Analysis of Functional Promoter of Camel FGF21 Gene and Identification of Small Compounds Targeting FGF21 Protein
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
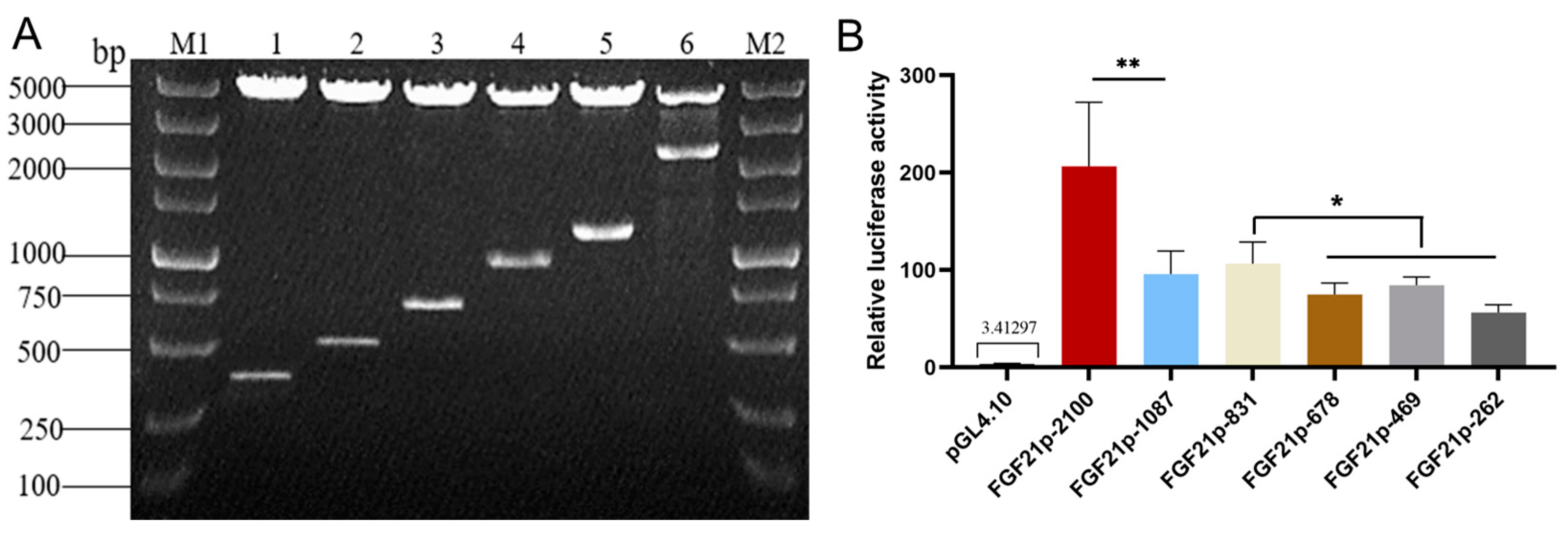
2.1. Promoter Cloning and Construction
2.2. Cell Culture and Luciferase Reporter Assay
2.3. Bioinformatics Analysis of Promoters
2.4. Binding Analysis of Small Compounds Targeting FGF21 Protein
2.5. In Silico ADMET Druggability Prediction
3. Results
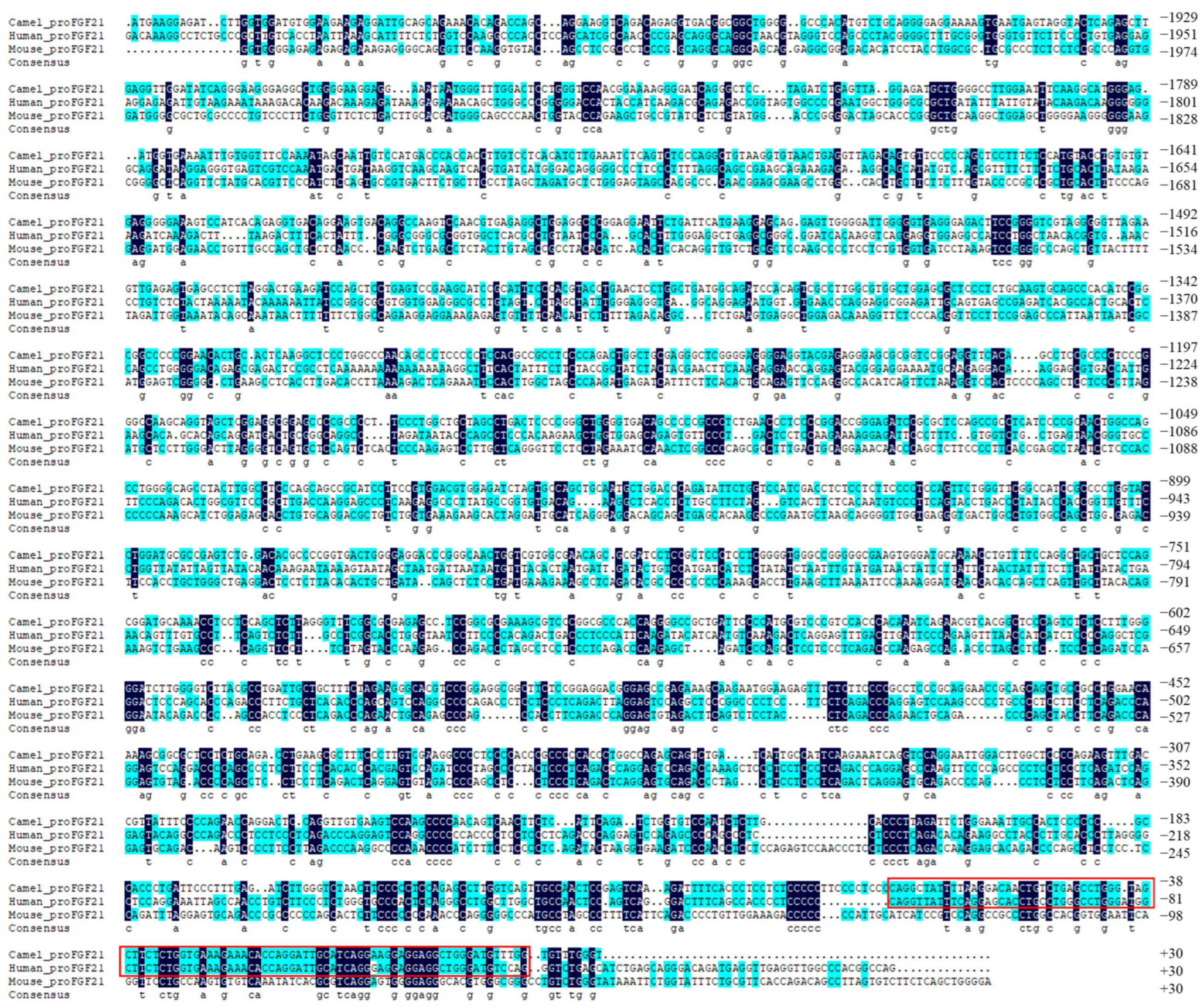
3.1. Alignment of FGF21 Promoter Sequence
3.2. Analysis of Core Active Region of FGF21 Promoter
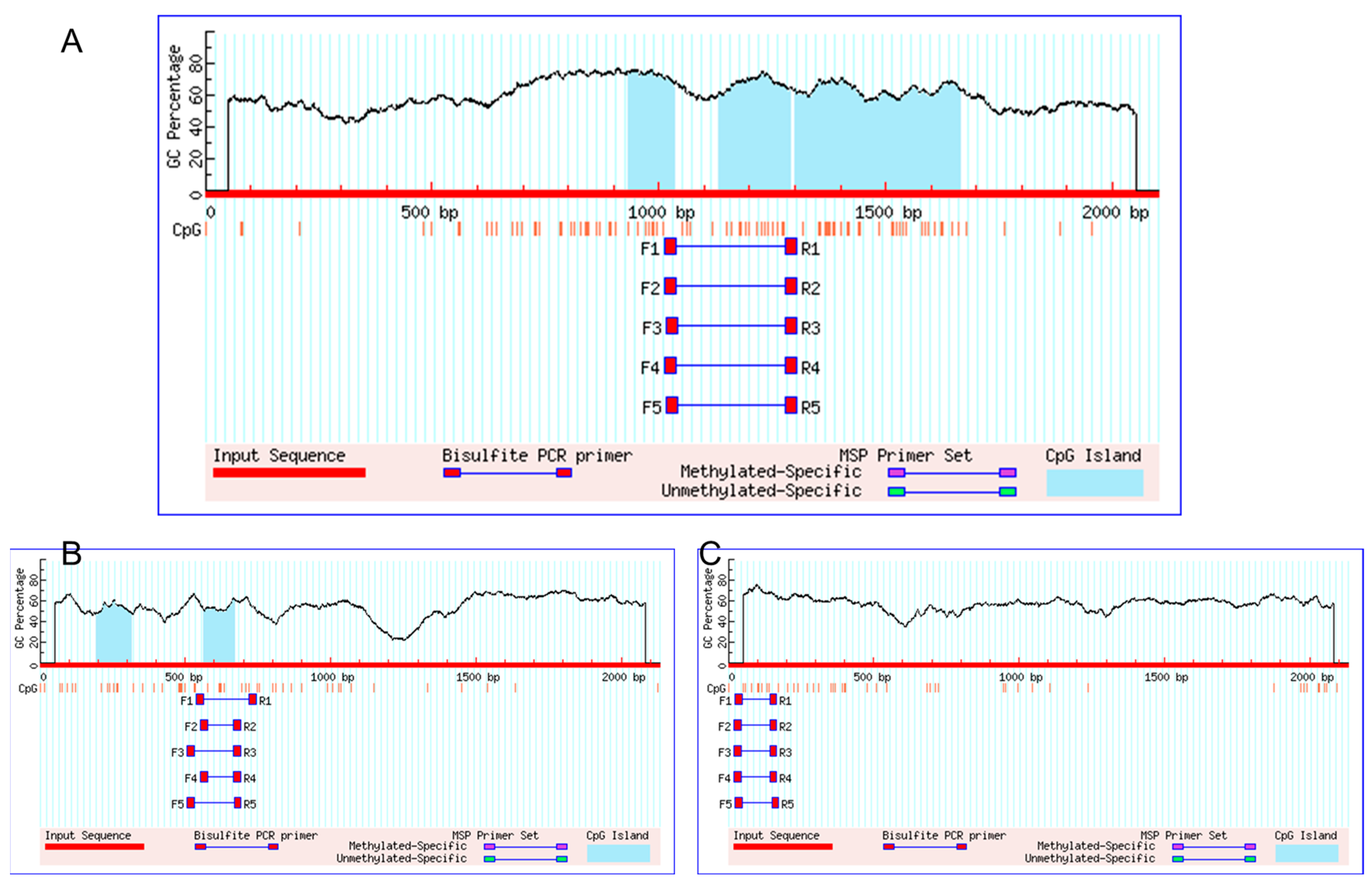
3.3. CGIs Prediction of FGF21 Promoter Sequence
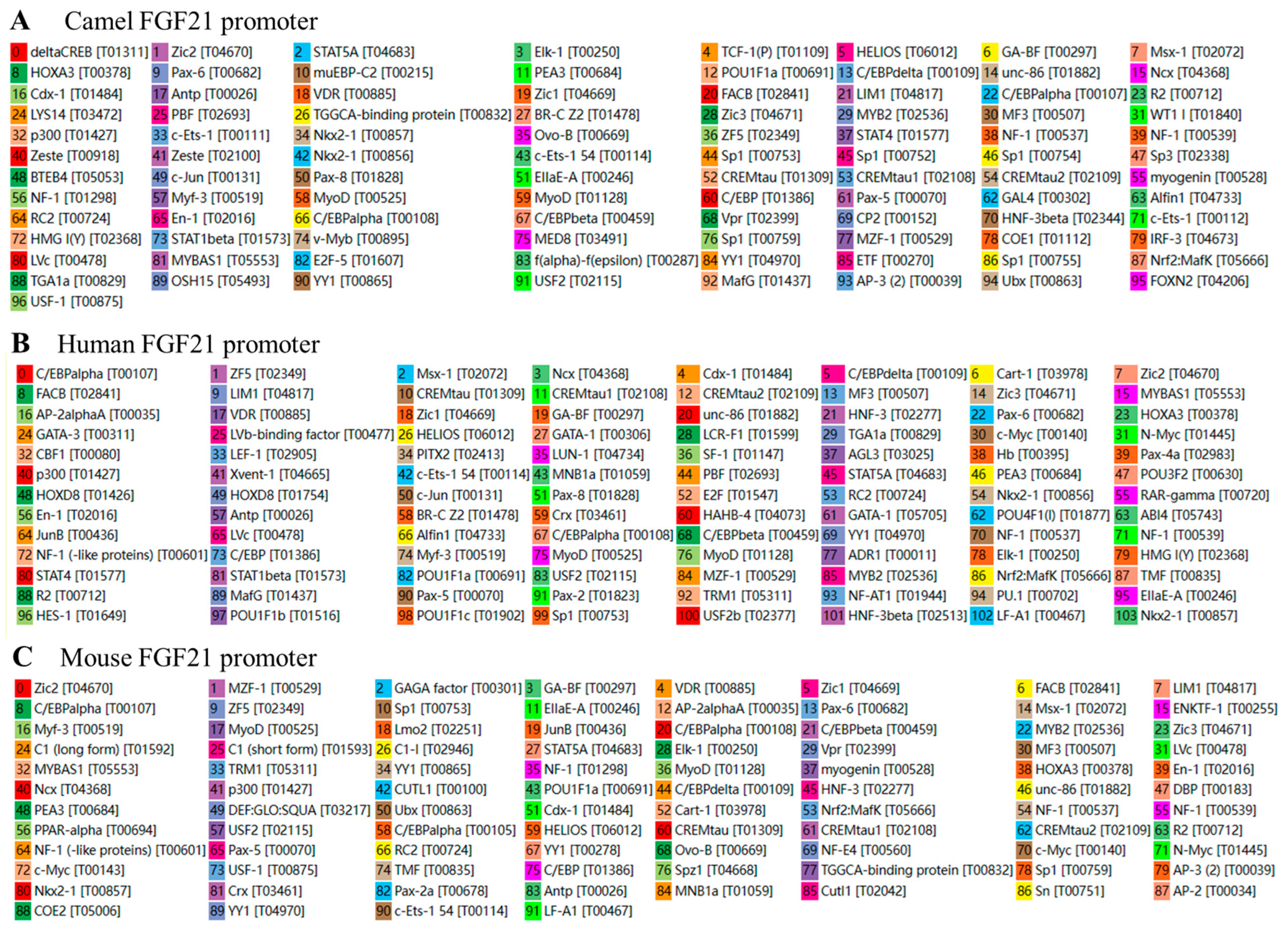
3.4. Analysis of Potential Transcription Factors Binding to FGF21 Promoter
3.5. Determination of the Core Active Region of Camel FGF21 Promoter
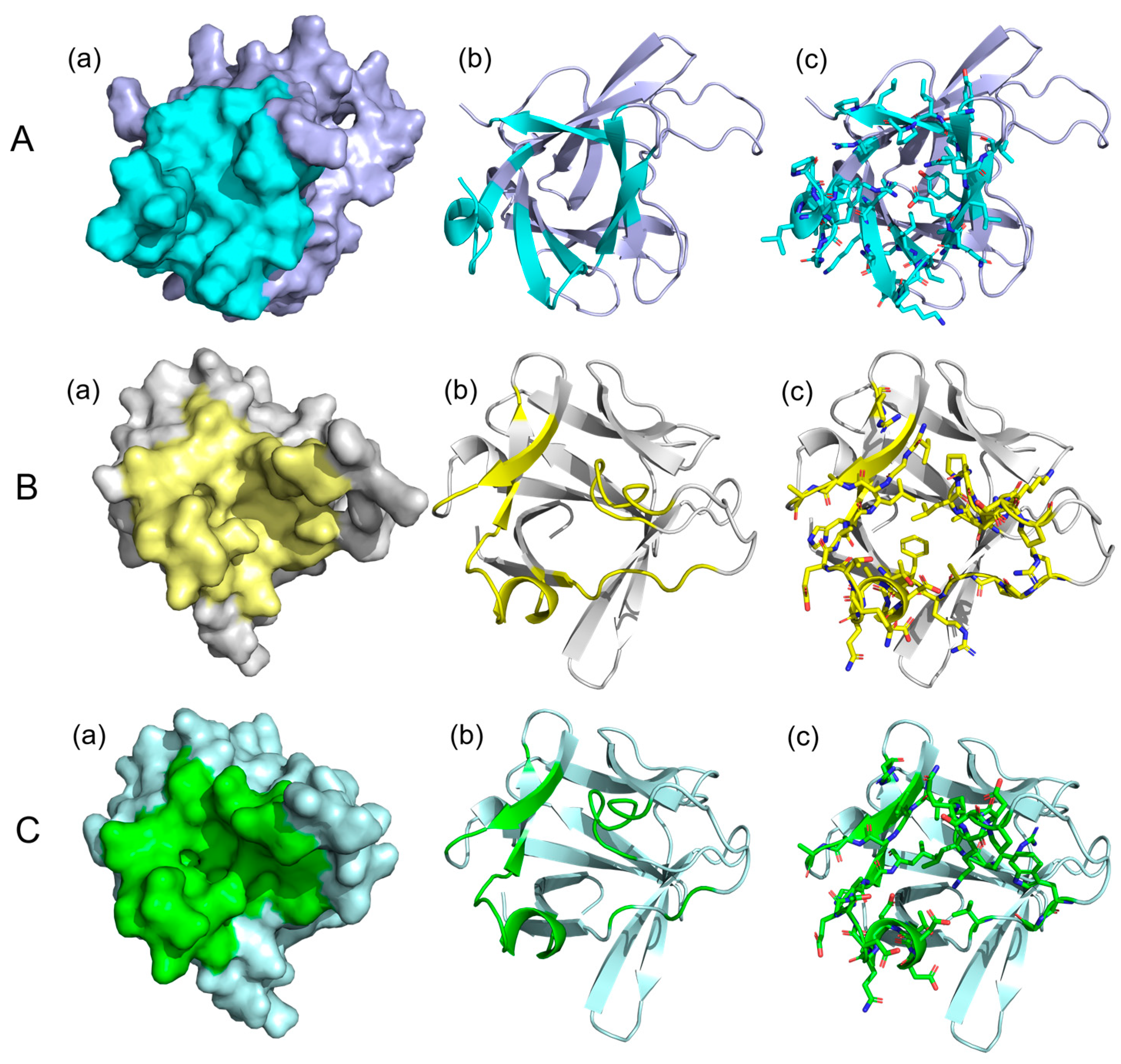
3.6. Analysis of Potential Drug-Binding Pocket of Camel FGF21 Protein
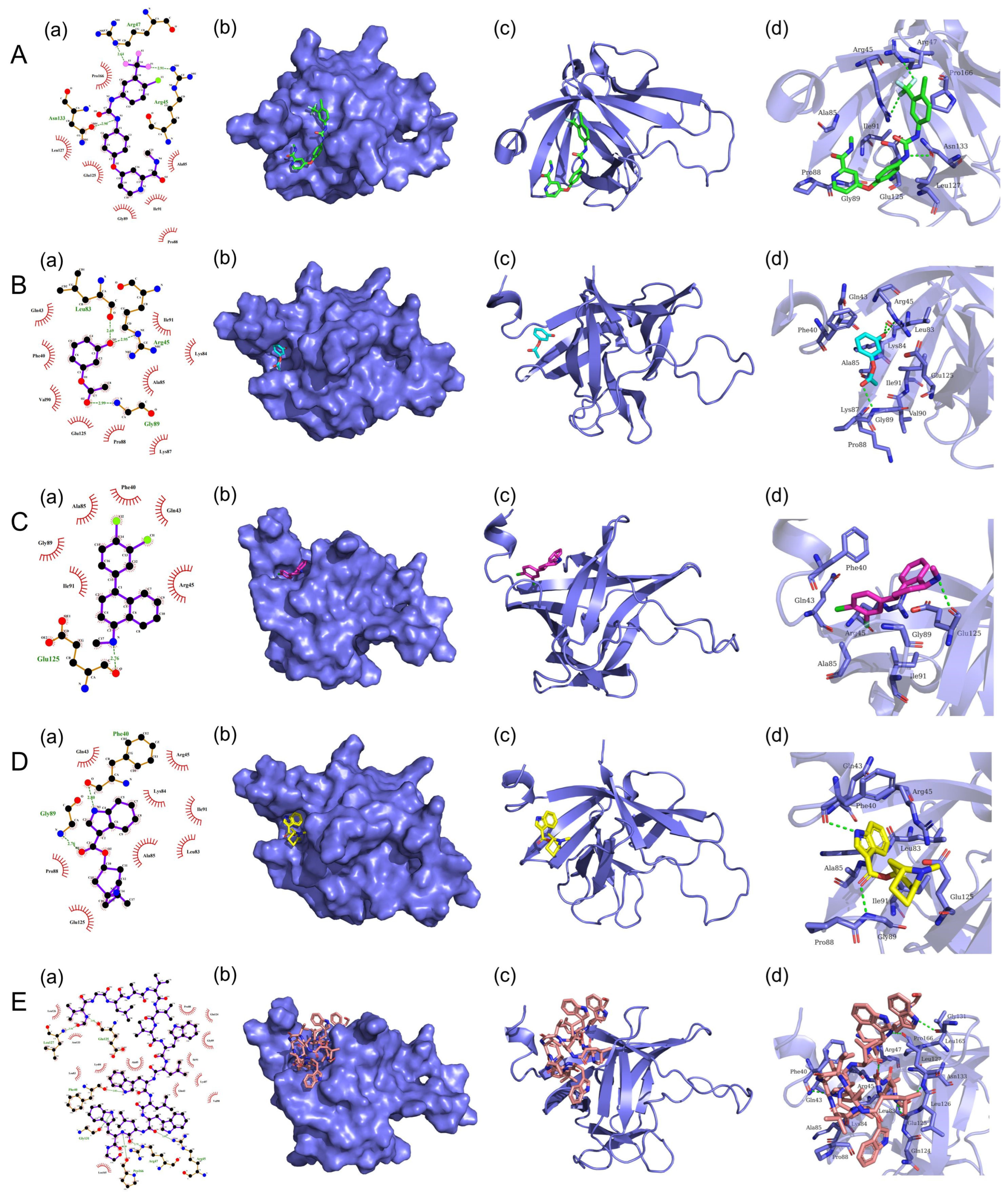
3.7. Screening of Potential Small Compounds Targeting FGF21 Protein
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kharitonenkov, A.; DiMarchi, R. Fibroblast growth factor 21 night watch: Advances and uncertainties in the field. J. Intern. Med. 2017, 281, 233–246. [Google Scholar] [CrossRef]
- Agrawal, A.; Parlee, S.; Perez-Tilve, D.; Li, P.; Pan, J.; Mroz, P.A.; Kruse Hansen, A.M.; Andersen, B.; Finan, B.; Kharitonenkov, A.; et al. Molecular elements in FGF19 and FGF21 defining KLB/FGFR activity and specificity. Mol. Metab. 2018, 13, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Claflin, K.E.; Flippo, K.H.; Sullivan, A.I.; Asghari, A.; Tadinada, S.M.; Jensen-Cody, S.O.; Abel, T.; Potthoff, M.J. Central FGF21 production regulates memory but not peripheral metabolism. Cell Rep. 2022, 40, 111239. [Google Scholar] [CrossRef] [PubMed]
- Staiger, H.; Keuper, M.; Berti, L.; Hrabe de Angelis, M.; Haring, H.U. Fibroblast Growth Factor 21-Metabolic Role in Mice and Men. Endocr. Rev. 2017, 38, 468–488. [Google Scholar] [CrossRef]
- Minard, A.Y.; Tan, S.X.; Yang, P.; Fazakerley, D.J.; Domanova, W.; Parker, B.L.; Humphrey, S.J.; Jothi, R.; Stockli, J.; James, D.E. mTORC1 Is a Major Regulatory Node in the FGF21 Signaling Network in Adipocytes. Cell Rep. 2016, 17, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Engel, J.A.; Jerlhag, E.; Svensson, L.; Smith, R.G.; Egecioglu, E. Blockade of growth hormone secretagogue receptor 1A signaling by JMV 2959 attenuates the NMDAR antagonist, phencyclidine-induced impairments in prepulse inhibition. Psychopharmacology 2015, 232, 4285–4292. [Google Scholar] [CrossRef]
- Rajaram, R.B.; Jayaraman, T.; Yoong, B.K.; Koh, P.S.; Loh, P.S.; Koong, J.K.; Khalil, A.A.; Hashim, N.; Jamaluddin, F.H.; Mahadeva, S. Non-alcoholic fatty liver disease and obesity among adult donors are major challenges to living-donor liver transplantation: A single-center experience. Asian J. Surg. 2022, 45, 441–447. [Google Scholar] [CrossRef]
- Zheng, S.; Wu, J.; Xiang, S.; Zang, Y.; Kong, D.; Wei, X.; Sun, W.; Li, W. An fgf21-like gene from swamp eel (Monopterus albus): Recombinant expression and its potential roles in glucose and lipid homeostasis. Comp. Biochem. Physiol. Part. A Mol. Integr. Physiol. 2022, 267, 111170. [Google Scholar] [CrossRef]
- Yanucil, C.; Kentrup, D.; Li, X.; Grabner, A.; Schramm, K.; Martinez, E.C.; Li, J.; Campos, I.; Czaya, B.; Heitman, K. FGF21-FGFR4 signaling in cardiac myocytes promotes concentric cardiac hypertrophy in mouse models of diabetes. Sci. Rep. 2022, 12, 7326. [Google Scholar] [CrossRef]
- Rühlmann, C.; Dannehl, D.; Brodtrück, M.; Adams, A.C.; Kuhla, A. Neuroprotective Effects of the FGF21 Analogue LY2405319. J. Alzheimer’s Dis. JAD 2021, 80, 1–13. [Google Scholar] [CrossRef]
- Weng, Y.; Ishino, T.; Sievers, A.; Talukdar, S.; Chabot, J.R.; Tam, A.; Duan, W.; Kerns, K.; Sousa, E.; He, T. Glyco-engineered Long Acting FGF21 Variant with Optimal Pharmaceutical and Pharmacokinetic Properties to Enable Weekly to Twice Monthly Subcutaneous Dosing. Sci. Rep. 2018, 8, 4241. [Google Scholar] [CrossRef]
- Hoter, A.; Rizk, S.; Naim, H.Y. Cellular and Molecular Adaptation of Arabian Camel to Heat Stress. Front. Genet. 2019, 10, 588. [Google Scholar] [CrossRef]
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Dao, L.T.M.; Galindo-Albarran, A.O.; Castro-Mondragon, J.A.; Andrieu-Soler, C.; Medina-Rivera, A.; Souaid, C.; Charbonnier, G.; Griffon, A.; Vanhille, L.; Stephen, T.; et al. Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat. Genet. 2017, 49, 1073–1081. [Google Scholar] [CrossRef]
- Andersson, R.; Gebhard, C.; Miguel-Escalada, I.; Hoof, I.; Bornholdt, J.; Boyd, M.; Chen, Y.; Zhao, X.; Schmidl, C.; Suzuki, T.; et al. An atlas of active enhancers across human cell types and tissues. Nature 2014, 507, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Andersson, R.; Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 2020, 21, 71–87. [Google Scholar] [CrossRef]
- Sethi, A.; Gu, M.; Gumusgoz, E.; Chan, L.; Yan, K.K.; Rozowsky, J.; Barozzi, I.; Afzal, V.; Akiyama, J.A.; Plajzer-Frick, I.; et al. Supervised enhancer prediction with epigenetic pattern recognition and targeted validation. Nat. Methods 2020, 17, 807–814. [Google Scholar] [CrossRef] [PubMed]
- Stanzione, F.; Giangreco, I.; Cole, J.C. Use of molecular docking computational tools in drug discovery. Prog. Med. Chem. 2021, 60, 273–343. [Google Scholar] [CrossRef]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef]
- Gao, Y.; Zhao, S.; Zhang, W.; Tang, H.; Yan, M.; Yong, F.; Bai, X.; Wu, X.; Zhang, Y.; Zhang, Q. Localization of FGF21 Protein and Lipid Metabolism-Related Genes in Camels. Life 2023, 13, 432. [Google Scholar] [CrossRef]
- Pollastri, M.P. Overview on the Rule of Five. Curr. Protoc. Pharmacol. 2010, 49, 9.12.1–9.12.8. [Google Scholar] [CrossRef]
- Fisher, F.M.; Maratos-Flier, E. Understanding the Physiology of FGF21. Annu. Rev. Physiol. 2016, 78, 223–241. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Szczurek, A.T.; Kelley, J.R.; Lastuvkova, A.; Turberfield, A.H.; Dimitrova, E.; Blackledge, N.P.; Klose, R.J. A CpG island-encoded mechanism protects genes from premature transcription termination. Nat. Commun. 2023, 14, 726. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.L.G.; Andricopulo, A.D. ADMET modeling approaches in drug discovery. Drug Discov. Today 2019, 24, 1157–1165. [Google Scholar] [CrossRef]
- BonDurant, L.D.; Potthoff, M.J. Fibroblast Growth Factor 21: A Versatile Regulator of Metabolic Homeostasis. Annu. Rev. Nutr. 2018, 38, 173–196. [Google Scholar] [CrossRef] [PubMed]
- Tezze, C.; Romanello, V.; Sandri, M. FGF21 as Modulator of Metabolism in Health and Disease. Front. Physiol. 2019, 10, 419. [Google Scholar] [CrossRef]
- Kim, J.H.; Lim, S.; Seo, M.; Choi, H.H.; Kim, D.; Mi, K.J.; Park, J.Y.; Choi, B.H.; Lee, J.K.; Kim, J.G. Long-Acting FGF21 Fusion Proteins and Pharmaceutical Composition Comprising Same. U.S. Patent 17/478,600, 6 January 2022. [Google Scholar]
- Angeloni, A.; Bogdanovic, O. Sequence determinants, function, and evolution of CpG islands. Biochem. Soc. Trans. 2021, 49, 1109–1119. [Google Scholar] [CrossRef]
- Yu, Q.; Liu, W.; Chen, Z.; Zhang, M. Specificity Protein 1: A Protein With a Two-Sided Role in Ischemic Stroke. Front. Cell. Neurosci. 2021, 15, 757670. [Google Scholar] [CrossRef]
- Laniel, M.A.; Poirier, G.G.; Guerin, S.L. Nuclear factor 1 interferes with Sp1 binding through a composite element on the rat poly(ADP-ribose) polymerase promoter to modulate its activity in vitro. J. Biol. Chem. 2001, 276, 20766–20773. [Google Scholar] [CrossRef]
- Gutmann, D.H.; Ferner, R.E.; Listernick, R.H.; Korf, B.R.; Wolters, P.L.; Johnson, K.J. Neurofibromatosis type 1. Nat. Rev. Dis. Primers 2017, 3, 17004. [Google Scholar] [CrossRef]
- Steiner, L. Helping GATA1 make complex decisions. Blood 2022, 139, 3457–3459. [Google Scholar] [CrossRef] [PubMed]
- Martins Pecanha, F.L.; Jaafar, R.; Werneck-de-Castro, J.P.; Apostolopolou, C.C.; Bhushan, A.; Bernal-Mizrachi, E. The Transcription Factor YY1 Is Essential for Normal DNA Repair and Cell Cycle in Human and Mouse beta-Cells. Diabetes 2022, 71, 1694–1705. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Barceinas, R.S.; Gehringer, F.; Ducker, C.; Saxton, J.; Shaw, P.E. ELK-1 ubiquitination status and transcriptional activity are modulated independently of F-Box protein FBXO25. J. Biol. Chem. 2021, 296, 100214. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.H.; Ye, Q.F.; Miao, X.Y.; Liu, X.; Huang, S.Q.; Xiong, L.; Wen, Y.; Zhang, Z.J. Current status of sorafenib nanoparticle delivery systems in the treatment of hepatocellular carcinoma. Theranostics 2021, 11, 5464–5490. [Google Scholar] [CrossRef]
- Hwang, H.Y.; Shim, J.S.; Kim, D.; Kwon, H.J. Antidepressant drug sertraline modulates AMPK-MTOR signaling-mediated autophagy via targeting mitochondrial VDAC1 protein. Autophagy 2021, 17, 2783–2799. [Google Scholar] [CrossRef] [PubMed]
- Naderi, R.; Shirpoor, A.; Samadi, M.; Pourheydar, B.; Moslehi, A. Tropisetron attenuates pancreas apoptosis in the STZ-induced diabetic rats: Involvement of SIRT1/NF-kappaB signaling. Pharmacol. Rep. 2020, 72, 1657–1665. [Google Scholar] [CrossRef]
- Protic, S.; Kalicanin, N.; Sencanski, M.; Prodanovic, O.; Milicevic, J.; Perovic, V.; Paessler, S.; Prodanovic, R.; Glisic, S. In Silico and In Vitro Inhibition of SARS-CoV-2 PL(pro) with Gramicidin D. Int. J. Mol. Sci. 2023, 24, 1955. [Google Scholar] [CrossRef]
- Jia, L.; Gao, H. Machine Learning for In Silico ADMET Prediction. Methods Mol. Biol. 2022, 2390, 447–460. [Google Scholar] [CrossRef]
- Goller, A.H.; Kuhnke, L.; Montanari, F.; Bonin, A.; Schneckener, S.; Ter Laak, A.; Wichard, J.; Lobell, M.; Hillisch, A. Bayer’s in silico ADMET platform: A journey of machine learning over the past two decades. Drug Discov. Today 2020, 25, 1702–1709. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Yong, F.; Ji, W.; Zhang, L.; Zhao, S.; Gao, Y. Construction and Characterization of Immortalized Fibroblast Cell Line from Bactrian Camel. Life 2023, 13, 1337. [Google Scholar] [CrossRef]
| Species | Location/bp | Sequence | Score |
|---|---|---|---|
| Camel | −445~−495 | TGCCGCCTGGAACAAAAGCGGCGCTCCTCTGCAGACCTGAAGCGCTTTCC | 0.90 |
| −562~−612 | CTGATTGCTGCTTTCTAGAAGGGCACGTCCCGGAGGCGGCTTCTCCGGAG | 0.81 | |
| Mouse | −18~−68 | CCTGTCTGGGTATAAATTCTGGTATTTCTGCGTTCACCAGACAGCCTTAG | 0.96 |
| −2049~−2099 | TTCCAAGGTGTACAGCCTCCGCCCTCCCGGCAGGGCAGGCAGCAGGAGGC | 0.82 |
| Transcription Factor | Camel | Mouse | Human | Transcription Factor | Camel | Mouse | Human |
|---|---|---|---|---|---|---|---|
| SP1 | + | + | + | GATA-1 | − | − | + |
| NF-1 | + | + | + | USF2 | + | + | + |
| TCF-1 | + | − | − | YY1 | + | + | + |
| c-Ets-1 | + | − | − | FOXN2 | + | − | − |
| Elk-1 | + | + | + | E2F | + | − | + |
| Pax-4 | + | + | + | MyoD | + | + | + |
| Drug Name | Relative Molecular Mass | H-Bond Donors | H-Bond Acceptors | Rotatable Bonds | Polar Surface Area (Å2) |
|---|---|---|---|---|---|
| Sorafenib | 464.825 | 3 | 3 | 6 | 92.35 |
| Resorcinol monoacetate | 152.149 | 1 | 2 | 2 | 46.53 |
| Sertraline | 306.23 | 1 | 1 | 2 | 12.03 |
| Tropisetron | 284.3529 | 1 | 2 | 3 | 45.33 |
| Gramicidin D | 1811.253 | 20 | 16 | 50 | 519.89 |
| Name | Rotatable Bonds | H-Bond Acceptors | H-Bond Donors | Consensus Log P | GI Absorption | log Kp (cm/s) |
|---|---|---|---|---|---|---|
| Sorafenib | 9 | 7 | 3 | 4.1 | Low | −6.25 |
| Resorcinol monoacetate | 2 | 3 | 1 | 1.32 | High | −6.35 |
| Sertraline | 2 | 1 | 1 | 4.55 | High | −4.77 |
| Tropisetron | 3 | 3 | 1 | 2.79 | High | −5.53 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yong, F.; Yan, M.; Zhang, L.; Ji, W.; Zhao, S.; Gao, Y. Analysis of Functional Promoter of Camel FGF21 Gene and Identification of Small Compounds Targeting FGF21 Protein. Vet. Sci. 2023, 10, 452. https://doi.org/10.3390/vetsci10070452
Yong F, Yan M, Zhang L, Ji W, Zhao S, Gao Y. Analysis of Functional Promoter of Camel FGF21 Gene and Identification of Small Compounds Targeting FGF21 Protein. Veterinary Sciences. 2023; 10(7):452. https://doi.org/10.3390/vetsci10070452
Chicago/Turabian StyleYong, Fang, Meilin Yan, Lili Zhang, Wangye Ji, Shuqin Zhao, and Yuan Gao. 2023. "Analysis of Functional Promoter of Camel FGF21 Gene and Identification of Small Compounds Targeting FGF21 Protein" Veterinary Sciences 10, no. 7: 452. https://doi.org/10.3390/vetsci10070452
APA StyleYong, F., Yan, M., Zhang, L., Ji, W., Zhao, S., & Gao, Y. (2023). Analysis of Functional Promoter of Camel FGF21 Gene and Identification of Small Compounds Targeting FGF21 Protein. Veterinary Sciences, 10(7), 452. https://doi.org/10.3390/vetsci10070452







