Biomolecular Analysis of Canine Distemper Virus Strains in Two Domestic Ferrets (Mustela putorius furo)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Case History and Samples Collection
2.2. RNA Extraction
2.3. Detection of Canine Distemper Virus RNA
2.4. CDV Sequence and Phylogenetic Analyses
2.5. Bacteriological, Parasitological, and Mycological Analyses
3. Results
3.1. Clinical Examination
3.2. CDV Detection and RFLP Analysis
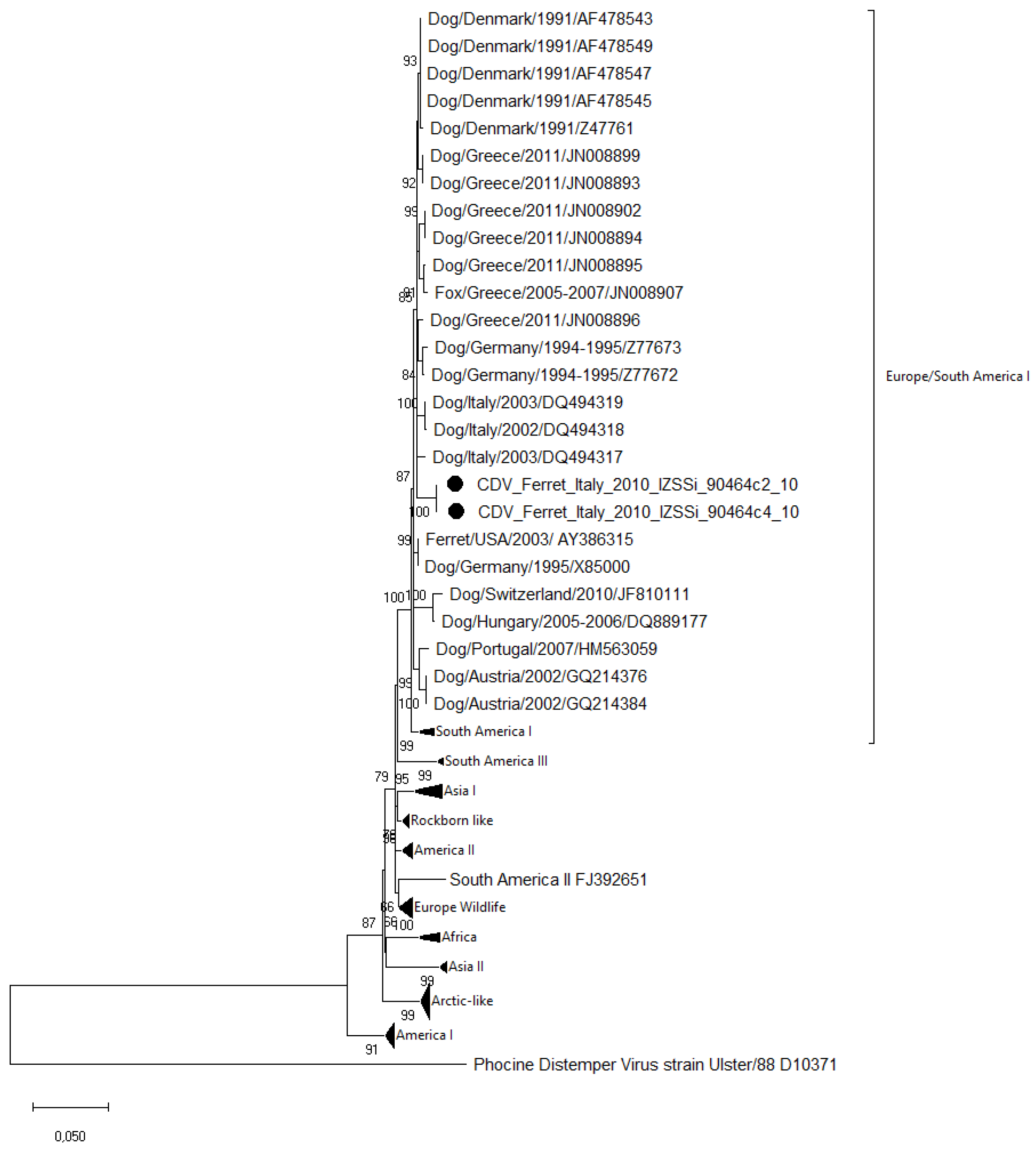
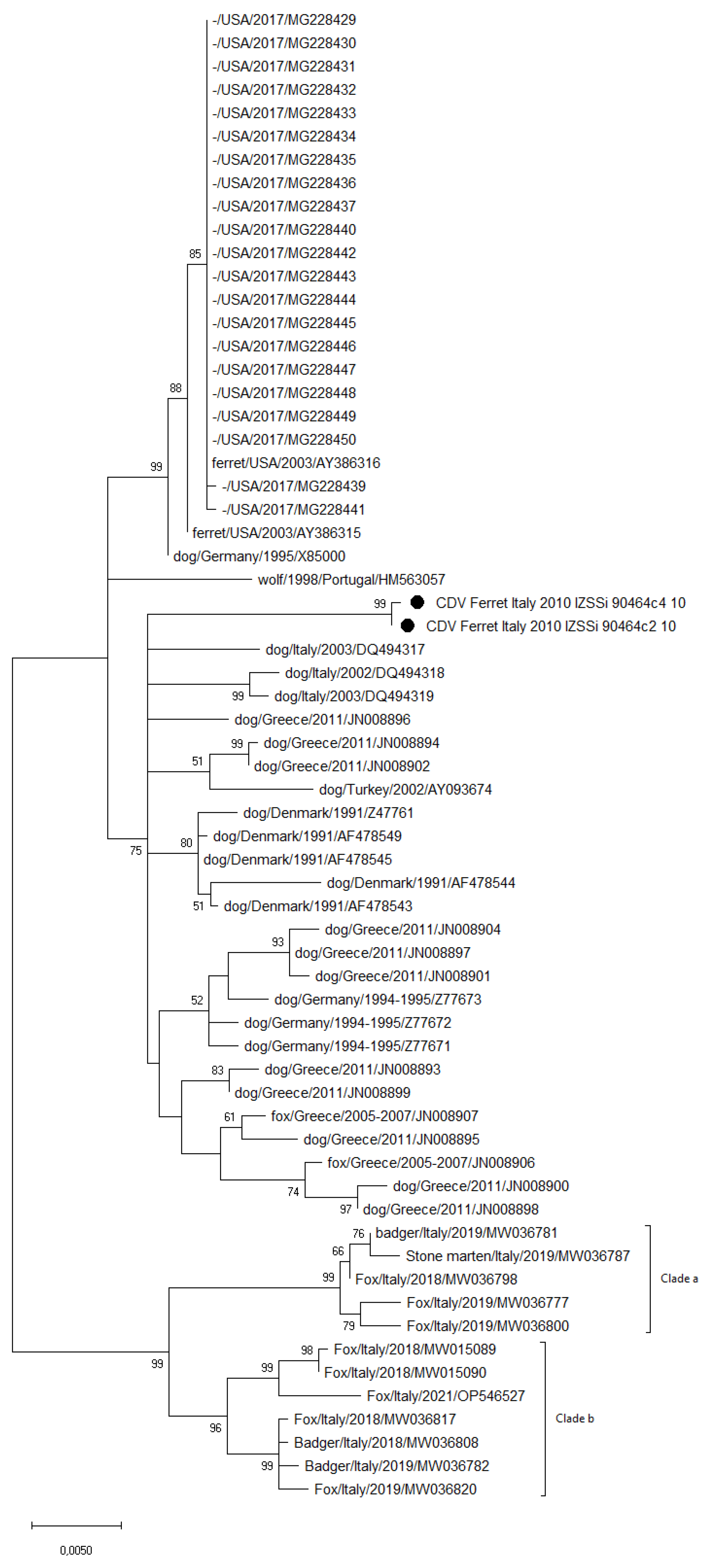
3.3. Sequence and Phylogenetic Analysis
3.4. Bacteriological, Parasitological, and Mycological Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rima, B.; Balkema-Buschmann, A.; Dundon, W.G.; Duprex, P.; Easton, A.; Fouchier, R.; Kurath, G.; Lamb, R.; Lee, B.; Rota, P.; et al. ICTV Virus Taxonomy Profile: Paramyxoviridae. J. Gen. Virol. 2019, 100, 1593–1594. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Elia, G.; Buonavoglia, C. Canine distemper virus. Vet. Clin. N. Am. Small Anim. Pract. 2008, 38, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Mira, F.; Purpari, G.; Di Bella, S.; Vicari, D.; Schirò, G.; Di Marco, P.; Macaluso, G.; Battilani, M.; Guercio, A. Update on canine distemper virus (CDV) strains of Arctic-like lineage detected in dogs in Italy. Vet. Ital. 2018, 54, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Balboni, A.; De Lorenzo Dandola, G.; Scagliarini, A.; Prosperi, S.; Battilani, M. Occurrence of different Canine distemper virus lineages in Italian dogs. Vet. Ital. 2014, 50, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Di Sabatino, D.; Lorusso, A.; Di Francesco, C.E.; Gentile, L.; Di Pirro, V.; Bellacicco, A.L.; Giovannini, A.; Di Francesco, G.; Marruchella, G.; Marsilio, F.; et al. Arctic lineage-canine distemper virus as a cause of death in appennine wolves (Canis lupus) in Italy. PLoS ONE 2014, 9, e82356. [Google Scholar] [CrossRef]
- Di Sabatino, D.; Di Francesco, G.; Zaccaria, G.; Malatesta, D.; Brugnola, L.; Marcacci, M.; Portanti, O.; De Massis, F.; Savini, G.; Teodori, L.; et al. Lethal distemper in badgers (Meles meles) following epidemic in dogs and wolves. Infect. Genet. Evol. 2016, 46, 130–137. [Google Scholar] [CrossRef]
- Peserico, A.; Marcacci, M.; Malatesta, D.; Di Domenico, M.; Pratelli, A.; Mangone, I.; D’Alterio, N.; Pizzurro, F.; Cirone, F.; Zaccaria, G.; et al. Diagnosis and characterization of canine distemper virus through sequencing by MinION nanopore technology. Sci. Rep. 2018, 9, 1714. [Google Scholar] [CrossRef]
- Von Messling, V.; Zimmer, G.; Herrler, G.; Haas, L.; Cattaneo, R. The hemagglutinin of canine distemper virus determines tropism and cytopathogenicity. J. Virol. 2001, 75, 6418–6427. [Google Scholar] [CrossRef]
- Granjeiro, M.D.B.; Kavasaki, M.L.; Morgado, T.O.; Pavelegini, L.A.D.; De Barros, M.A.; Fontana, C.; De Assis Bianchini, M.; De Oliveira Souza, A.; Santos, A.R.G.L.O.; Lunardi, M.; et al. First report of a canine morbillivirus infection in a giant anteater (Myrmecophaga tridactyla) in Brazil. Vet. Med. Sci. 2020, 6, 606–611. [Google Scholar] [CrossRef]
- Pavlacik, L.; Celer, V.; Koubek, P.; Literak, I. Prevalence of canine distemper virus in wild mustelids in the Czech Republic and a case of canine distemper in young stone martens. Vet. Med. Czech 2007, 52, 69–73. [Google Scholar] [CrossRef]
- George, A.M.; Wille, M.; Wang, J.; Anderson, K.; Cohen, S.; Moselen, J.; Yang Lee, L.Y.; Suen, W.W.; Bingham, J.; Dalziel, A.E.; et al. A novel and highly divergent Canine Distemper Virus lineage causing distemper in ferrets in Australia. BioRxiv 2021, 576, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Avota, E.; Koethe, S.; Sibylle, S.S. Membrane dynamics and interactions in measles virus dendritic cell infections. Cell. Microbiol. 2013, 15, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Koethe, S.; Avota, E.; Sibylle, S.S. Measles virus transmission from dendritic cells to t cells: Formation of synapse-like interfaces concentrating viral and cellular components. J. Virol. 2012, 86, 9773–9781. [Google Scholar] [CrossRef] [PubMed]
- Lempp, C.; Spitzbarth, I.; Puff, C.; Cana, A.; Kegler, K.; Techangamsuwan, S.; Baumgärtner, W.; Seehusen, F. New aspects of the pathogenesis of canine distemper leukoencephalitis. Viruses 2014, 6, 2571–2601. [Google Scholar] [CrossRef]
- Rendon-Marin, S.; da Fontoura, B.R.; Canal, C.W.; Ruiz-Saenz, J. Tropism and molecular pathogenesis of canine distemper virus. Virol. J. 2019, 16, 30. [Google Scholar] [CrossRef]
- Weckworth, J.K.; Davis, B.W.; Dubovi, E.; Fountain-Jones, N.; Packer, C.; Cleaveland, S.; Craft, M.E.; Eblate, E.; Schwartz, M.; Mills, L.S.; et al. Cross-species transmission and evolutionary dynamics of canine distemper virus during a spillover in African lions of Serengeti National Park. Mol. Ecol. 2020, 29, 4308–4321. [Google Scholar] [CrossRef]
- Beineke, A.; Baumgartner, W.; Wohlsein, P. Cross-species transmission of canine distemper virus-an update. One Health 2015, 1, 49–59. [Google Scholar] [CrossRef]
- Nikolin, V.M.; Olarte-Castillo, X.A.; Osterrieder, N.; Hofer, H.; Dubovi, E.; Mazzoni, C.J.; Brunner, E.; Goller, K.V.; Fyumagwa, R.D.; Moehlman, P.D.; et al. Canine distemper virus in the Serengeti ecosystem: Molecular adaptation to different carnivore species. Mol. Ecol. 2017, 26, 2111–2130. [Google Scholar] [CrossRef]
- Pope, J.P.; Miller, D.L.; Riley, M.C.; Anis, E.; Wilkes, R.P. Characterization of a novel Canine distemper virus causing disease in wildlife. J. Vet. Diagn. Investig. 2016, 28, 506–513. [Google Scholar] [CrossRef]
- Martinez-Gutierrez, M.; Ruiz-Saenz, J. Diversity of susceptible hosts in canine distemper virus infection: A systematic review and data synthesis. BMC Vet. Res. 2016, 12, 78. [Google Scholar] [CrossRef]
- Jo, W.K.; Peters, M.; Kydyrmanov, A.; van de Bildt, M.W.G.; Kuiken, T.; Osterhaus, A.; Ludlow, M. The Canine Morbillivirus strain associated with an epizootic in Caspian Seals provides new insights into the evolutionary history of this virus. Viruses 2019, 11, e894. [Google Scholar] [CrossRef] [PubMed]
- Ripple, W.J.; Estes, J.A.; Beschta, R.L.; Wilmers, C.C.; Ritchie, E.G.; Hebblewhite, M.; Berger, J.; Elmhagen, B.; Letnic, M.; Nelson, M.P.; et al. Status and ecological effects of the world’s largest carnivores. Science 2014, 343, 1241484. [Google Scholar] [CrossRef] [PubMed]
- Viana, M.; Cleaveland, S.; Matthiopoulos, J.; Halliday, J.; Packer, C.; Craft, M.E.; Hampson, K.; Czupryna, A.; Dobson, A.P.; Dubovi, E.J.; et al. Dynamics of a morbillivirus at the domestic-wildlife interface: Canine distemper virus in domestic dogs and lions. Proc. Natl. Acad. Sci. USA 2015, 112, 1464–1469. [Google Scholar] [CrossRef] [PubMed]
- Krumm, S.A.; Yan, D.; Hovingh, E.S.; Evers, T.J.; Enkirch, T.; Reddy, G.P.; Sun, A.; Saindane, M.T.; Arrendale, R.F.; Painter, G.; et al. An orally available, small-molecule polymerase inhibitor shows efficacy against a lethal morbillivirus infection in a large animal model. Sci. Transl. Med. 2014, 6, 232ra52. [Google Scholar] [CrossRef] [PubMed]
- Wyllie, S.E.; Kelman, M.; Ward, M.P. Epidemiology and clinical presentation of canine distemper disease in dogs and ferrets in Australia, 2006–2014. Aust. Vet. J. 2016, 94, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Karki, M.; Rajak, K.K.; Singh, R.P. Canine morbillivirus (CDV): A review on current status, emergence and the diagnostics. Virusdisease 2022, 33, 309–321. [Google Scholar] [CrossRef]
- Yi, L.; Cheng, S.; Xu, H.; Wang, J.; Cheng, Y.; Yang, S.; Luo, B. Development of a combined canine distemper virus specific RT-PCR protocol for the differentiation of infected and vaccinated animals (DIVA) and genetic characterization of the hemagglutinin gene of seven Chinese strains demonstrated in dogs. J. Virol. Methods 2012, 179, 281–287. [Google Scholar] [CrossRef]
- Demeter, Z.; Palade, E.A.; Hornyák, A.; Rusvai, M. Controversial results of the genetic analysis of a canine distemper vaccine strain. Vet. Microbiol. 2010, 142, 420–426. [Google Scholar] [CrossRef]
- Deem, S.L.; Spelman, L.H.; Yates, R.A.; Montali, R.J. Canine distemper in terrestrial carnivores: A review. JZWM 2000, 31, 441–451. [Google Scholar]
- Barret, T.; Visser, I.K.J.; Mamaev, L.; Goatley, L.; van Bressem, M.F.; Osterhaus, A.D.M.E. Dolphin and Porpoise Morbilliviruses are genetically distinct from Phocine Distemper Virus. Virology 1993, 193, 1010–1012. [Google Scholar] [CrossRef]
- Demeter, Z.; Lakatos, B.; Palade, E.A.; Kozma, T.; Forgách, P.; Rusvai, M. Genetic diversity of Hungarian canine distemper virus strains. Vet. Microbiol. 2007, 122, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Trogu, T.; Castelli, A.; Canziani, S.; Tolini, C.; Carrera, M.; Sozzi, E.; Lelli, D.; Tosi, G.; Fiorentini, L.; Di Donato, A.; et al. Detection and Molecular Characterization of Canine Distemper Virus in Wildlife from Northern Italy. Pathogens 2022, 11, 1557. [Google Scholar] [CrossRef]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [CrossRef]
- Trogu, T.; Canziani, S.; Salvato, S.; Bianchi, A.; Bertoletti, I.; Gibelli, L.R.; Alborali, G.L.; Barbieri, I.; Gaffuri, A.; Sala, G.; et al. Canine Distemper Outbreaks in Wild Carnivores in Northern Italy. Viruses 2021, 13, 99. [Google Scholar] [CrossRef]
- Alfano, F.; Lanave, G.; Lucibelli, M.G.; Miletti, G.; D’Alessio, N.; Gallo, A.; Auriemma, C.; Amoroso, M.G.; Lucente, M.S.; De Carlo, E.; et al. Canine Distemper Virus in Autochtonous and Imported Dogs, Southern Italy (2014–2021). Animals 2022, 12, 2852. [Google Scholar] [CrossRef]
- Martella, V.; Cirone, F.; Elia, G.; Lorusso, E.; Decaro, N.; Campolo, M.; Desario, C.; Lucente, M.S.; Bellacicco, A.L.; Blixenkrone-Møller, M.; et al. Heterogeneity within the hemagglutinin genes of canine distemper virus (CDV) strains detected in Italy. Vet. Microbiol. 2006, 116, 301–309. [Google Scholar] [CrossRef]
- Martella, V.; Pratelli, A.; Cirone, F.; Zizzo, N.; Decaro, N.; Tinelli, A.; Foti, M.; Buonavoglia, C. Detection and genetic characterization of canine distemper virus (CDV) from free-ranging red foxes in Italy. Mol. Cell. Probes 2002, 16, 77–83. [Google Scholar] [CrossRef]
- Ke, G.M.; Ho, C.H.; Chiang, M.J.; Sanno-Duanda, B.; Chung, C.S.; Lin, M.Y.; Shi, Y.Y.; Yang, M.H.; Tyan, Y.C.; Liao, P.C.; et al. Phylodynamic analysis of the canine distemper virus hemagglutinin gene. BMC Vet. Res. 2015, 11, 164. [Google Scholar] [CrossRef] [PubMed]
- Sawatsky, B.; von Messling, V. Canine distemper viruses expressing a hemagglutinin without N-glycans lose virulence but retain immunosuppression. J. Virol. 2010, 84, 2753–2761. [Google Scholar] [CrossRef] [PubMed]
- Di Francesco, C.E.; Smoglica, C.; Di Pirro, V.; Cafini, F.; Gentile, L.; Marsilio, F. Molecular Detection and Phylogenetic Analysis of Canine Distemper Virus in Marsican Brown Bear (Ursus arctos marsicanus). Animals 2022, 12, 1826. [Google Scholar] [CrossRef] [PubMed]
- Ricci, I.; Cersini, A.; Manna, G.; Marcario, G.A.; Conti, R.; Brocherel, G.; Grifoni, G.; Eleni, C.; Scicluna, M.T. A Canine Distemper Virus Retrospective Study Conducted from 2011 to 2019 in Central Italy (Latium and Tuscany Regions). Viruses 2021, 13, 272. [Google Scholar] [CrossRef]
- Bianco, A.; Zecchin, B.; Fusaro, A.; Schivo, A.; Ormelli, S.; Bregoli, M.; Citterio, C.V.; Obber, F.; Dellamaria, D.; Trevisiol, K.; et al. Two waves of canine distemper virus showing different spatio-temporal dynamics in Alpine wildlife (2006–2018). Infect. Genet. Evol. 2020, 84, 104359. [Google Scholar] [CrossRef]
- Balboni, A.; Savini, F.; Scagliarini, A.; Berti, E.; Naldi, M.; Urbani, L.; Fontana, M.C.; Carra, E.; Gibelli, L.R.M.; Gobbo, F.; et al. Natural distemper infection in stone martens (Martes foina): From infection to neutralizing antibodies. Res. Vet. Sci. 2021, 138, 196–200. [Google Scholar] [CrossRef]
- Ndiana, L.A.; Lanave, G.; Vasinioti, V.; Desario, C.; Martino, C.; Colaianni, M.L.; Pellegrini, F.; Camarda, A.; Berjaoui, S.; Sgroi, G.; et al. Detection and Genetic Characterization of Canine Adenoviruses, Circoviruses, and Novel Cycloviruses from Wild Carnivores in Italy. Front. Vet. Sci. 2022, 9, 851987. [Google Scholar] [CrossRef]
- Perpiñán, D.; Ramis, A.; Tomás, A.; Carpintero, E.; Bargalló, F. Outbreak of canine distemper in domestic ferrets (Mustela putorius furo). Vet. Rec. 2008, 163, 246–250. [Google Scholar] [CrossRef]
- Evermann, J.F.; Leathers, C.W.; Gorham, J.R.; McKeirnan, A.J.; Appel, M.J. Pathogenesis of two strains of lion (Panthera leo) morbillivirus in ferrets (Mustela putorius furo). Vet. Pathol. 2001, 38, 311–316. [Google Scholar] [CrossRef]
- Beineke, A.; Puff, C.; Seehusen, F.; Baumgärtner, W. Pathogenesis and immunopathology of systemic and nervous canine distemper. Vet. Immunol. Immunopathol. 2009, 127, 1–18. [Google Scholar] [CrossRef]
- Kauffman, C.A.; Bergman, A.G.; O’Connor, R.P. Distemper virus infection in ferrets: An animal model of measles-induced immunosuppression. Clin. Exp. Immunol. 1982, 47, 617–625. [Google Scholar] [PubMed]
- Stephensen, C.B.; Welter, J.; Thaker, S.R.; Taylor, J.; Tartaglia, J.; Paoletti, E. Canine distemper virus (CDV) infection of ferrets as a model for testing morbillivirus vaccine strategies: NYVAC- and ALVAC-based CDV recombinants protect against symptomatic infection. J. Virol. 1997, 71, 1506–1513. [Google Scholar] [CrossRef] [PubMed]
- Welter, J.; Taylor, J.; Tartaglia, J.; Paoletti, E.; Stephensen, C.B. Vaccination against canine distemper virus infection in infant ferrets with and without maternal antibody protection, using recombinant attenuated poxvirus vaccines. J. Virol. 2000, 74, 6358–6367. [Google Scholar] [CrossRef] [PubMed]
- Zehnder, A.M.; Hawkins, M.G.; Kiski, M.A.; Luff, J.A.; Benak, J.; Lowenstine, L.J.; White, S.D. An unusual presentation of canine distemper virus infection in a domestic ferret (Mustela putorius furo). Vet. Dermatol. 2008, 19, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Von Messling, V.; Springfeld, C.; Devaux, P.; Cattaneo, R. A ferret model of canine distemper virus virulence and immunosuppression. J. Virol. 2003, 77, 12579–12591. [Google Scholar] [CrossRef] [PubMed]
- Weckworth, J.K.; Davis, B.W.; Roelke-Parker, M.E.; Wilkes, R.P.; Packer, C.; Eblate, E.; Schwartz, M.K.; Mills, L.S. Identifying Candidate Genetic Markers of CDV Cross-Species Pathogenicity in African Lions. Pathogens 2020, 9, 872. [Google Scholar] [CrossRef] [PubMed]
- Origgi, F.C.; Plattet, P.; Sattler, U.; Robert, N.; Casaubon, J.; Mavrot, F.; Pewsner, M.; Wu, N.; Giovannini, S.; Oevermann, A.; et al. Emergence of ca-nine distemper virus strains with modified molecular signature and enhanced neuronal tropism leading to high mortality in wild carnivores. Vet. Pathol. 2012, 49, 913–929. [Google Scholar] [CrossRef]
- Wilkes, R.P. Canine Distemper Virus in Endangered Species: Species Jump, Clinical Variations, and Vaccination. Pathogens 2022, 12, 57. [Google Scholar] [CrossRef]
- Loots, A.K.; Mokgokong, P.S.; Mitchell, E.; Venter, E.H.; Kotze, A.; Dalton, D.L. Phylogenetic analysis of canine distemper virus in South African wildlife. PLoS ONE 2018, 13, e0199993. [Google Scholar] [CrossRef]
- Willi, B.; Spiri, A.M.; Meli, M.L.; Grimm, F.; Beatrice, L.; Riond, B.; Bley, T.; Jordi, R.; Dennler, M.; Hofmann-Lehmann, R. Clinical and molecular investigation of a canine distemper out-break and vector-borne infections in a group of rescue dogs imported from Hungary to Switzerland. BMC Vet. Res. 2015, 11, 154. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, C.; Ding, H.; Cao, Y.; Sun, Z.; Wu, H.; Wang, L.; He, W.; Huang, B.; Xi, X.; et al. A highly virulent canine distemper virus strain isolated from vaccinated mink in China. Virus Genes 2021, 57, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Wade, L.L. Vaccination of Ferrets for Rabies and Distemper. Vet. Clin. N. Am. Exot. Anim. Pract. 2018, 21, 105–114. [Google Scholar] [CrossRef] [PubMed]
| Method | Primer Sequence (5′-3′) | Target | Size | Reference |
|---|---|---|---|---|
| RT-PCR | DMV 1: ATG TTT ATG ATC ACA GCG GT’ DMV 2: ATT GGG TTG CAC CAC TTG TC | P gene | 429 bp | [30] |
| RT-PCR/RFLP | B-for: AGG CCG TAC ATC ACC AAG TC B-rev: TGG TAA GCC ATC CGG AGT TC | H gene | 1110 bp | [31] |
| Sequence analysis | C-for: AAC TTA GGG CTC AGG TAG TC C-rev: AGA TGG ACC TCA GGG TAT AG | H gene | 2023 bp | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guercio, A.; Mira, F.; Di Bella, S.; Gucciardi, F.; Lastra, A.; Purpari, G.; Castronovo, C.; Pennisi, M.; Di Marco Lo Presti, V.; Rizzo, M.; et al. Biomolecular Analysis of Canine Distemper Virus Strains in Two Domestic Ferrets (Mustela putorius furo). Vet. Sci. 2023, 10, 375. https://doi.org/10.3390/vetsci10060375
Guercio A, Mira F, Di Bella S, Gucciardi F, Lastra A, Purpari G, Castronovo C, Pennisi M, Di Marco Lo Presti V, Rizzo M, et al. Biomolecular Analysis of Canine Distemper Virus Strains in Two Domestic Ferrets (Mustela putorius furo). Veterinary Sciences. 2023; 10(6):375. https://doi.org/10.3390/vetsci10060375
Chicago/Turabian StyleGuercio, Annalisa, Francesco Mira, Santina Di Bella, Francesca Gucciardi, Antonio Lastra, Giuseppa Purpari, Calogero Castronovo, Melissa Pennisi, Vincenzo Di Marco Lo Presti, Maria Rizzo, and et al. 2023. "Biomolecular Analysis of Canine Distemper Virus Strains in Two Domestic Ferrets (Mustela putorius furo)" Veterinary Sciences 10, no. 6: 375. https://doi.org/10.3390/vetsci10060375
APA StyleGuercio, A., Mira, F., Di Bella, S., Gucciardi, F., Lastra, A., Purpari, G., Castronovo, C., Pennisi, M., Di Marco Lo Presti, V., Rizzo, M., & Giudice, E. (2023). Biomolecular Analysis of Canine Distemper Virus Strains in Two Domestic Ferrets (Mustela putorius furo). Veterinary Sciences, 10(6), 375. https://doi.org/10.3390/vetsci10060375





