Simple Summary
RNA N6-methyladenosine (m6A) modification plays an important role in RNA processing, transport, translation, and stem cell differentiation. However, the function of METTL3 in quails’ skeletal muscle remains unknown. In this study, we cloned the full-length coding sequence of the quail METTL3 and examined its control of myoblast proliferation and differentiation, as well as the downstream genes and signaling pathways that it regulates. Our findings shed light on the regulatory network that governs poultry skeletal muscle growth and development.
Abstract
Methyltransferase 3 (METTL3), which has been demonstrated to play a crucial role in a variety of biological processes, is the key enzyme for catalyzing m6A modification in RNA. However, the complete protein sequence of METTL3 in quail has not been annotated, and its function in skeletal muscle of quails remains unknown. In the current study, the full-length coding sequence of the quail METTL3 was obtained through the 3′ rapid amplification of cDNA ends (3’ RACE) and its homology with that of other species was predicted based on a generated phylogenetic tree. A Cell Counting Kit-8 assay and flow cytometry in a quail myoblast cell line (QM7) demonstrated that METTL3 promotes myoblast proliferation. The overexpression of METTL3 in QM7 cells significantly increased the expression levels of the myoblast differentiation markers myogenin (MYOG), myogenic differentiation 1 (MYOD1), and myocyte enhancer factor 2C (MEF2C), further demonstrating that METTL3 promotes myoblast differentiation. Additionally, transcriptome sequencing following METTL3 overexpression revealed that METTL3 controls the expression of various genes involved in RNA splicing and the regulation of gene expression, as well as pathways such as the MAPK signaling pathway. Taken together, our findings demonstrated that METTL3 plays a vital function in quail myoblast proliferation and differentiation and that the METTL3-mediated RNA m6A modification represents an important epigenetic regulatory mechanism in poultry skeletal muscle development.
1. Introduction
Skeletal muscle growth and development are closely associated with livestock and poultry meat production, with muscle quantity being the most economically important trait in the poultry industry [1]. Quail, a type of poultry, is mainly used for egg and meat production in China (http://www.moa.gov.cn/xw/zwdt/202005/t20200529_6345528.htm, accessed on 29 May 2020). With the increase in meat consumption in this country, quail meat production performance has received increasing attention from breeders. The quail industry has broad development prospects owing to the small size, short breeding cycle, and high nutritional value of the birds [2].
N6-methyladenosine (m6A) modification is the most common form of RNA modification in eukaryotic cells [3]. Recent studies have shown that RNA m6A modification plays an important role in RNA processing, transport, and translation, as well as in stem cell differentiation [4,5,6,7,8,9]. The catalysis of m6A is performed by several enzymes, including methyltransferase 3 (METTL3) and METTL16 [10,11]. METTL3 recognizes specific RNA sequences and catalyzes the formation of m6A, thereby affecting RNA stability, metabolism, and, consequently, its post-transcriptional regulation [12]. m6A modification plays an important role in embryonic development, cell differentiation, and nervous system development, and has also been implicated in tumor occurrence and progression [13].
Several lines of evidence indicate that the METTL3-mediated regulation of RNA metabolism plays a role in skeletal muscle development. For instance, the loss of Mettl3 in mice results in abnormal muscle development and impaired muscle repair [14]. In murine and bovine myoblasts, METTL3 promotes cell proliferation, while contradictory results exist regarding its role in cell differentiation [15,16,17,18]. In C2C12 cells, METTL3 was reported to enhance myogenic differentiation by mediating the m6A RNA modification of the key myogenic factor MYOD and by regulating the Notch signaling pathway [17,19]. Conversely, METTL3 was shown to suppress differentiation by regulating ERK signaling through the METTL3/METTL14/MNK2 axis [15]. In addition to its effects on functional genes, METTL3 has also been reported to promote the expression of muscle-specific miRNAs (miR-1a, miR-133a, miR-133b, miR-206) post-transcriptionally, thereby inhibiting the translation n of their target genes and suppressing muscle differentiation [16]. In primary bovine myotubes, the METTL3-m6A-YTHDF1 axis stabilizes MEF2C RNA and increases its translation, which promotes cell differentiation [20]. Moreover, METTL3 enhances myotube differentiation by downregulating the expression of the DNA demethylase TET1 [21]. Combined, these findings suggest that METTL3 plays a multidimensional regulatory role in myoblast differentiation; however, the underlying mechanisms remain poorly understood.
Investigating the genetic mechanisms underlying skeletal muscle development in poultry, including understanding the regulatory role of METTL3 in poultry skeletal muscle development, is vital for improving the economic benefits of the poultry industry. The aim of this study was to clone the full-length coding sequence (CDS) of quail METTL3, clarify its expression pattern during quail skeletal muscle development, and elucidate its regulatory role, including the potential underlying mechanisms in cell proliferation and differentiation using the QM7 cell line.
2. Materials and Methods
2.1. Animals and Cells
A total of 220 fertilized Korean Longcheng quail eggs were purchased from Hengyan Poultry Industry (Yichun, China). The eggs were incubated in a Hao Hua De incubator (Nanchang, China) at 38 °C with 60% relative humidity and were turned 12 times per day. The leg muscle tissue of quail embryos was collected daily from day 7 of incubation until hatching. Specifically, the legs were dissected from the embryos once they had been removed from the eggshell and the skin, bone, and claws were removed. The tissues were then frozen in liquid nitrogen and stored at −80 °C. The sex of each bird was determined by the PCR amplification of the chromodomain helicase DNA binding protein 1 gene (CHD1) using the primers listed in Table 1. The amplification was performed with Taq Master Mix (Dye Plus, Vazyme, Nanjing, China) in a T100 Thermal Cycler (Bio-Rad, Hercules, CA, USA) using the following program (20 μL reaction mixture): 95 °C for 3 min, followed by 36 cycles of 95 °C for 30 s, an annealing temperature for 30 s, and 72 °C for 60 s, with a final extension step at 72 °C for 5 min.

Table 1.
The information of primers and small interfering RNAs.
The QM7 cell line used in this study was obtained from our college. The cells were cultured in M199 medium (Cytiva, MA, USA) containing 10% fetal bovine serum (Gibco, Grand Island, NE, USA), 10% tryptose phosphate broth solution (Sigma, St. Louis, MO, USA), and 0.5% penicillin/streptomycin (Gibco) at 37 °C with 5% CO2 in a humidified cell culture incubator (BB150; Thermo Scientific, Waltham, MA, USA). At 90%~100% confluence, the cells were detached in 1 mL of 0.25% trypsin solution for 1 min, followed by the addition of 4 mL of complete medium to halt digestion. The cells were then dissociated into single-cell suspensions by gentle tapping and collected by centrifugation at 1000 rpm for 5 min for sub-culture or plating. After achieving 90% confluency, the QM7 were induced to differentiation with M199 medium supplemented with 2% horse serum. Following the induction of differentiation, QM7 cells were collected daily up to the fifth day (DM1–DM5).
2.2. RNA Extraction and 3′ Rapid Amplification of cDNA Ends (RACE) PCR
RNA was isolated from tissues and cells using the FastPure Cell/Tissue Total RNA Isolation Kit (Vazyme). First-strand cDNA for 3′ RACE was synthesized using the SMART RACE cDNA Amplification Kit (Clontech, Otsu, Japan) with the RNA from quail leg muscle as a template. The gene-specific primers were designed based on the NCBI reference sequence XM_015850477.2 using Oligo 7 and were synthesized by TsingKe (Changsha, China) (Table 1). RACE-PCR was performed using the KOD FX (Toyobo, Osaka, Japan) touch-down PCR program as follows: initial denaturation at 94 °C for 2 min; 5 cycles of 98 °C for 10 s and 74 °C for 2 min; 5 cycles of 98 °C for 10 s and 72 °C for 2 min; 5 cycles of 98 °C for 10 s and 70 °C for 2 min; and 25 cycles of 98 °C for 10 s and 68 °C for 2 min, followed by a final extension at 68 °C for 7 min and storage at 4 °C. The PCR products were purified using the FastPure Gel DNA Extraction Mini Kit (Vazyme), according to the manufacturer’s instructions, and were then cloned into the pJET 1.2/Blunt vector (Thermo Scientific) for Sanger sequencing (Tsingke). The sequencing results were assembled and analyzed using DNASTAR SeqMan (https://www.dnastar.com/, accessed on 2 July 2021) and the phylogenetic tree was constructed using DNAstar MegAlign.
2.3. cDNA Synthesis and Fluorescence Quantitative PCR (qPCR)
Total RNA extracted from cells or tissues was reverse transcribed using the HiScript II 1st Strand cDNA Synthesis Kit with gDNA wiper (Vazyme), according to the manufacturer’s instructions. The synthesized cDNA was diluted at a ratio of 1:4 and used for subsequent qPCR analysis. qPCR was performed using 2× T5 Fast qPCR Mix (Tsingke) on an ABI Q5 Real-Time PCR System (Thermo Scientific). The qPCR steps were as follows (10 µL reaction volume): 95 °C for 3 min, followed by 40 cycles of 95 °C for 10 s and annealing temperature for 1 min, and melt-curve analysis was performed at 65–95 °C. Three technical replicates were performed for each sample, and the quantification results were calculated using the 2−∆∆Ct with quail GAPDH serving as the internal control. The results are presented as means ± S.E.M. The primers were designed using NCBI Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/, accessed on 10 January 2022) and the sequences are shown in Table 1.
2.4. Vector Construction and Small Interfering RNA (siRNA) Synthesis
The assembled METTL3 CDS was synthesized by Tsingke and inserted into the pEGFP-C1-3 × flag vector at the BglII and HindIII restriction sites. The Endo-Free Plasmid DNA Mini Kit II (Omega, Guangzhou, China) was used for plasmid purification (METTL3 and negative control). siRNAs targeting quail METTL3 were designed using BLOCK-iT RNAi Designer (https://rnaidesigner.thermofisher.com/rnaiexpress/sort.do, accessed on 16 January 2022) and synthesized by Tsingke. The target sequences are shown in Table 1. Cell transfection was performed with Lipo8000 according to the manufacturer’s instructions (Beyotime, Shanghai, China). The amount of transfection reagent used was 5/8 of the recommended amount.
2.5. Western Blotting
Western blotting was performed as previously described [22], with minor modifications. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer containing phenylmethanesulfonyl fluoride (PMSF) (Beyotime) for 5 min, centrifuged at 12,000 rpm for 10 min at 4 °C, and the supernatant was then collected. Protein was separated by SDS–PAGE, blotted onto nitrocellulose membranes (Merck Millipore, Billerica, MA, USA) and labeled with primary and secondary antibodies. The primary antibody was anti-DYKDDDDK (1:10,000; M20008, Abmart, Shanghai, China) and the secondary antibody was HRP-conjugated goat anti-mouse lgG (1:20,000; Signalway Antibody, MD, USA). The gel was exposed, and images were captured on an Amersham Imager 600 (GE Healthcare, Uppsala, Sweden).
2.6. Cell Cycle Analysis
QM7 cells were seeded in 12-well plates and transfected with pEGFP-C1-3 × flag-METTL3, pEGFP-C1-3 × flag, si-METTL3, or si-NC (negative control) using Lipo8000. After 6 h of transfection, the cells were refreshed with complete medium and harvested after 42 h. Cell cycle stage analysis was performed by flow cytometry as previously described [23].
2.7. Cell Counting Kit-8 (CCK-8) Assay
QM7 cells were seeded in 96-well plates and transfected with pEGFP-C1-METTL3, pEGFP-C1, si-METTL3, or si-NC, and cell proliferation was evaluated every 12 h using CCK-8 reagent (Meilunbio, Dalian, China). Specifically, 10 μL of CCK-8 solution was added to each well, and the cells were incubated for 2 h at 37 °C in a CO2 cell culture incubator. The absorbance of each well at 450 nm was measured using a microplate reader (Infinite 200 PRO, Tecan, Männedorf, Switzerland). After the assay, the cells were washed two or three times with PBS and then replenished with complete medium.
2.8. RNA-seq and Data Analysis
QM7 cells were seeded in 12-well plates, transfected with pEGFP-C1-METTL3 or pEGFP-C1 for 6 h, and then cultured in fresh complete medium for 42 h. RNA was subsequently extracted from the cells using the FastPure Cell/Tissue Total RNA Isolation Kit. Total RNA quantity and quality were determined using a Nanodrop ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA) and a Bioanalyzer 2100 (Agilent, CA, USA), respectively. Poly(A) RNA sequencing libraries were generated and paired-end (PE150) sequenced using an Illumina Novaseq 6000 System (LC Bio, Hangzhou, China) according to standard protocols. Clean reads were mapped to the Coturnix japonica reference genome (https://www.ncbi.nlm.nih.gov/genome/113?genome_assembly_id=265191, accessed on 20 June 2022) and subjected to subsequent analysis following previously described methods [24]. Gene set enrichment analysis (GSEA) was conducted as previously described [25]. Pathways with |normalized enriched scores (NESs)| > 1, NES p-values < 0.05, and false discovery rate (FDR) q-values < 0.05 were considered significantly enriched.
3. Results
3.1. Quail Sex Determination
The quail CHD1 gene is located on the sex chromosome. Here, the agarose gel electrophoresis of CHD1 amplicons identified 63 female quail embryos (two bands (638 and 470 bp) and 55 male quail embryos (one band (638 bp)) (Figure 1A, Table 2).
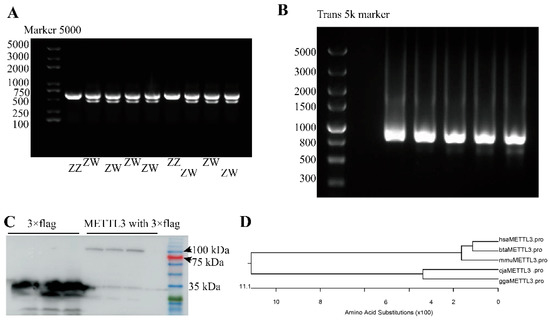
Figure 1.
Cloning and sequence analysis of the coding sequence (CDS) of quail METTL3. (A) PCR products of CHD1 gene amplification. The products observed in different lanes are derived from distinct quail individuals. (B) Agarose gel electrophoresis of 3′ RACE-PCR products. The products observed in different lanes represent the same product obtained from various PCR tubes. (C) The western blotting analysis of the METTL3-3 × flag fusion protein and the negative control. The initial three lanes depict the protein products of blank vector with 3 × flag and eGFP-fused, whereas the latter three lanes represent the protein products of quail METTL3 fused with 3 × Flag and eGFP. (D) The phylogenetic tree generated for METTL3. (Please find the WB full membrane and the full images of agarose gel electrophoresis of Figure 1 in Figure S1).

Table 2.
Count of quail embryos by sex.
3.2. Cloning and Sequence Analysis of the Full-Length CDS of METTL3
Before constructing the expression vector, we compared the size of the METTL3 protein of quail in the NCBI database (XP_015705963.1, 477 aa) with that of chicken (XP_040510970.1, 574 aa), human (NP_062826.2, 580 aa), mouse (NP_062695.2, 580 aa), and cattle (NP_001095708.1, 580 aa) and found it to be smaller. This suggested that the CDS of quail METTL3 was incomplete; therefore, we subsequently amplified the full length of METTL3 using RACE-PCR and obtained an 889-bp sequence (Figure 1B). After removing the part that overlapped with the known sequence, we obtained a new 3′ sequence of 384 bp for quail METTL3 (Supplementary File S1). The splicing of the sequence obtained by 3′ RACE yielded the new RNA for METTL3, which contained a CDS region of 1683 bp that was predicted to encode a protein of 560 aa (Supplementary File S1). Based on the results of Western blotting analysis, it was determined that the size of the quail METTL3 protein after fusion with 3 × flag and eGFP was approximately 100 kDa. In comparison, the size of the 3 × flag protein fused with eGFP in the empty vector was approximately 30 kDa. Thus, we can infer that the size of the quail METTL3 protein is approximately 70 kDa (Figure 1C). The predicted protein was closer in length and size to the METTL3 proteins of chicken, human, mouse, and cattle, while the protein sequence of quail METTL3 was closer to that of chicken than to that of humans, mice, and cattle METTL3, which was expected (Figure 1D). These observations demonstrated that the obtained CDS of quail METTL3 was the full-length CDS.
3.3. Expression Patterns of Differentiation Markers and METTL3 during Quail Embryo Leg Muscle Development
To investigate the key events of quail myoblast differentiation, we extracted total RNA from leg muscles at various stages of embryonic development and used qPCR to determine the expression patterns of the differentiation marker genes MYOG and MYOD and the muscle-inhibiting factor MSTN. In order to identify patterns of gene expression, four female quail individuals were used at each time point. The results showed that MYOG expression remained low at E7 and E8, significantly increased from E8 to E9, remained at a high level from E9 to E10, and then gradually decreased from E10 (Figure 2A). MYOD expression gradually increased from E7 to E10 and significantly decreased from E10 (Figure 2B). MSTN expression remained low at E7 and E8, significantly increased from E8 to E9, and, subsequently, gradually decreased from E9 (Figure 2C). The expression level of METTL3 during quail leg muscle development was relatively low at E7 and E8, peaked at E9, and gradually decreased during subsequent embryonic development (Figure 2D). These results suggested that METTL3 expression gradually decreases during quail leg muscle differentiation.
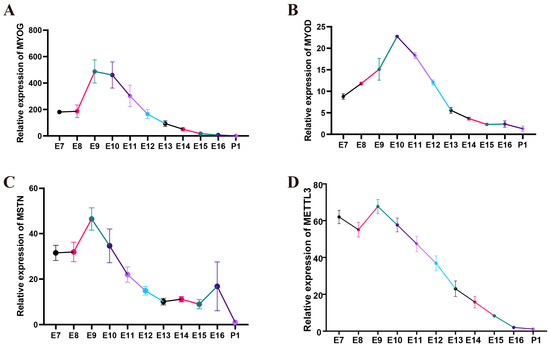
Figure 2.
The spatiotemporal expression of skeletal muscle differentiation marker genes and METTL3 in quail leg muscle. (A–D) Spatiotemporal expression of MYOG, MYOD, MSTN, and METTL3 from E7 to P1 in leg muscle. E7–E16 represents embryonic day 7 to embryonic day 16, and P1 represents post-hatch day 1. Four female quails were used at each time point. Different colors in the figure indicate the different developmental stages of quail leg muscle.
3.4. The Effects of METTL3 on QM7 Cell Proliferation
To investigate the role of METTL3 in QM7 cell proliferation, we first validated the efficiency of METTL3 overexpression and knockdown in QM7 cells. Quantitative analysis showed that the expression of the METTL3 gene in cells transfected with the overexpression vector (pEGFP-C1-METTL3) was significantly higher (an approximately 150-fold increase, p < 0.001) than that in the control group (Figure 3A). Of the three siRNAs we designed, si-1 achieved an efficiency of over 50% in knocking down METTL3 gene expression (Figure 3B) and was thus used in subsequent METTL3 knockdown experiments. The flow cytometric analysis of the cell cycle in QM7 cells overexpressing METTL3 showed that, compared with the negative controls, there was a significant decrease (p < 0.001) in the proportion of cells in the G1 phase and a significant increase (p < 0.01) in that of cells in the S phase compared with that in the negative control group (Figure 3C). Conversely, the knockdown of METTL3 resulted in a significant decrease (p < 0.01) in the proportion of cells in the G2 phase (Figure 3D), indicating that METTL3 can accelerate cell cycle progression. CCK-8 assay results showed that METTL3 overexpression significantly increased (p < 0.001) the proliferative ability of QM7 cells (Figure 3E), while the knockdown of METTL3 exerted the opposite effect (p < 0.01) (Figure 3F). These results indicate that the METTL3 gene enhances cell proliferation in QM7 cells.
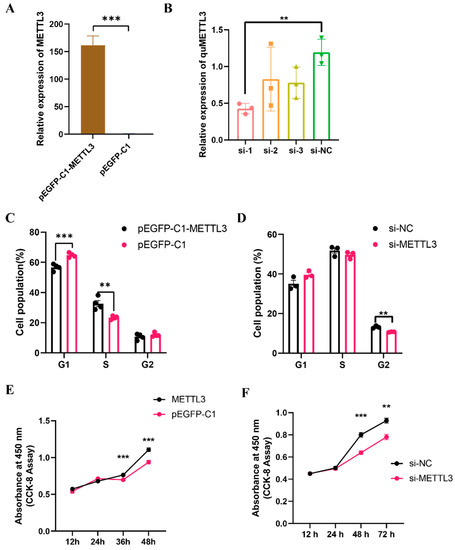
Figure 3.
METTL3 enhances the proliferative ability of QM7 cells. (A,B) The relative expression level of METTL3 after the overexpression (A) and knockdown (B) of METTL3. (C) Cell cycle analysis of QM7 cells after the transfection of pEGFP-C1-METTL3 or pEGFP-C1. (D) Cell cycle analysis of QM7 cells after the transfection of siRNA targeting METTL3 (si-1) or the negative control (si-NC). (E,F) CCK-8 assay in QM7 cells after the overexpression or knockdown of METTL3. ** indicates p < 0.01; *** indicates p < 0.001.
3.5. The Regulatory Effect of METTL3 Protein in QM7 Cell Differentiation
The mRNA expression level of METTL3 gradually decreased during the proliferation phase (GM) and various stages of differentiation (DM1–DM5) in QM7 cells (Figure 4A), suggesting that METTL3 plays an important role in myoblast differentiation. The overexpression of METTL3 significantly increased the expression levels of myogenic and fusion marker genes (MYOG, MYOD, and MEF2C) (Figure 4B), whereas the opposite was seen with METTL3 knockdown (Figure 4C). These results indicated that METTL3 can promote differentiation in QM7 cells.
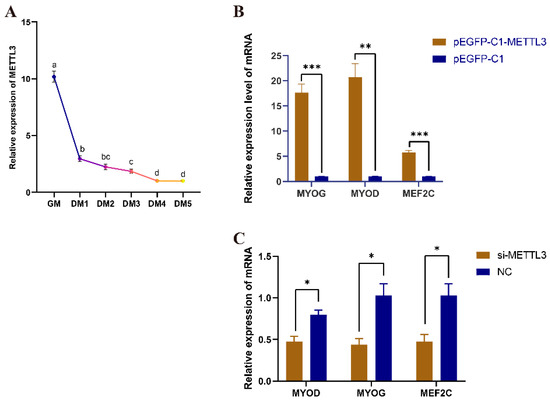
Figure 4.
METTL3 promotes the differentiation of QM7 cells. (A) The expression level of METTL3 before and after QM7 cell differentiation. Different colors indicate the different differentiation stages of QM7. The significant differences between the two groups are indicated by the distinct letters (p < 0.05). (B) The expression levels of MYOG, MYOD, and MEF2C after the overexpression of METTL3. (C) The expression levels of MYOG, MYOD, and MEF2C after the knockdown of METTL3. GM represents growth medium and the cells undergo proliferation, whereas the differentiation medium (DM) phases, DM1–DM5, represent the first to fifth day of differentiation. * indicates p < 0.05; ** indicates p < 0.01; *** indicates p < 0.001.
3.6. The Effects of METTL3 Overexpression on the Transcriptome of QM7 Cells
After determining the effect of METTL3 overexpression in QM7 cells, cells in the overexpression and negative control groups were subjected to poly(A)-tail library sequencing. Differential gene expression analysis identified a total of 138 differentially expressed genes, 80 of which were upregulated and 58 downregulated (Figure 5A, Table S1). KEGG pathway enrichment analysis showed that the differentially expressed genes were significantly enriched in 14 signaling pathways, including those related to lipid metabolism, vascular smooth muscle contraction, and MAPK signaling pathway-fly (Figure 5B, Table S2). LOC107314843, LOC107320189, LOC107314846, receptor activity modifying protein 2 (RAMP2), and RAMP3 were enriched on vascular smooth muscle contraction. Docking protein 5 (DOK5) was enriched on MAPK signaling pathway-fly. It is noteworthy that the gene exhibiting the highest expression level and most significant differential expression among all genes was the Heat shock protein beta-9 (HSPB9). In addition, GSEA was used to analyze the overall effect of METTL3 on the transcriptome of QM7 cells. The results indicated that significant changes occurred in the MANNOSE_TYPE_O_GLYCAN_BIOSYNTHEIS, RIBOSOME, and PENTOSE_PHOSPHATE_PATHWAY signaling pathways (|NES| > 1, NOM.pval < 0.05, FDR.qval < 0.05, Figure 5C, Table S3). In the RIBOSOME signaling pathway, the expression of RPL and RPS family genes related to ribosome function, RNA processing, and translation was significantly altered (Table S4).
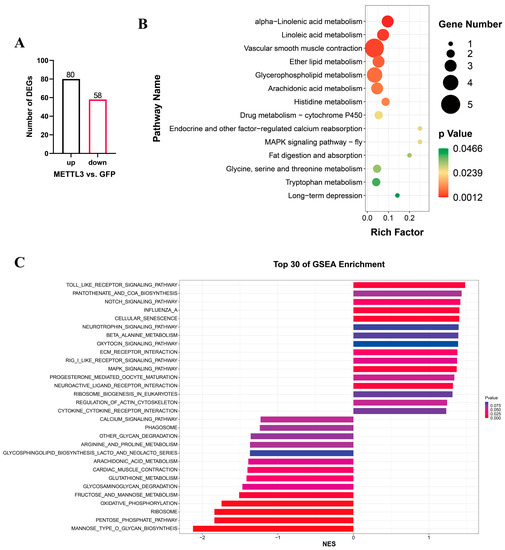
Figure 5.
RNA-seq analysis in QM7 cells overexpressing METTL3. (A) The number of differentially expressed genes (DEGs) after METTL3 overexpression. (B) KEGG pathway enrichment analysis relating to the differentially expressed genes. (C) The top 30 enriched KEGG pathways based on GSEA with the RNA-seq data.
4. Discussion
The m6A modification mediated by METTL3 is known to play multiple roles in muscle development [26]; however, whether it exerts a regulatory function in avian myoblast proliferation and differentiation is unknown. In this study, we performed 3′ RACE using METTL3 gene-specific primers designed based on the predicted sequence of quail METTL3 mRNA in the NCBI database (XM_015850477.2). The 3′ RACE product was cloned, sequenced, and spliced, yielding the predicted mRNA sequence of METTL3. However, despite multiple primer design attempts, we were unable to obtain results for 5′ RACE. Sequence analysis showed that the predicted 1823 bp METTL3 mRNA containing a CDS of 1683 bp that was predicted to encode a 560 aa amino acid protein similar in length to METTL3 proteins from humans, mice, cows, and chickens. Western blotting analysis indicated that the protein had a molecular mass of approximately 100 kDa with the 3× flag-eGFP and approximately 70 kDa without the 3× flag-eGFP, which was similar to that seen for METTL3 proteins from other species. Homology analysis results indicated that the obtained CDS represented the full-length CDS of quail METTL3 and was, therefore, used in subsequent cellular experiments.
Upon proliferation, migration, and contact with other cells, myogenic cells undergo a cessation of proliferation and transition into a state of cell cycle arrest. They then express the myogenic regulatory factors MYOG and MYOD to promote differentiation into myocytes, which migrate, recognize each other, and ultimately fuse into myotubes [27]. MSTN is a muscle growth inhibitor the expression of which initially increases, and then decreases, during avian muscle differentiation [28,29]. To better understand the changes in METTL3 expression during quail leg muscle development, we first explored the spatiotemporal expression patterns of differentiation marker genes in quail leg muscle to identify developmental turning points. MYOG, MYOD, and MSTN all showed a trend of an initial increase in expression, followed by a decrease, with expression peaking at E9 and E10. This suggested that quail leg muscle differentiation begins at E8, peaks at E10, and is mostly completed by E13. The expression of METTL3 in undifferentiated quail leg muscle was relatively high and gradually decreased starting at E9, indicating that METTL3 expression decreases with increasing muscle differentiation. This result was consistent with the spatiotemporal expression patterns of METTL3 in the hindlimbs of mice, suggesting that a decrease in METTL3 expression is necessary for quail skeletal muscle differentiation [15].
Studies have demonstrated that METTL3 regulates myoblast proliferation and differentiation by modulating multiple genes and pathways through m6A RNA modification [18,21]. METTL3 enhances cell proliferation by elevating the m6A modification level of p21 and p27 mRNA, which shortens their half-lives [18]. METTL3 also promotes myoblast differentiation by increasing the stability of the mRNA of myogenic regulatory factors such as MyHC, MYOD, and MYOG through m6A modification [18]. In addition, METTL3 was shown to promote the differentiation of C2C12 cells by regulating the Notch signaling pathway [17]. Furthermore, METTL3 can suppress the expression of the DNA demethylase TET1 through m6A modification, which, in turn, promotes METTL3 expression by mediating the demethylation of the METTL3 DNA region, leading to the regulation of the m6A modification of the entire genome and transcriptome and the promotion of myoblast differentiation [21]. In bovine myoblasts, the addition of the methylation inhibitor 3-Deazaadenosine (DAA) reduces the expression of MEF2C protein, and interference with METTL3 also inhibits the expression of MEF2C protein, indicating that RNA m6A modification helps to stabilize MEF2C mRNA, promote its protein expression, and, thus, promote myogenic differentiation [20]. In contrast, in C2C12 cells, METTL3 can inhibit myoblast differentiation by increasing the expression of muscle-specific miRNAs [16]. In complex with METTL14, METTL3 also mediates the m6A modification of MNK2, thereby exerting regulatory effects on the ERK signaling pathway. The knockout of both METTL3 and METTL14 promotes myoblast differentiation [15]. We speculate that METTL3 regulates myoblast differentiation through multiple dimensions, which may be due to the overexpression and interference efficiency during experimentation in different papers. The effects of genes on the same process may vary depending on the species, culture conditions, and regulatory dimensions. Different experiments adopt varying treatments, transfection reagents, and gene expression methods, which can result in inconsistent experimental results. The most reliable outcomes can be obtained by knocking out a specific gene in vivo level. METTL3’s regulation of m6A modification is characterized by changes in the overall transcriptome modification level, with knockdown being less effective than overexpression. Here, we found that, in QM7 cells, METTL3 accelerates the cell cycle, promotes proliferation, and enhances differentiation.
To further investigate the potential mechanisms underlying the METTL3-mediated regulation of QM7 cell proliferation, we overexpressed METTL3 and then performed an RNA-seq analysis, with pEGFP-C1-transfected cells serving as a negative control. RNA-seq data analysis revealed significant changes in the expression of 138 genes in METTL3-overexpressing cells. KEGG pathway enrichment analysis showed that the differentially expressed genes were significantly enriched in the MAPK signaling pathway-fly. GSEA similarly showed significant changes in the expression of genes in the MAPK signaling pathway. Cellular processes, such as cell differentiation, cell division, cell proliferation, metabolism, and apoptosis, are regulated by the MAPK signaling pathway [30]. MAPK signaling plays a crucial role in myogenesis [31]. These observations are consistent with a previous RNA-seq analysis on C2C12 cells overexpressing METTL3 in which a significant enrichment of differentially expressed genes in the MAPK signaling pathway was reported [15]. Interestingly, our RNA-seq results, as well as previous studies in C2C12 cells overexpressing METTL3, showed the enrichment of partially overlapping signaling pathways such as the notch signaling pathway and ECM receptor interaction, suggesting that METTL3 regulates multiple genes and signaling pathways. The RIBOSOME pathway, which plays an important role in cell cycle regulation, RNA binding, and mRNA translation [32], was also significantly enriched in METTL3-overexpressing QM7 cells in our study. The regulation of the RIBOSOME pathway by METTL3 may be one dimension of its multidimensional regulatory function. Notably, our RNA-seq analysis of QM7 cells overexpressing METTL3 showed primarily the enrichment of regulatory genes and pathways associated with cell proliferation. The mechanisms underlying the METTL3-mediated regulation of QM7 cell differentiation require further exploration.
5. Conclusions
Gene cloning and sequence analysis showed that the quail METTL3 protein has high homology with its mammalian and chicken counterparts. Additionally, quail METTL3 was found to promote the proliferation and differentiation of skeletal muscle cells.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/vetsci10040300/s1, Supplementary File S1: the sequence of 3′RACE product and the splicing mRNA of quail METTL3; Table S1: the list of the differentially expressed genes in METTL3 overexpression group vs. negative control group; Table S2: the KEGG analysis with all differentially expressed genes in METTL3 overexpression group vs. negative control group; Table S3: the GSEA KEGG analysis of the gene expression profile in METTL3 overexpression group vs. negative control group; Table S4: the GSEA details of RIBOSOME pathway; Figure S1: the WB full membrane and the full images of agarose gel electrophoresis of Figure 1.
Author Contributions
Conceptualization, B.C. and J.X.; methodology, J.L.; software, S.L. (Shuibing Liu); validation, J.L., W.Z. and H.J.; formal analysis, B.C.; investigation, B.C.; resources, S.L. (Sanfeng Liu); data curation, B.C.; writing—original draft preparation, B.C.; writing—review and editing, W.L.; visualization, B.C.; supervision, S.L. (Sanfeng Liu) and B.C.; project administration, S.L. (Sanfeng Liu); funding acquisition, S.L. (Sanfeng Liu). All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by the Department of Agriculture and Rural Affairs of Jiangxi Province. The project names and grant numbers are the Technology System of Modern Agricultural Poultry Industry of Jiangxi Province (JXARS-09) and Jiangxi Joint Key Project of Quail Improvement (2022JXCQZY02).
Institutional Review Board Statement
The animal experiments in this study were conducted in accordance with Jiangxi Agricultural University’s ethical standards and regulations (JXAULL-2017002). The current experiment was approved with the authorization code JXAU#2022D004.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the RNA sequencing data and processing files can be found on the China National Center for Bioinformation website, under accession number PRJCA015359. The information will be released on 4 March 2025.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Baéza, E.; Guillier, L.; Petracci, M. Review: Production Factors Affecting Poultry Carcass and Meat Quality Attributes. Animal 2022, 16, 100331. [Google Scholar] [CrossRef] [PubMed]
- Shafik, B.M.; Kamel, E.R.; Mamdouh, M.; Elrafaay, S.; Nassan, M.A.; El-Bahy, S.M.; El-Tarabany, M.S.; Manaa, E.A. Performance, Blood Lipid Profile, and the Expression of Growth Hormone Receptor (GHR) and Insulin-like Growth Factor-1 (IGF-1) Genes in Purebred and Crossbred Quail Lines. Animals 2022, 12, 1245. [Google Scholar] [CrossRef] [PubMed]
- Sendinc, E.; Shi, Y. RNA M6A Methylation across the Transcriptome. Mol. Cell 2023, 83, 428–441. [Google Scholar] [CrossRef] [PubMed]
- Livneh, I.; Moshitch-Moshkovitz, S.; Amariglio, N.; Rechavi, G.; Dominissini, D. The M6A Epitranscriptome: Transcriptome Plasticity in Brain Development and Function. Nat. Rev. Neurosci. 2020, 21, 36–51. [Google Scholar] [CrossRef]
- Shulman, Z.; Stern-Ginossar, N. The RNA Modification N6-Methyladenosine as a Novel Regulator of the Immune System. Nat. Immunol. 2020, 21, 501–512. [Google Scholar] [CrossRef]
- Vu, L.P.; Cheng, Y.; Kharas, M.G. The Biology of m6A RNA Methylation in Normal and Malignant Hematopoiesis. Cancer Discov. 2019, 9, 25–33. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Chen, J. M6A Modification in Coding and Non-Coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell 2020, 37, 270–288. [Google Scholar] [CrossRef]
- Frye, M.; Harada, B.T.; Behm, M.; He, C. RNA Modifications Modulate Gene Expression during Development. Science 2018, 361, 1346–1349. [Google Scholar] [CrossRef]
- Boulias, K.; Greer, E.L. Biological Roles of Adenine Methylation in RNA. Nat. Rev. Genet. 2023, 24, 143–160. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, Writing and Erasing MRNA Methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.-S.; Yang, Y.-G. Dynamic Transcriptomic m6A Decoration: Writers, Erasers, Readers and Functions in RNA Metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Xu, W.; Li, J.; He, C.; Wen, J.; Ma, H.; Rong, B.; Diao, J.; Wang, L.; Wang, J.; Wu, F.; et al. METTL3 Regulates Heterochromatin in Mouse Embryonic Stem Cells. Nature 2021, 591, 317–321. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The Role of M6A Modification in the Biological Functions and Diseases. Signal Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef]
- Petrosino, J.M.; Hinger, S.A.; Golubeva, V.A.; Barajas, J.M.; Dorn, L.E.; Iyer, C.C.; Sun, H.-L.; Arnold, W.D.; He, C.; Accornero, F. The M6A Methyltransferase METTL3 Regulates Muscle Maintenance and Growth in Mice. Nat. Commun. 2022, 13, 168. [Google Scholar] [CrossRef]
- Xie, S.-J.; Lei, H.; Yang, B.; Diao, L.-T.; Liao, J.-Y.; He, J.-H.; Tao, S.; Hu, Y.-X.; Hou, Y.-R.; Sun, Y.-J.; et al. Dynamic M6A MRNA Methylation Reveals the Role of METTL3/14-M6A-MNK2-ERK Signaling Axis in Skeletal Muscle Differentiation and Regeneration. Front. Cell Dev. Biol. 2021, 9, 744171. [Google Scholar] [CrossRef]
- Diao, L.-T.; Xie, S.-J.; Lei, H.; Qiu, X.-S.; Huang, M.-C.; Tao, S.; Hou, Y.-R.; Hu, Y.-X.; Sun, Y.-J.; Zhang, Q.; et al. METTL3 Regulates Skeletal Muscle Specific MiRNAs at Both Transcriptional and Post-Transcriptional Levels. Biochem. Biophys. Res. Commun. 2021, 552, 52–58. [Google Scholar] [CrossRef]
- Liang, Y.; Han, H.; Xiong, Q.; Yang, C.; Wang, L.; Ma, J.; Lin, S.; Jiang, Y.-Z. METTL3-Mediated M6A Methylation Regulates Muscle Stem Cells and Muscle Regeneration by Notch Signaling Pathway. Stem. Cells Int. 2021, 2021, 9955691. [Google Scholar] [CrossRef]
- Zhao, T.; Zhao, R.; Yi, X.; Cai, R.; Pang, W. METTL3 Promotes Proliferation and Myogenic Differentiation through M6A RNA Methylation/YTHDF1/2 Signaling Axis in Myoblasts. Life Sci. 2022, 298, 120496. [Google Scholar] [CrossRef]
- Kudou, K.; Komatsu, T.; Nogami, J.; Maehara, K.; Harada, A.; Saeki, H.; Oki, E.; Maehara, Y.; Ohkawa, Y. The Requirement of Mettl3-Promoted MyoD MRNA Maintenance in Proliferative Myoblasts for Skeletal Muscle Differentiation. Open Biol. 2017, 7, 170119. [Google Scholar] [CrossRef]
- Yang, X.; Ning, Y.; Raza, S.H.; Mei, C.; Zan, L. MEF2C Expression Is Regulated by the Post-Transcriptional Activation of the METTL3-M6A-YTHDF1 Axis in Myoblast Differentiation. Front. Vet. Sci. 2022, 9, 900924. [Google Scholar] [CrossRef]
- Yang, X.; Mei, C.; Raza, S.H.A.; Ma, X.; Wang, J.; Du, J.; Zan, L. Interactive Regulation of DNA Demethylase Gene TET1 and M6A Methyltransferase Gene METTL3 in Myoblast Differentiation. Int. J. Biol. Macromol. 2022, 223, 916–930. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Yu, J.; Guo, L.; Byers, M.S.; Wang, Z.; Chen, X.; Xu, H.; Nie, Q. Circular RNA CircHIPK3 Promotes the Proliferation and Differentiation of Chicken Myoblast Cells by Sponging MiR-30a-3p. Cells 2019, 8, 177. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, S.; Zhang, W.; Hu, X.; Mao, H.; Liu, S.; Chen, B. Transcriptome RNA Sequencing Reveals That Circular RNAs Are Abundantly Expressed in Embryonic Breast Muscle of Duck. Vet. Sci. 2023, 10, 75. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Liu, W.; Zhou, Y.; Zhang, Y.; Wu, J.; Wang, R.; Shao, L. Stage-Specific Requirement for METTL3-Dependent M6A Modification during Dental Pulp Stem Cell Differentiation. J. Transl. Med. 2022, 20, 605. [Google Scholar] [CrossRef] [PubMed]
- Bertocchi, M.; Sirri, F.; Palumbo, O.; Luise, D.; Maiorano, G.; Bosi, P.; Trevisi, P. Exploring Differential Transcriptome between Jejunal and Cecal Tissue of Broiler Chickens. Animals 2019, 9, 221. [Google Scholar] [CrossRef]
- Zhang, D.; Wu, S.; Zhang, X.; Ren, S.; Tang, Z.; Gao, F. Coordinated Transcriptional and Post-Transcriptional Epigenetic Regulation during Skeletal Muscle Development and Growth in Pigs. J. Anim. Sci. Biotechnol. 2022, 13, 146. [Google Scholar] [CrossRef]
- Li, M.; Zhou, X.; Chen, Y.; Nie, Y.; Huang, H.; Chen, H.; Mo, D. Not All the Number of Skeletal Muscle Fibers Is Determined Prenatally. BMC Dev. Biol. 2015, 15, 42. [Google Scholar] [CrossRef]
- Tao, Q.; Chen, Y.; Bai, D.-P.; Mai, L.; Fan, Q.-M.; Shi, Y.-Z.; Chen, C.; Li, A. Differential Expression of MSTN, IGF2BP1, and FABP2 across Different Embryonic Ages and Sexes in White Muscovy Ducks. Gene 2022, 829, 146479. [Google Scholar] [CrossRef]
- Gu, L.H.; Xu, T.S.; Huang, W.; Xie, M.; Shi, W.B.; Sun, S.D.; Hou, S.S. Developmental Characteristics of Pectoralis Muscle in Pekin Duck Embryos. Genet. Mol. Res. 2013, 12, 6733–6742. [Google Scholar] [CrossRef]
- Danquah, A.; de Zelicourt, A.; Colcombet, J.; Hirt, H. The Role of ABA and MAPK Signaling Pathways in Plant Abiotic Stress Responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef]
- Wei, Y.; Qi, T.; Cao, S.; Zhang, W.; Yu, F.; Zeng, H.; Weng, J. LncRNA XLOC_015548 Affects the Proliferation and Differentiation of Myoblasts via the MAPK Signaling Pathway. Exp. Biol. Med. 2023, 153537022311519. [Google Scholar] [CrossRef]
- Wilson, D.N.; Doudna Cate, J.H. The Structure and Function of the Eukaryotic Ribosome. Cold Spring Harb. Perspect. Biol. 2012, 4, a011536. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).