. Microbiol. 1995, 33, 684–689. [Google Scholar] [CrossRef]
- Kennedy, M.A.; Brenneman, K.; Millsaps, R.K.; Black, J.; Potgieter, L.N.D. Correlation of genomic detection of feline coronavirus with various diagnostic assays for feline infectious peritonitis. J. Vet. Diagn. Investig. 1998, 10, 93–97. [Google Scholar] [CrossRef]
- Barker, E.N.; Stranieri, A.; Helps, C.R.; Porter, E.L.; Davidson, A.D.; Day, M.J.; Knowles, T.; Kipar, A.; Tasker, S. Limitations of using feline coronavirus spike protein gene mutations to diagnose feline infectious peritonitis. Vet. Res. 2017, 48, 60. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.-Y.; Chueh, L.-L.; Lin, C.-N.; Su, B.-L. Clinicopathological findings and disease staging of feline infectious peritonitis: 51 cases from 2003 to 2009 in Taiwan. J. Feline Med. Surg. 2011, 13, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, K.; Binder, C.; Hirschberger, J.; Cole, D.; Reinacher, M.; Schroo, S.; Frost, J.; Egberink, H.; Lutz, H.; Hermanns, W. Comparison of different tests to diagnose feline infectious peritonitis. J. Vet. Intern. Med. 2003, 17, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Felten, S.; Weider, K.; Doenges, S.; Gruendl, S.; Matiasek, K.; Hermanns, W.; Mueller, E.; Matiasek, L.; Fischer, A.; Weber, K.; et al. Detection of feline coronavirus spike gene mutations as a tool to diagnose feline infectious peritonitis. J. Feline Med. Surg. 2017, 19, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Felten, S.; Matiasek, K.; Leutenegger, C.M.; Sangl, L.; Herre, S.; Dörfelt, S.; Fischer, A.; Hartmann, K. Diagnostic value of detecting feline coronavirus RNA and spike gene mutations in cerebrospinal fluid to confirm feline infectious peritonitis. Viruses 2021, 13, 186. [Google Scholar] [CrossRef]
- Doenges, S.J.; Weber, K.; Dorsch, R.; Fux, R.; Hartmann, K. Comparison of real-time reverse transcriptase polymerase chain reaction of peripheral blood mononuclear cells, serum and cell-free body cavity effusion for the diagnosis of feline infectious peritonitis. J. Feline Med. Surg. 2017, 19, 344–350. [Google Scholar] [CrossRef]
- Longstaff, L.; Porter, E.; Crossley, V.J.; Hayhow, S.E.; Helps, C.R.; Tasker, S. Feline coronavirus quantitative reverse transcriptase polymerase chain reaction on effusion samples in cats with and without feline infectious peritonitis. J. Feline Med. Surg. 2017, 19, 240–245. [Google Scholar] [CrossRef]
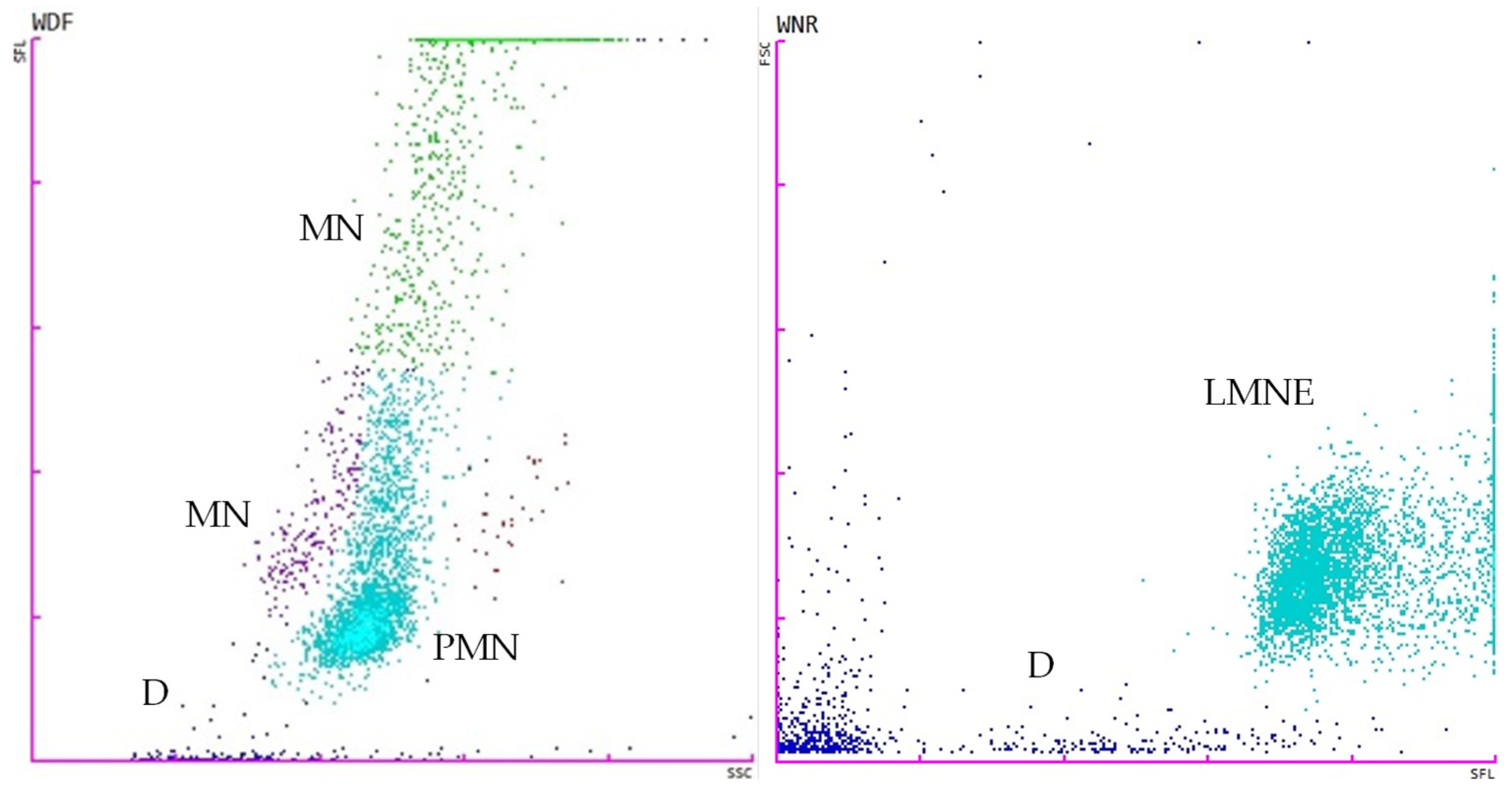
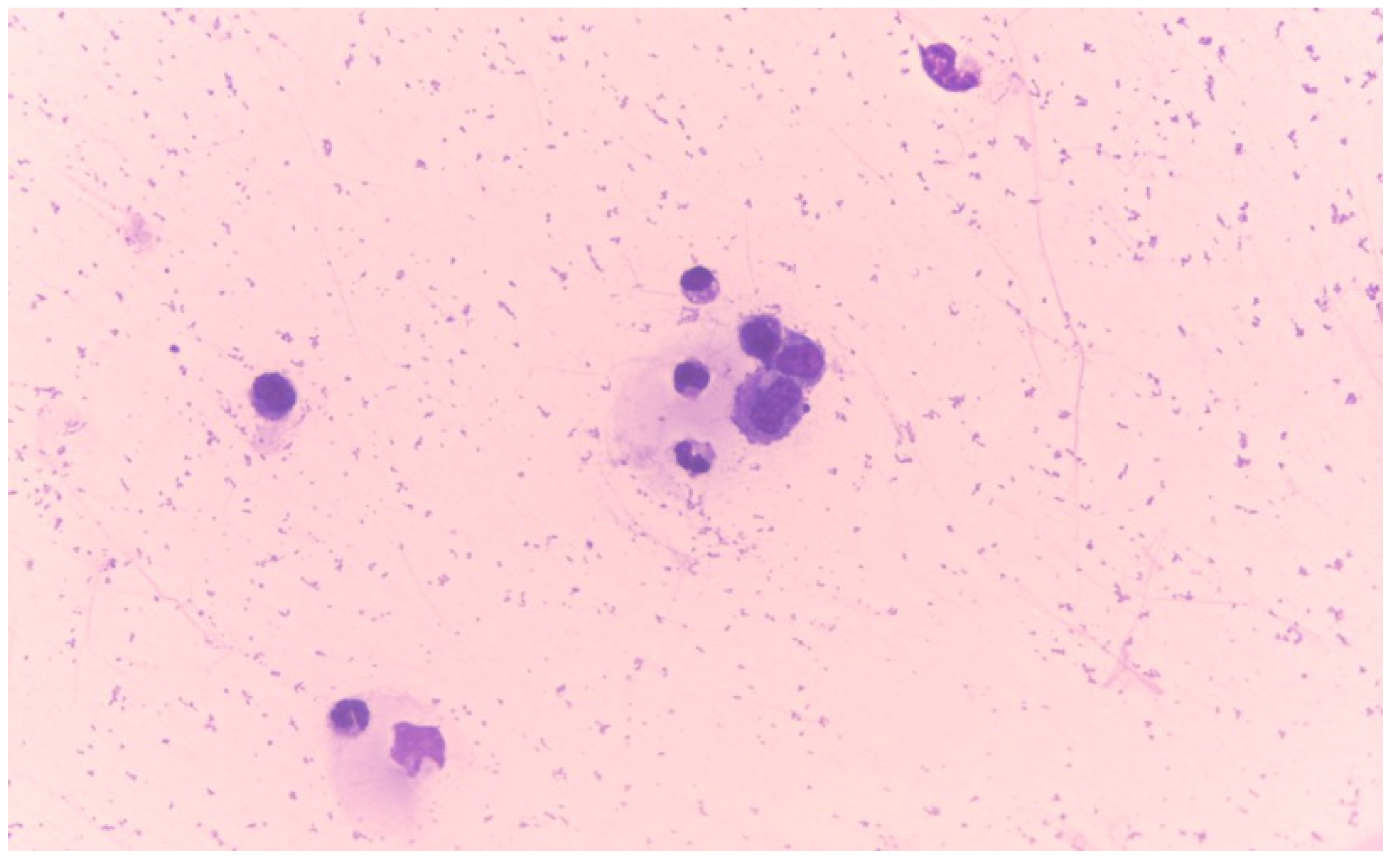
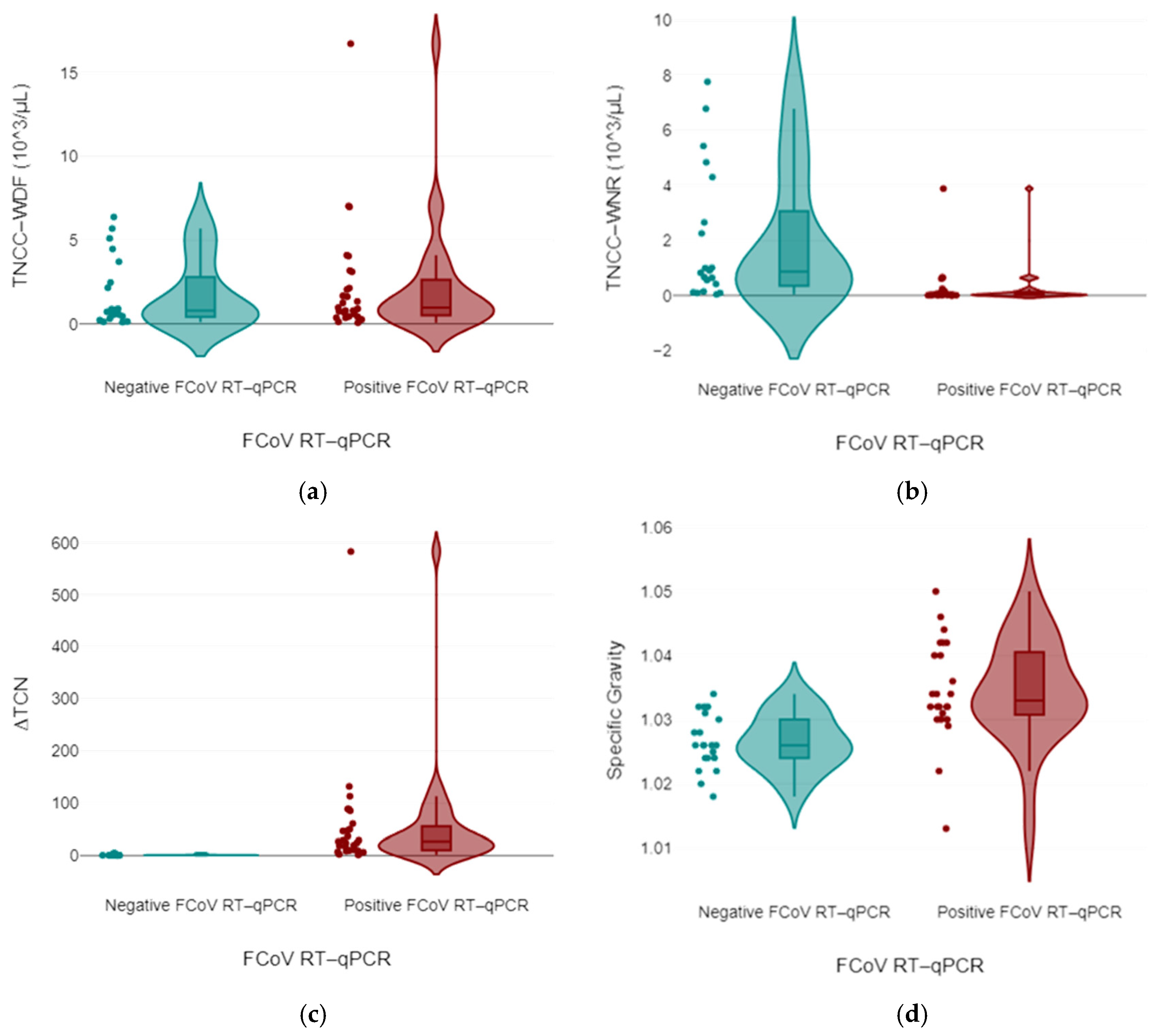

| Type | Total Protein (g/dL) | Total Cell Count (cells/μL) |
|---|---|---|
| Transudate | <2.5 | <1500 |
| Modified transudate | 2.5–7.5 | 1000–7000 |
| Exudate | >3.0 | >7000 |
| Cycles | Temperature | Time | Notes |
|---|---|---|---|
| 1 | 50 °C | 20 min | Reverse transcription |
| 1 | 95 °C | 2 min | Polymerase activation |
| 40 | 95 °C | 5 s | Denaturation |
| 60 °C | 30 s | Annealing/Extension |
| Feline Coronavirus RT-qPCR | |||
|---|---|---|---|
| Sex | Negative | Positive | Total |
| Female | 11 (22%) | 7 (14%) | 18 (36%) |
| Male | 12 (24%) | 20 (40%) | 32 (64%) |
| Total | 23 (46%) | 27 (54%) | 50 (100%) |
| Age Group | |||||||
|---|---|---|---|---|---|---|---|
| Kitten (<1 Year Old) | Young (1 to <2 Years Old) | Adult (2 to <6 Years Old) | Senior (6 to <11 Years Old) | Old (≥11 Years Old) | Total | ||
| Feline Coronavirus RT-qPCR | Negative | 4 (8%) | 6 (12%) | 5 (10%) | 3 (6%) | 5 (10%) | 23 (46%) |
| Positive | 9 (18%) | 6 (12%) | 8 (16%) | 4 (8%) | 0 (0%) | 27 (54%) | |
| Total | 13 (26%) | 12 (24%) | 13 (26%) | 7 (14%) | 5 (10%) | 50 (100%) | |
| Rivalta Test | ||||||
|---|---|---|---|---|---|---|
| Negative | Positive | Total | ||||
| n | % Within Rivalta Test | n | % Within Rivalta Test | n | ||
| Feline Coronavirus RT-qPCR | Negative | 16 (32%) | 69.6% (specificity) | 7 (14%) | 30.4% | 23 (46%) |
| Positive | 1 (2%) | 3.7% | 26 (52%) | 96.3% (sensitivity) | 27 (54%) | |
| Total | 17 (34%) | 33 (66%) | 50 (100%) | |||
| Effusion Type | ||||
|---|---|---|---|---|
| Modified Transudate | Exudate | Total | ||
| Feline Coronavirus RT-qPCR | Negative | 21 (42%) | 2 (4%) | 23 (46%) |
| Positive | 25 (50%) | 2 (4%) | 27 (54%) | |
| Total | 46 (92%) | 4 (8%) | 50 (100%) | |
| Feline Coronavirus RT-qPCR in Effusions | ||||
|---|---|---|---|---|
| Effusion | Negative | Positive | ||
| Parameter (Units) | Minimum | Maximum | Median (IQR) | Median (IQR) |
| Specific gravity (SG) | 1.01 | 1.05 | 1.03 | 1.03 |
| TP (g/dL) | 2.80 | 10.76 | 4.00 | 5.64 |
| ALB (g/dL) | 1.02 | 2.47 | 1.93 | 1.65 |
| GLOB (g/dL) | 1.25 | 9.11 | 2.00 | 4.15 |
| ALB/GLOB ratio | 0.18 | 1.5 | 0.93 | 0.35 |
| WBC-BF (103/μL) | 0.05 | 341,612 | 0.63 | 1.11 |
| TC-BF (103/μL) | 0.05 | 343,632 | 0.64 | 1.12 |
| TNCC-WDF (103/μL) | 0.06 | 335,752 | 0.88 | 0.97 |
| TNCC-WNR (103/μL) | 0.00 | 342,445 | 1.00 | 0.03 |
| ∆TNC | 0.30 | 583 | 0.93 | 27.04 |
| ∆TNC | Sensitivity (%) | 95% CI | Specificity (%) | 95% CI |
|---|---|---|---|---|
| ≥1.115 | 96.30 | 81.0–99.9 | 86.96 | 66.4–97.2 |
| ≥2.081 | 96.30 | 81.0–99.9 | 91.30 | 72.0–98.9 |
| ≥2.506 | 92.59 | 75.7–99.1 | 91.30 | 72.0–98.9 |
| ≥4.903 | 92.59 | 75.7–99.1 | 100 | 85.2–100 |
| FIP Effusion Index | Sensitivity (%) | 95% CI | Specificity (%) | 95% CI |
|---|---|---|---|---|
| ≥1.637 | 100 | 87.2–100 | 86.96 | 66.4–97.2 |
| ≥4.968 | 96.30 | 81.0–99.9 | 91.30 | 72.0–98.9 |
| ≥5.059 | 96.30 | 81.0–99.9 | 95.65 | 78.1–99.9 |
| ≥7.542 | 96.30 | 81.0–99.9 | 100 | 85.2–100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopes, R.; Sampaio, F.; Carvalho, H.L.d.; Garcês, A.; Fernandes, C.; Neves, C.V.; Brito, A.S.d.; Marques, T.; Sousa, C.; Silva, A.R.; et al. Feline Infectious Peritonitis Effusion Index: A Novel Diagnostic Method and Validation of Flow Cytometry-Based Delta Total Nucleated Cells Analysis on the Sysmex XN-1000V®. Vet. Sci. 2024, 11, 563. https://doi.org/10.3390/vetsci11110563
Lopes R, Sampaio F, Carvalho HLd, Garcês A, Fernandes C, Neves CV, Brito ASd, Marques T, Sousa C, Silva AR, et al. Feline Infectious Peritonitis Effusion Index: A Novel Diagnostic Method and Validation of Flow Cytometry-Based Delta Total Nucleated Cells Analysis on the Sysmex XN-1000V®. Veterinary Sciences. 2024; 11(11):563. https://doi.org/10.3390/vetsci11110563
Chicago/Turabian StyleLopes, Ricardo, Filipe Sampaio, Hugo Lima de Carvalho, Andreia Garcês, Cátia Fernandes, Carolina Vitória Neves, Alexandre Sardinha de Brito, Tiago Marques, Carlos Sousa, Ana Rita Silva, and et al. 2024. "Feline Infectious Peritonitis Effusion Index: A Novel Diagnostic Method and Validation of Flow Cytometry-Based Delta Total Nucleated Cells Analysis on the Sysmex XN-1000V®" Veterinary Sciences 11, no. 11: 563. https://doi.org/10.3390/vetsci11110563
APA StyleLopes, R., Sampaio, F., Carvalho, H. L. d., Garcês, A., Fernandes, C., Neves, C. V., Brito, A. S. d., Marques, T., Sousa, C., Silva, A. R., Martins, Â., Cardoso, L., Coelho, A. C., & Duarte, E. L. (2024). Feline Infectious Peritonitis Effusion Index: A Novel Diagnostic Method and Validation of Flow Cytometry-Based Delta Total Nucleated Cells Analysis on the Sysmex XN-1000V®. Veterinary Sciences, 11(11), 563. https://doi.org/10.3390/vetsci11110563
















