Antibacterial, Antifungal, and Anticancer Effects of Camel Milk Exosomes: An In Vitro Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Milk Sampling
2.2. Exosome Isolation and Characterization
2.3. Microbial Analysis
2.4. Cytotoxicity by MTT Assay
2.5. Experimental Design
2.6. Caspase 3 Activity Assay
2.7. Detection of Intracellular Reactive Oxygen Species
2.8. Gene Expression Analysis by qPCR
2.9. Statistical Analysis
3. Results
3.1. Characterization of Exosomes Derived from Camel Milk
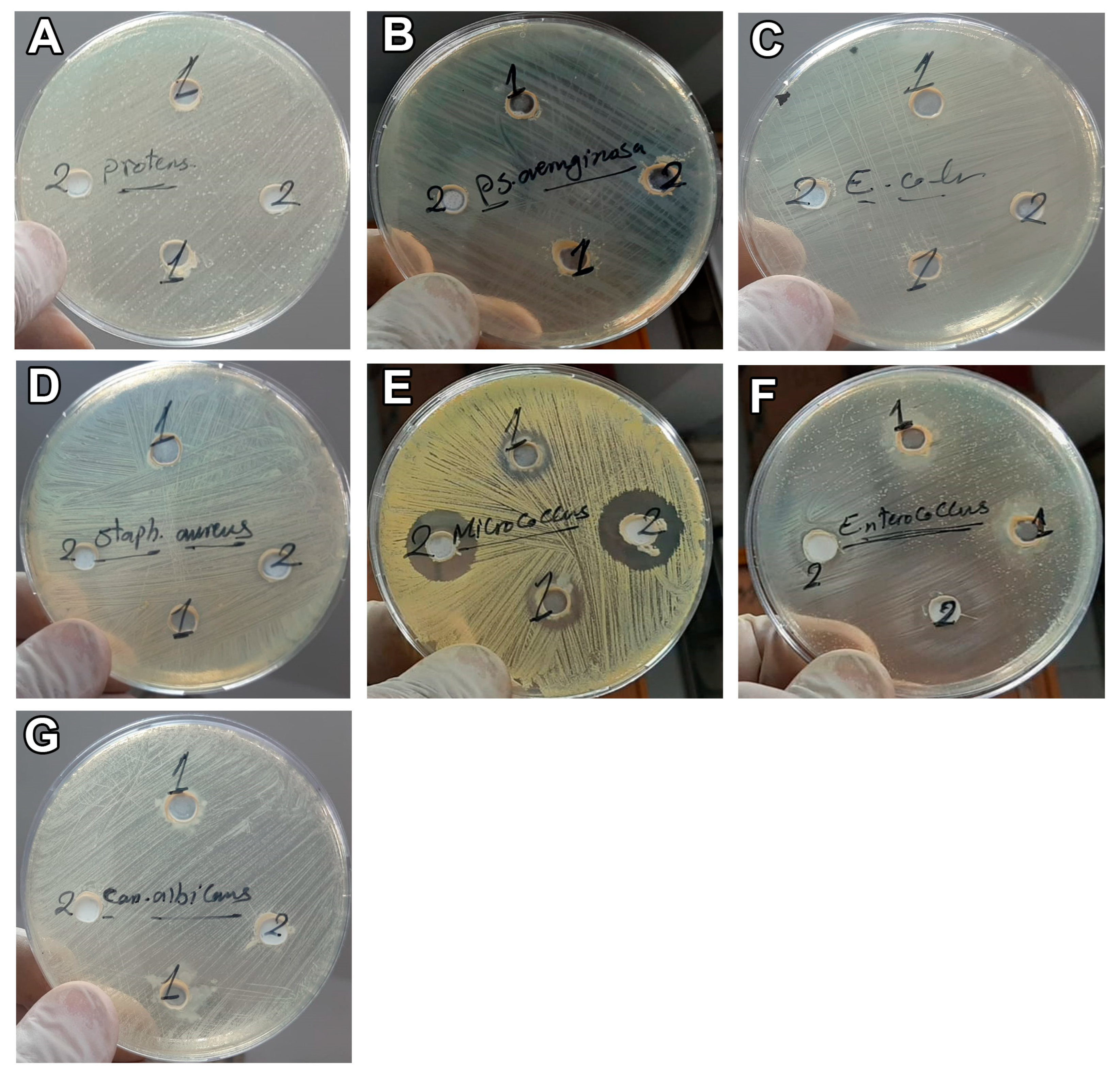
3.2. Antimicrobial Effect of Camel Milk Exosomes
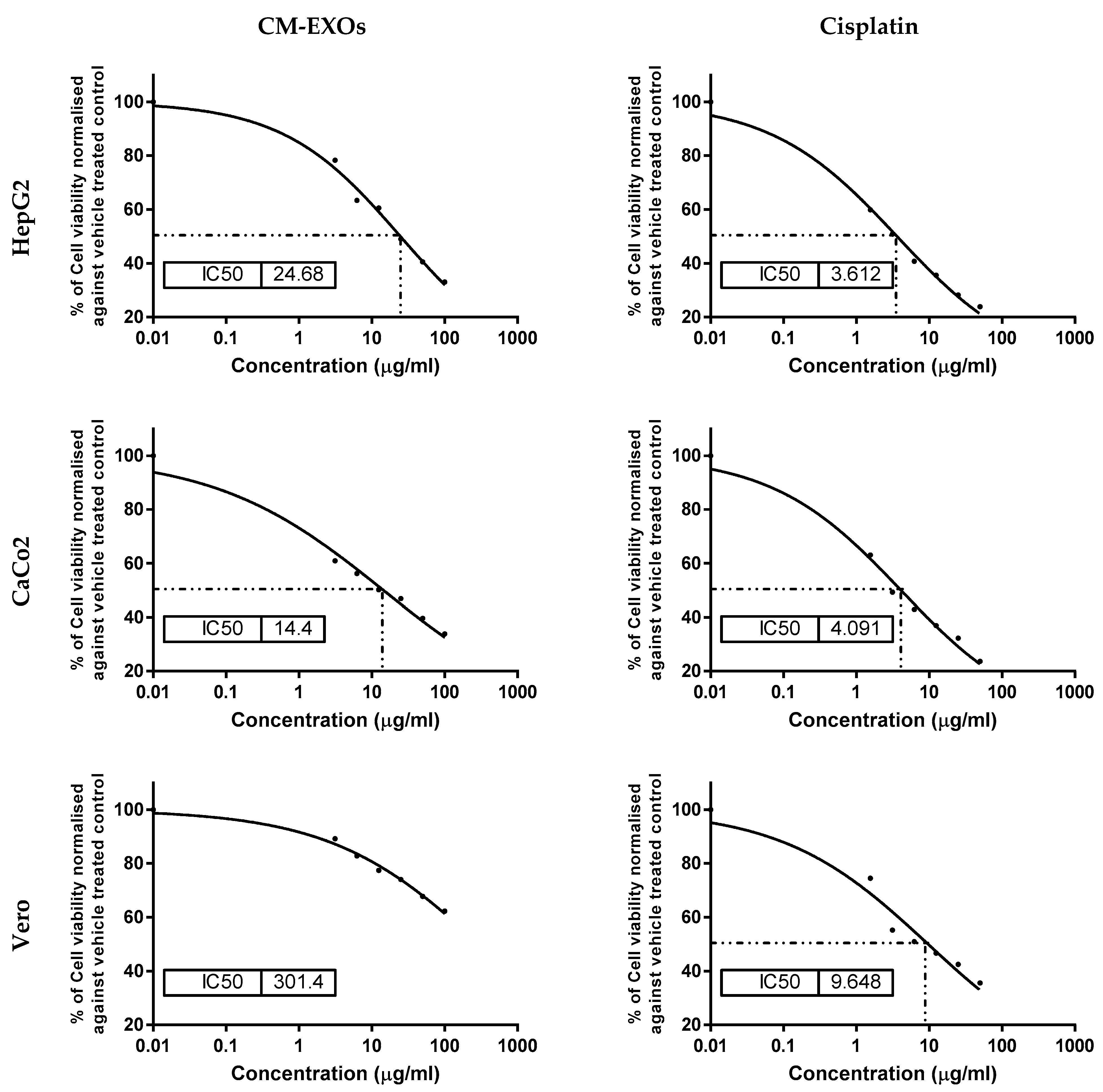
3.3. CM-EXOs Cytotoxic Effect on Cancer Cells
3.4. CM-EXOs Inhibited Cancer Cell Proliferation via Apoptosis
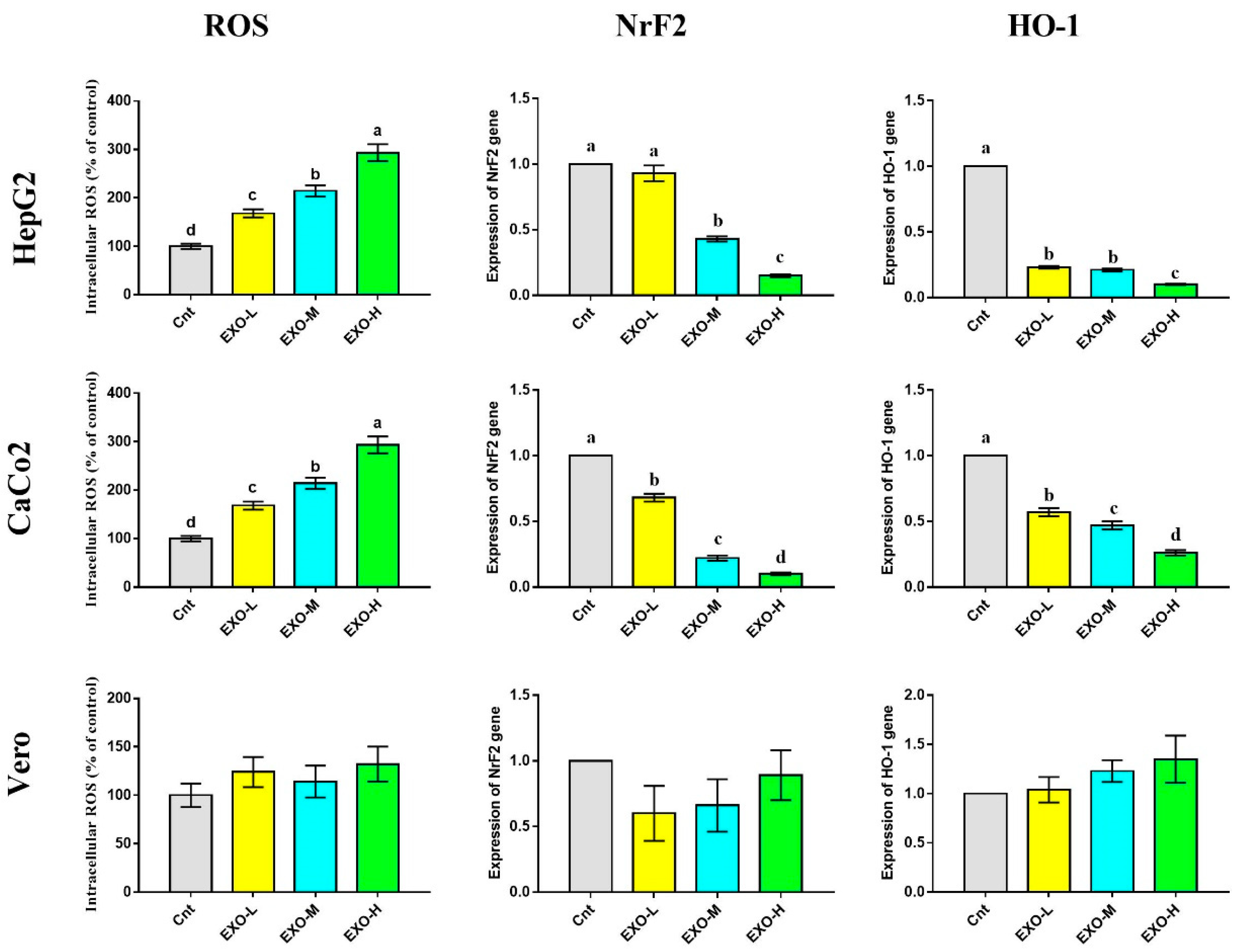
3.5. CM-EXOs Triggered Oxidative Stress in Cancer Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Simeneh, K. Characterization of Camelus Dromedarius in Ethiopia: Production Systems, Reproductive Performances and Infertility Problems. Ph.D. Thesis, Addis Ababa Uiversity, Addis Ababa, Ethiopia, 2015. [Google Scholar]
- Gader, A.G.M.A.; Alhaider, A.A. The unique medicinal properties of camel products: A review of the scientific evidence. J. Taibah Univ. Med. Soc. 2016, 11, 98–103. [Google Scholar]
- Yadav, A.K.; Kumar, R.; Priyadarshini, L.; Singh, J. Composition and medicinal properties of camel milk: A Review. Asian J. Dairy Food Res. 2015, 34, 83–91. [Google Scholar] [CrossRef]
- Singh, R.; Mal, G.; Kumar, D.; Patil, N.V.; Pathak, K.M.L. Camel Milk: An Important Natural Adjuvant. Agri. Res. 2017, 6, 327–340. [Google Scholar] [CrossRef]
- Al-Majali, A.M.; Ismail, Z.B.; Al-Hami, Y.; Nour, A.Y. Lactoferrin concentration in milk from camels (Camelus dromedarius) with and without subclinical mastitis. Int. J. Appl. Res. Vet. Med. 2007, 5, 120. [Google Scholar]
- Benkerroum, N.; Mekkaoui, M.; Bennani, N.; Hidane, K. Antimicrobial activity of camel’s milk against pathogenic strains of Escherichia coli and Listeria monocytogenes. Int. J. Dairy Technol. 2004, 57, 39–43. [Google Scholar] [CrossRef]
- Jrad, Z.; El Hatmi, H.; Adt, I.; Khorchani, T.; Degraeve, P.; Oulahal, N. Antimicrobial activity of camel milk casein and its hydrolysates. Acta Aliment. 2015, 44, 609–616. [Google Scholar] [CrossRef]
- Mal, G.; Sena, D.S.; Jain, V.; Sahani, M. Therapeutic value of camel milk as a nutritional supplement for multiple drug resistant (MDR) tuberculosis patients. Isr. J. Vet. Med. 2006, 61, 88. [Google Scholar]
- Yassin, M.H.; Soliman, M.M.; Mostafa, S.A.-E.; Ali, H.A.-M. Antimicrobial effects of camel milk against some bacterial pathogens. J. Food Nut. Res. 2015, 3, 162–168. [Google Scholar] [CrossRef]
- Badawy, A.A.; El-Magd, M.A.; AlSadrah, S.A. Therapeutic Effect of Camel Milk and Its Exosomes on MCF7 Cells In Vitro and In Vivo. Integr. Cancer Ther. 2018, 7, 1235–1246. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.; Steiner, L.; Yagil, R. Camel milk: Disease control and dietary laws. J. Health Sci. 2013, 1, 48–53. [Google Scholar]
- Musaad, A.M.; Faye, B.; Al-Mutairi, S.E. Seasonal and physiological variation of gross composition of camel milk in Saudi Arabia. Emir. J. Food Agric. 2013, 25, 618–624. [Google Scholar] [CrossRef]
- Habib, H.M.; Ibrahim, W.H.; Schneider-Stock, R.; Hassan, H.M. Camel milk lactoferrin reduces the proliferation of colorectal cancer cells and exerts antioxidant and DNA damage inhibitory activities. Food Chem. 2013, 141, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Korashy, H.M.; El Gendy, M.A.; Alhaider, A.A.; El-Kadi, A.O. Camel milk modulates the expression of aryl hydrocarbon receptor-regulated genes, Cyp1a1, Nqo1, and Gsta1, in murine hepatoma Hepa 1c1c7 cells. J. Biomed. Biotechnol. 2012, 2012, 782642. [Google Scholar] [CrossRef]
- Mincheva-Nilsson, L.; Baranov, V. The role of placental exosomes in reproduction. Am. J. Reprod. Immunol. 2010, 63, 520–533. [Google Scholar] [CrossRef]
- Van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: A common pathway for a specialized function. J. Biochem. 2006, 140, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Aalberts, M.; van Dissel-Emiliani, F.M.; van Adrichem, N.P.; van Wijnen, M.; Wauben, M.H.; Stout, T.A.; Stoorvogel, W. Identification of distinct populations of prostasomes that differentially express prostate stem cell antigen, annexin A1, and GLIPR2 in humans. Biol. Reprod. 2012, 86, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filen, J.J.; Lahesmaa, R.; Norman, M.; Neve, E.P.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta 2012, 1820, 940–948. [Google Scholar] [CrossRef]
- Pieters, B.C.; Arntz, O.J.; Bennink, M.B.; Broeren, M.G.; van Caam, A.P.; Koenders, M.I.; van Lent, P.L.; van den Berg, W.B.; de Vries, M.; van der Kraan, P.M. Commercial cow milk contains physically stable extracellular vesicles expressing immunoregulatory TGF-β. PLoS ONE 2015, 10, e0121123. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C. Milk—A nutrient system of mammalian evolution promoting mTORC1-dependent translation. Int. J. Mol. Sci. 2015, 16, 17048–17087. [Google Scholar] [CrossRef] [PubMed]
- Yassin, A.M.; Abdel Hamid, M.I.; Farid, O.A.; Amer, H.; Warda, M. Dromedary milk exosomes as mammary transcriptome nano-vehicle: Their isolation, vesicular and phospholipidomic characterizations. J. Adv. Res. 2016, 7, 749–756. [Google Scholar] [CrossRef]
- Othman, R.; Badawy, A.; Alruwaili, M.; El-Magd, M. Camel Milk Exosomes Potentiate The Anticancer Effect of Doxorubicin on Multidrug-Resistant Human Leukemia Hl60 Cells in Vitro and in Vivo. Pak. J. Med. Health Sci. 2021, 15, 3313–3320. [Google Scholar] [CrossRef]
- Ibrahim, H.M.; Mohammed-Geba, K.; Tawfic, A.A.; El-Magd, M.A. Camel milk exosomes modulate cyclophosphamide-induced oxidative stress and immuno-toxicity in rats. Food Funct. 2019, 10, 7523–7532. [Google Scholar] [CrossRef] [PubMed]
- Badawy, A.A.; Othman, R.Q.A.; El-Magd, M.A. Effect of combined therapy with camel milk-derived exosomes, tamoxifen, and hesperidin on breast cancer. Mol. Cell. Toxicol. 2021, 17, 543–552. [Google Scholar] [CrossRef]
- Ali, M.; Elsayed, G.R.; Salama, M.F.; El-Magd, M.A. Camel milk exosomes had a selective anticancer effect on PANC1 cells and a proliferative effect on H6c7 cells. KFS. Vet. Med. J. 2022, 20, 1–5. [Google Scholar] [CrossRef]
- El-Kattawy, A.M.; Algezawy, O.; Alfaifi, M.Y.; Noseer, E.A.; Hawsawi, Y.M.; Alzahrani, O.R.; Algarni, A.; Kahilo, K.A.; El-Magd, M.A. Therapeutic potential of camel milk exosomes against HepaRG cells with potent apoptotic, anti-inflammatory, and anti-angiogenesis effects for colostrum exosomes. Biomed. Pharmacother. 2021, 143, 112220. [Google Scholar] [CrossRef]
- Tong, L.; Hao, H.; Zhang, X.; Zhang, Z.; Lv, Y.; Zhang, L.; Yi, H. Oral Administration of Bovine Milk-Derived Extracellular Vesicles Alters the Gut Microbiota and Enhances Intestinal Immunity in Mice. Mol. Nut. Food Res. 2020, 64, 1901251. [Google Scholar] [CrossRef]
- Zhou, F.; Paz, H.A.; Sadri, M.; Cui, J.; Kachman, S.D.; Fernando, S.C.; Zempleni, J. Dietary bovine milk exosomes elicit changes in bacterial communities in C57BL/6 mice. Am. J. Physiol.-Gastrointest. Liver Physiol. 2019, 317, G618–G624. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Chen, X.; Zheng, X.; Zhu, H.; Qi, Q.; Liu, S.; Zhang, H.; Che, J. Latest Trend of Milk Derived Exosomes: Cargos, Functions, and Applications. Front. Nutr. 2021, 8, 747294. [Google Scholar] [CrossRef]
- Mahmoodzadeh Hosseini, H.; Ali Imani Fooladi, A.; Reza Nourani, M.; Ghanezadeh, F. The role of exosomes in infectious diseases. Inflam. Allergy Targets 2013, 12, 29–37. [Google Scholar] [CrossRef]
- Wang, J.; Yao, Y.; Chen, X.; Wu, J.; Gu, T.; Tang, X. Host derived exosomes-pathogens interactions: Potential functions of exosomes in pathogen infection. Biomed. Pharmacother. 2018, 108, 1451–1459. [Google Scholar] [CrossRef] [PubMed]
- Baddela, V.S.; Nayan, V.; Rani, P.; Onteru, S.K.; Singh, D. Physicochemical Biomolecular Insights into Buffalo Milk-Derived Nanovesicles. Appl. Biochem. Biotechnol. 2016, 178, 544–557. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Chatli, M.K.; Singh, R.; Mehta, N.; Kumar, P. Antioxidant and antimicrobial activity of camel milk casein hydrolysates and its fractions. Small Rumin. Res. 2016, 139, 20–25. [Google Scholar] [CrossRef]
- Schuh, C.M.A.P.; Aguayo, S.; Zavala, G.; Khoury, M. Exosome-like vesicles in Apis mellifera bee pollen, honey and royal jelly contribute to their antibacterial and pro-regenerative activity. J. Exp. Biol. 2019, 222. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, T.J.; Cleary, T.G. Effect of lactoferrin on enteric pathogens. Biochimie 2009, 91, 30–34. [Google Scholar] [CrossRef]
- Jahani, S.; Shakiba, A.; Jahani, L. The Antimicrobial effect of lactoferrin on Gram-negative and Gram-positive bacteria. Int. J. Inf. 2015, 2, e27954. [Google Scholar] [CrossRef]
- Moradian, F.; Sharbafi, R.; Rafiei, A. Lactoferrin, Isolation, Purification and Antimicrobial Effects. J. Med. Bioeng. 2014, 3, 203–206. [Google Scholar] [CrossRef]
- Korashy, H.M.; Maayah, Z.H.; Abd-Allah, A.R.; El-Kadi, A.O.; Alhaider, A.A. Camel milk triggers apoptotic signaling pathways in human hepatoma HepG2 and breast cancer MCF7 cell lines through transcriptional mechanism. J. Biomed. Biotechnol. 2012, 2012, 593195. [Google Scholar] [CrossRef] [PubMed]
- Shariatikia, M.; Behbahani, M.; Mohabatkar, H. Anticancer activity of cow, sheep, goat, mare, donkey and camel milks and their caseins and whey proteins and in silico comparison of the caseins. Mol. Biol. Res. Commun. 2017, 6, 57–64. [Google Scholar] [PubMed]
- Homayouni-Tabrizi, M.; Asoodeh, A.; Soltani, M. Cytotoxic and antioxidant capacity of camel milk peptides: Effects of isolated peptide on superoxide dismutase and catalase gene expression. J. Food Drug Anal. 2017, 25, 567–575. [Google Scholar] [CrossRef]
- Ross, M.; Atalla, H.; Karrow, N.; Mallard, B.A. The bioactivity of colostrum and milk exosomes of high, average, and low immune responder cows on human intestinal epithelial cells. J. Dairy Sci. 2021, 104, 2499–2510. [Google Scholar] [CrossRef] [PubMed]
- García-Montoya, I.A.; Cendón, T.S.; Arévalo-Gallegos, S.; Rascón-Cruz, Q. Lactoferrin a multiple bioactive protein: An overview. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2012, 1820, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shi, Z.; Wang, X.; Mu, S.; Xu, X.; Shen, L.; Li, P. Protective effects of bovine milk exosomes against oxidative stress in IEC-6 cells. Eur. J. Nutr. 2020, 10, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Patel, M.; Williams, S.; Arora, H.; Sims, B. Human breast milk-derived exosomes attenuate cell death in intestinal epithelial cells. Innate Immun. 2018, 24, 278–284. [Google Scholar] [CrossRef] [PubMed]



| Gene | Forward (5′ ------ 3′) | Reverse (5′ ------ 3′) | Accession Number | Length (bp) |
|---|---|---|---|---|
| Bax | TGCTTCAGGGTTTCATCCAG | GGCGGCAATCATCCTCTG | NM_001291428.2 | 170 |
| Bcl2 | GAACTGGGGGAGGATTGTGG | CATCCCAGCCTCCGTTATCC | NM_000633.3 | 164 |
| NrF2 | CAGCGACGGAAAGAGTATG | TGGGCAACCTGGGAGTAG | NM_006164.5 | 200 |
| HO-1 | CGGGCCAGCAACAAAGTG | AGTGTAAGGACCCATCGGAGAA | NM_002133.3 | 107 |
| β actin | CACCAACTGGGACGACAT | ACAGCCTGGATAGCAACG | NM_001101.5 | 189 |
| Strain | CM | CM-EXOs | CN | MCZ |
|---|---|---|---|---|
| S. aureus | 11 | NiL | 18 | - |
| E. faecalis | 16 | NiL | 12 | - |
| M. luteus | 27 | NiL | 18 | - |
| E. coli | NiL | NiL | 14 | - |
| P. mirabilis | NiL | NiL | 15 | - |
| P. aeruginosa | NiL | NiL | 10 | - |
| C. albicans | NiL | NiL | - | 10 |
| Strain | CM | CM-EXOs |
|---|---|---|
| S. aureus | 100.00 ± 2.28 a | 93.73 ± 1.74 b |
| E. faecalis | 100.00 ± 2.35 a | 91.97 ± 2.06 b |
| M. luteus | 100.00 ± 2.10 a | 90.87 ± 2.23 b |
| E. coli | 82.95 ± 1.64 b | 97.69 ± 1.49 a |
| P. mirabilis | 79.15 ± 1.17 b | 95.5 ± 2.20 a |
| P. aeruginosa | 75.51 ± 1.09 b | 90.50 ± 2.39 a |
| C. albicans | 83.34 ± 1.50 a | 68.39 ± 1.27 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaban, A.M.; Raslan, M.; Sharawi, Z.W.; Abdelhameed, M.S.; Hammouda, O.; El-Masry, H.M.; Elsayed, K.N.M.; El-Magd, M.A. Antibacterial, Antifungal, and Anticancer Effects of Camel Milk Exosomes: An In Vitro Study. Vet. Sci. 2023, 10, 124. https://doi.org/10.3390/vetsci10020124
Shaban AM, Raslan M, Sharawi ZW, Abdelhameed MS, Hammouda O, El-Masry HM, Elsayed KNM, El-Magd MA. Antibacterial, Antifungal, and Anticancer Effects of Camel Milk Exosomes: An In Vitro Study. Veterinary Sciences. 2023; 10(2):124. https://doi.org/10.3390/vetsci10020124
Chicago/Turabian StyleShaban, Amira M., Mai Raslan, Zeina Walid Sharawi, Mohamed Sayed Abdelhameed, Ola Hammouda, Hossam M. El-Masry, Khaled N. M. Elsayed, and Mohammed A. El-Magd. 2023. "Antibacterial, Antifungal, and Anticancer Effects of Camel Milk Exosomes: An In Vitro Study" Veterinary Sciences 10, no. 2: 124. https://doi.org/10.3390/vetsci10020124
APA StyleShaban, A. M., Raslan, M., Sharawi, Z. W., Abdelhameed, M. S., Hammouda, O., El-Masry, H. M., Elsayed, K. N. M., & El-Magd, M. A. (2023). Antibacterial, Antifungal, and Anticancer Effects of Camel Milk Exosomes: An In Vitro Study. Veterinary Sciences, 10(2), 124. https://doi.org/10.3390/vetsci10020124







