An ELISA to Detect Antibodies to Bovine Alphaherpesviruses 1 and 5 and Bubaline Alphaherpesvirus 1 in Cattle Sera
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Samples
2.2. Cells
2.3. Viruses
2.4. Serum Neutralization Tests (SN)
2.5. Antigen Preparation
2.6. Optimization of the Assays
2.7. Preparative, “Single Antigens” ELISAs (sAgELISAs)
2.8. Multiple Antigen ELISA (mAgELISA)
2.9. Determination of the Cut-Off Points of sAgELISAs and mAgELISA
2.10. Sensitivity and Statistical Analyses
3. Results
Comparisons between SN, sAgELISA, and mAgELISA
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- ICTV—International Committee on Taxonomy of Virus. ICTV Report Chapters. Herpesviridae. Family: Herpesviridae. Subfamily Alphaherpesvirinae. Genus: Varicellovirus. 2023. Available online: https://ictv.global/report/chapter/herpesviridae/herpesviridae/varicellovirus (accessed on 2 January 2023).
- Oliveira, M.T.; Campos, F.S.; Dias, M.M.; Velho, F.A.; Freneau, G.E.; Brito, W.M.; Rijsewijk, F.A.M.; Franco, A.C.; Roehe, P.M. Detection of bovine herpesvirus 1 and 5 in semen from Brazilian bulls. Theriogenology 2010, 75, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.C.; Roehe, P.M.; Varela, A.P.M. Herpesviridae; Flores, E.F., Virologia, V., Eds.; UFSM: Santa Maria, Brazil, 2012; pp. 503–570. [Google Scholar]
- St. George, T.D.; Philpott, M. Isolation of IBR virus from the prepuce of water buffalo bulls in Australia. Aust. Vet. J. 1972, 48, 126. [Google Scholar] [PubMed]
- Petrini, S.; Amoroso, M.G.; Perugini, G.; Gianfelici, P.; Corrado, F.; Bazzucchi, M.; Paniccià, M.; Casciari, C.; Fortunati, M.; Giammarioli, M.; et al. Rilievo del Bubaline herpesvirus 1 (BuHV-1) in un allevamento di bufali nel centro Italia. Large Anim. Rev. 2012, 18, 113–116. [Google Scholar]
- Amoroso, F.; Corrado, E.; De Carlo, M.G.; Lucibelli, A.; Martucciello, A.; Guarino, G. Bubaline herpesvirus 1 associated with abortion in a Mediterranean water buffalo. Res. Vet. Sci. 2013, 94, 813–816. [Google Scholar] [CrossRef] [PubMed]
- Thiry, J.; Keuser, V.; Muylkens, B.; Meurens, F.; Gogev, S.; Vanderplasschen, A.; Thiry, E. Ruminant alphaherpesviruses related to bovine herpesvirus 1. Vet. Res. 2006, 37, 169–190. [Google Scholar] [CrossRef]
- Thiry, J.; Widén, F.; Grégoire, F.; Linden, A.; Belák, S.; Thiry, E. Isolation and characterization of a ruminant alphaherpesvirus closely related to bovine herpesvirus 1 in a free-ranging red deer. Vet. Res. 2007, 3, 26. [Google Scholar] [CrossRef]
- Nogarol, C.; Bertolotti, L.; De Carlo, E.; Masoero, L.; Caruso, C.; Profiti, M.; Martucciello, A.; Galiero, G.; Cordioli, P.; Lelli, D.; et al. Expression and antigenic characterization of bubaline herpesvirus 1 (BuHV-1) glycoprotein E and its potential application in the epidemiology and control of alphaherpesvirus infections in Mediterranean water buffalo. J. Virol. Methods 2014, 207, 16–21. [Google Scholar] [CrossRef]
- Scheffer, C.M.; Varela, A.P.; Cibulski, S.P.; Schmidt, C.; Campos, F.S.; Paim, W.P.; Santos, R.N.D.; Teixeira, T.F.; Loiko, M.R.; Tochetto, C.; et al. Genome sequence of bubaline alphaherpesvirus 1 (BuHV-1) isolated in Australia in 1972. Arch. Virol. 2017, 162, 1169–1176. [Google Scholar] [CrossRef]
- Bulach, D.M.; Studdert, M.J. Comparative genome mapping of bovine encephalitis herpesvirus, bovine herpesvirus 1, and buffalo herpesvirus. Arch. Virol. 1990, 113, 17–34. [Google Scholar] [CrossRef]
- Varela, A.P.M.; Holz, C.L.; Cibulski, S.P.; Teixeira, T.F.; Antunes, D.A.; Franco, A.C.; Oliveira, M.T.; Campos, F.S.; Dezen, D.; Cenci, A.; et al. Neutralizing antibodies to bovine herpesvirus types 1 (BoAHV-1) and 5 (BoAHV-5) and its subtypes. Vet. Microbiol. 2010, 142, 254–260. [Google Scholar] [CrossRef]
- Reid, H.W.; Nettleton, P.F.; Pow, I.; Sinclair, J.A. Experimental infection of red deer (Cervus elaphus) and cattle with a herpesvirus isolated from red deer. Vet. Rec. 1986, 118, 156–158. [Google Scholar] [CrossRef]
- Nettleton, P.F.; Thiry, E.; Reid, H.; Pastoret, P.P. Herpesvirus infections in Cervidae. Rev. Sci. Tech. 1988, 7, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Tolari, F.; White, H.; Nixon, P. Isolation and reactivation of bovid herpesvirus 1 in goats. Microbiologica 1991, 13, 67–71. [Google Scholar]
- Engels, M.; Palatini, M.; Metzler, A.E.; Probst, U.; Kihm, U.; Ackermann, M. Interactions of bovine and caprine herpesviruses with the natural and foreign hosts. Vet. Microbiol. 1992, 33, 69–78. [Google Scholar] [CrossRef]
- Six, A.; Banks, M.; Engels, M.; Bascuñana, C.R.; Ackermann, M. Latency and reactivation of bovine herpesvirus 1 (BHV-1) in goats and of caprine herpesvirus 1 (CapHV-1) in calves. Arch. Virol. 2001, 146, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Freshney, R.I. Culture of Animal Cells: A Manual of Basic Technique and Specialized Applications, 7th ed.; Wiley Blackwe: Hoboken, NJ, USA, 2016; 736p. [Google Scholar]
- Madin, S.H.; York, R.J.; McKercher, D.G. Isolation of the infectious bovine rhinotracheitis virus. Science 1956, 124, 721–722. [Google Scholar] [CrossRef]
- Batista, H.B.C.R.; Schmidt, E.; Spilki, F.R.; Franco, A.C.; Roehe, P.M. Herpesvírus bovinos (BoAHV-1.1 e BoAHV-1.2b) em forma infecciosa em encéfalos de bovinos submetidos ao diagnóstico de raiva no estado do Rio Grande do Sul. Arq. Bras. Med. Vet. Zoot. 2010, 62, 1023–1028. [Google Scholar] [CrossRef]
- Roehe, P.M.; Silva, T.C.; Nardi, N.B.; Oliveira, L.G.; Rosa, J.C.A. Diferenciação entre o vírus da rinotraqueíte infecciosa bovina (BHV-1) e o herpesvírus da encefalite bovina (BHV-5) com anticorpos monoclonais. Pesq. Vet. Bras. 1997, 17, 41–44. [Google Scholar] [CrossRef]
- Franco, A.C.; Rijsewijk, F.A.M.; Flores, E.F.; Weiblen, R.; Roehe, P.M. Construction and characterization of a glycoprotein E deletion mutant of bovine herpesvirus type 1.2 strain isolated in Brazil. Braz. J. Microbiol. 2002, 33, 274–278. [Google Scholar] [CrossRef]
- Souza, V.F.; Melo, S.V.; Esteves, P.A.; Schmidt, C.S.; Gonçalves, D.A.; Schaefer; Silva, T.C.; Almeida, R.S.; Vicentini, F.; Franco, A.C.; et al. Caracterização de herpesvírus bovinos tipos 1 (BHV-1) e 5 (BHV-5) com anticorpos monoclonais. Pesq. Vet. Bras. 2002, 22, 13–18. [Google Scholar] [CrossRef]
- Mackinnon, A. A spreadsheet for the calculation of comprehensive statistics for the assessment of diagnostic tests and inter-rater agreement. Comput. Biol. Med. 2000, 30, 127–134. [Google Scholar] [CrossRef]
- Ackermann, M.; Bélak, S.; Bitsch, V.; Edwards, S.; Moussa, A.; Rockborn, G.; Thiry, E. Round table on infectious bovine rhinotracheitis/infectious vulvovaginitis virus infection diagnosis and control. Vet. Microbiol. 1990, 23, 361–363. [Google Scholar] [CrossRef]
- Perrin, B.; Calvo, T.; Cordioli, P.; Coudert, M.; Edwards, S.; Eloit, M.; Guérin, B.; Kramps, J.A.; Lenihan, P.; Paschaleri, E.; et al. Selection of European Union standard reference sera for use in the serological diagnosis of infectious bovine rhinotracheitis. Rev. Sci. Tech. 1996, 13, 947–960. [Google Scholar] [CrossRef]
- De Wit, J.J.; Hage, J.J.; Brinkhof, J.; Westenbrink, F. A comparative study of serological tests of use in the bovine herpesvirus 1 eradication programs in the Netherlands. Vet. Microbiol. 1998, 61, 153–163. [Google Scholar] [CrossRef]
- Van Oirschot, J.T.; Kaashoek, M.J.; Maris-Veldhuis, M.A.; Weerdmeester, K.; Rijsewijk, F.A. An enzyme-linked immunosorbent assay to detect antibodies against glycoprotein gE of bovine herpesvirus 1 allows differentiation between infected and vaccinated cattle. J. Virol. Methods 1997, 67, 23–34. [Google Scholar] [CrossRef]
- Teixeira, M.F.B.; Esteves, P.A.; Schmidt, C.S.; Spilki, F.R.; Da Silva, T.C.; Dotta, M.A.; Roehe, P.M. ELISA de bloqueio monoclonal para o diagnóstico sorológico de infecções pelo herpesvírus bovino tipo 1 (BHV-1). Pesq. Vet. Bras. 2001, 21, 33–37. [Google Scholar] [CrossRef]
- Parreño, V.; Romera, S.A.; Makek, L.; Rodriguez, D.; Malacari, D.; Maidana, S.; Compaired, D.; Combessies, G.; Vena, M.M.; Garaicoechea, L.; et al. Validation of an indirect ELISA to detect antibodies against BoAHV-1in bovine and guinea pig serum sample using ISO/IEC17025 standards. J. Virol. Methods 2010, 169, 143–153. [Google Scholar] [CrossRef] [PubMed]
- D’Arce, R.C.; Almeida, R.S.; Silva, T.C.; Franco, A.C.; Spilki, F.; Roehe, P.M.; Arns, C.W. Restriction endonuclease and monoclonal antibody analysis of Brazilian isolates of bovine herpesviruses types 1 and 5. Vet. Microbiol. 2002, 88, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Kramps, J.A.; Magdalena, J.; Quak, J.; Weerdmeester, K.; Kaashoek, M.J.; Maris-Veldhuis, M.A.; Rijsewijk, F.A.; Keil, G.; van Oirschot, J.T. A simple, specific, and highly sensitive blocking enzyme-linked immunosorbent assay for the detection of antibodies to bovine herpesvirus 1. J. Clin. Microbiol. 1994, 32, 2175–2181. [Google Scholar] [CrossRef] [PubMed]
- Scicluna, M.T.; Saralli, G.; Bruni, G.; Sala, M.; Cocumelli, C.; Caciolo, D.; Condoleo, R.U.; Autorino, G.L. Epidemiological situation of herpesvirus infections in buffalo herds: Bubaline Herpesvirus 1 or Bovine Herpesvirus 1? Ital. J. Anim. Sci. 2007, 6, 845–849. [Google Scholar] [CrossRef]
- Vieira, S.; Brito, W.D.; Souza, W.J.; Alfaia, B.T.; Linhares, D.C.L. Anticorpos para o herpesvírus bovino 1 (BHV-1) em bovinos do estado de Goiás. Ciênc. Anim. Bras. 2003, 4, 131–137. [Google Scholar]
- Holz, C.L.; Cibulski, S.P.; Teixeira, T.F.; Batista, H.B.C.R.; Dezen, D.; Campos, F.S.; Varela, A.P.M.; Roehe, P.M. Serum neutralization with different types and subtypes of bovine herpesvirus 1 and 5. Pesq. Vet. Bras. 2010, 30, 515–522. [Google Scholar] [CrossRef]
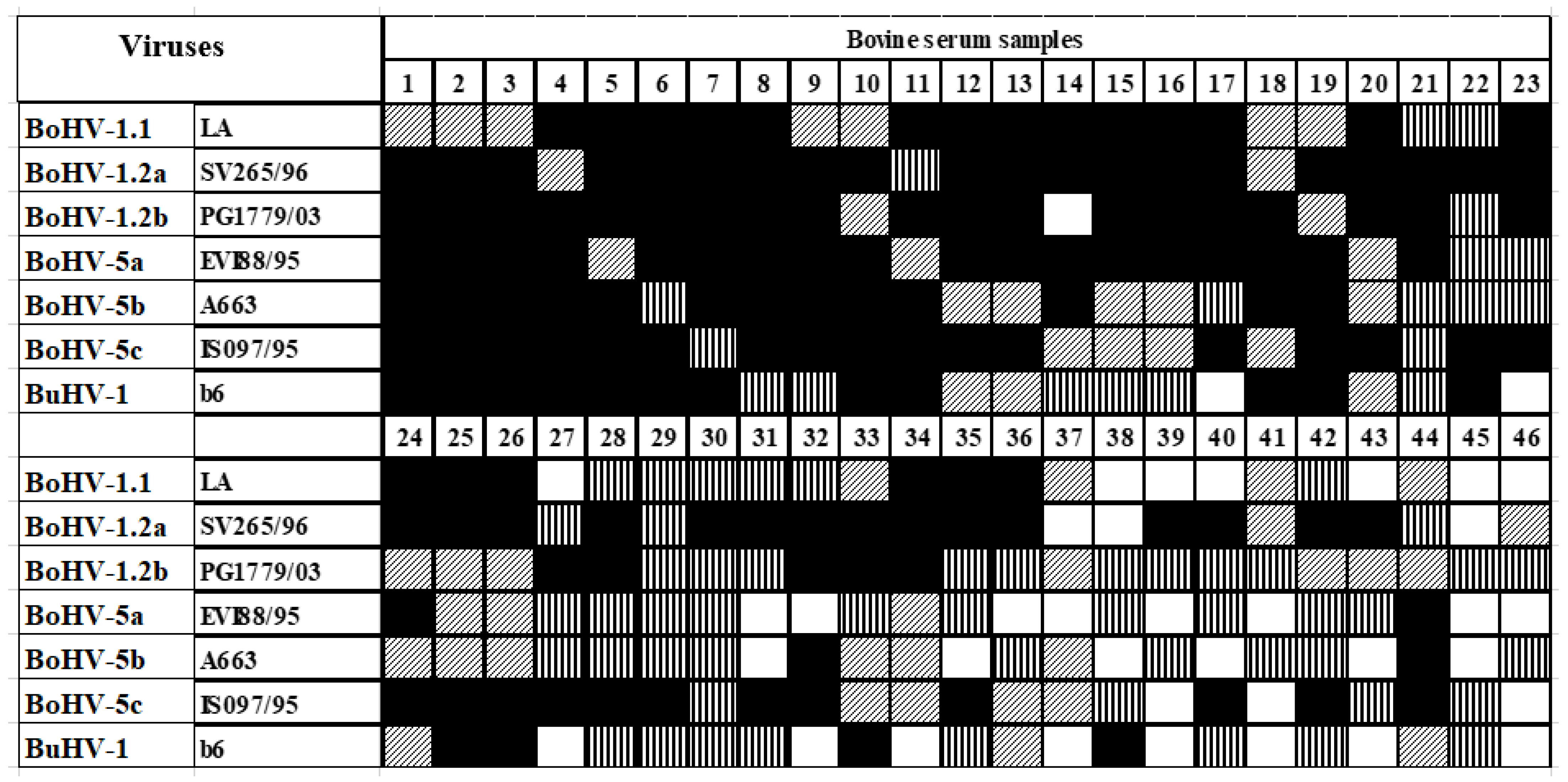
| Virus Strains | SN | sAgELISAs | ||
|---|---|---|---|---|
| Number of Positive Sera | Sensitivity a | Number of Positive Sera | Sensitivity a | |
| BoAHV-1.1 (LA) | 236 | 91.1 | 237 | 91.5 |
| BoAHV-1.2a (SV265/96) | 244 | 94.2 | 247 | 95.4 |
| BoAHV-1.2b (PG1779/03) | 241 | 93.1 | 241 | 93.1 |
| BoAHV-5a (EVI88/95) | 239 | 92.3 | 233 | 90.0 |
| BoAHV-5b (A663) | 235 | 90.7 | 232 | 89.6 |
| BoAHV-5c (ISO97/95) | 242 | 93.4 | 249 | 96.1 |
| BuAHV-1 (B6) | 237 | 91.5 | 229 | 88.4 |
| LA + SV265/96 | 248 | 95.8 | 251 | 96.9 |
| LA + PG1779/03 | 249 | 96.1 | 246 | 95.0 |
| LA + EVI88/95 | 251 | 96.9 | 240 | 92.7 |
| LA + A663 | 249 | 96.1 | 239 | 92.3 |
| LA + ISO97/95 | 250 | 96.5 | 251 | 96.9 |
| LA + B6 | 249 | 96.1 | 240 | 92.7 |
| SV265/96 + PG1779/03 | 250 | 96.5 | 254 | 98.1 |
| SV265/96 + EVI88/95 | 249 | 96.1 | 251 | 96.9 |
| SV265/96 + A663 | 249 | 96.1 | 251 | 96.9 |
| SV265/96 + ISO97/95 | 249 | 96.1 | 257 | 99.2 |
| SV265/96 + B6 | 250 | 96.5 | 252 | 97.3 & |
| PG1779/03 + EVI88/95 | 250 | 96.5 | 243 | 93.8 |
| PG1779/03 + A663 | 247 | 95.4 | 243 | 93.8 |
| PG1779/03 + ISO97/95 | 251 | 96.9 | 255 | 98.5 & |
| PG1779/03 + B6 | 249 | 96.1 | 246 | 95.0 |
| EVI88/95 + A663 | 246 | 95.0 | 237 | 91.5 |
| EVI88/95 + ISO97/95 | 249 | 96.1 | 252 | 97.3 & |
| EVI88/95 + B6 | 247 | 95.4 | 239 | 92.3 |
| A663 + ISO97/95 | 249 | 96.1 | 251 | 96.9 |
| A663 + B6 | 247 | 95.4 | 238 | 91.9 |
| ISO97/95 + B6 | 249 | 96.1 | 252 | 97.3 & |
| LA + SV265/96 + PG1779/03 | 253 | 97.7 & | 255 | 98.5 & |
| LA + SV265/96 + EVI88/95 | 252 | 97.3 & | 253 | 97.7 & |
| LA + SV265/96 + A663 | 252 | 97.3 & | 252 | 97.3 & |
| LA + SV265/96 + ISO97/95 | 252 | 97.3 & | 257 | 99.2 & |
| LA + SV265/96 + B6 | 253 | 97.7 & | 253 | 97.7 & |
| LA + PG1779/03 + EVI88/95 | 255 | 98.5 & | 247 | 95.4 |
| LA + PG1779/03 + A663 | 253 | 97.7 & | 247 | 95.4 |
| LA + PG1779/03 + ISO97/95 | 255 | 98.5 & | 256 | 98.8 & |
| LA + PG1779/03 + B6 | 253 | 97.7 & | 249 | 96.1 |
| LA + EVI88/95 + A663 | 254 | 98.1 & | 242 | 93.4 |
| LA + EVI88/95 + ISO97/95 | 254 | 98.1 & | 253 | 97.7 & |
| LA + EVI88/95 + B6 | 254 | 98.1 & | 243 | 93.8 |
| LA + A663 + ISO97/95 | 255 | 98.5 & | 252 | 97.3 & |
| LA + A663 + B6 | 255 | 98.5 & | 242 | 93.4 |
| LA + ISO97/95 + B6 | 254 | 98.1 & | 253 | 97.7 & |
| LA + SV265/96 + PG1779/03 + EVI88/95 | 255 | 98.5 & | 256 | 98.8 |
| LA + SV265/96 + PG1779/03 + A663 | 254 | 98.1 & | 256 | 98.8 |
| LA + SV265/96 + PG1779/03 + ISO97/95 | 255 | 98.5 & | 259 | 100 & |
| LA + SV265/96 + PG1779/03 + B6 | 255 | 98.5 & | 257 | 99.2 |
| LA + PG1779/03 + EVI88/95 + A663 | 255 | 98.5 & | 248 | 95.8 |
| LA + PG1779/03 + EVI88/95 + ISO97/95 | 257 | 99.2 & | 256 | 98.8 & |
| LA + PG1779/03 + EVI88/95 + B6 | 257 | 99.2 & | 250 | 96.5 |
| LA + EVI88/95 + A663 + ISO97/95 | 257 | 99.2 & | 254 | 98.1 & |
| LA + EVI88/95 + A663 + B6 | 257 | 99.2 & | 245 | 94.6 |
| LA + A663 + ISO97/95 + B6 | 258 | 99.6 & | 254 | 98.1 & |
| SV265/96 + PG1779/03 + EVI88/95 + A663 | 252 | 97.3 & | 256 | 98.8 & |
| SV265/96 + PG1779/03 + EVI88/95 + ISO97/95 | 254 | 98.1 & | 259 | 100 & |
| SV265/96 + PG1779/03 + EVI88/95 + B6 | 254 | 98.1 & | 258 | 99.6 & |
| SV265/96 + EVI88/95 + A663 + ISO97/95 | 254 | 98.1 & | 259 | 100 & |
| SV265/96 + EVI88/95 + A663 + B6 | 254 | 98.1 & | 256 | 98.8 & |
| SV265/96 + A663 + ISO97/95 + B6 | 255 | 98.5 & | 259 | 100 & |
| PG1779/03 + EVI88/95 + A663 + ISO97/95 | 254 | 98.1 & | 256 | 98.8 & |
| PG1779/03 + EVI88/95 + A663 + B6 | 254 | 98.1 & | 249 | 96.1 |
| PG1779/03 + A663 + ISO97/95 + B6 | 255 | 98.5 & | 258 | 99.6 & |
| EVI88/95 + A663 + ISO97/95 + B6 | 255 | 98.5 & | 255 | 98.5 & |
| LA + SV265/96 + PG1779/03 + EVI88/95 + A663 | 255 | 98.5 & | 257 | 99.2 & |
| LA + SV265/96 + PG1779/03 + EVI88/95 + ISO97/95 ** | 257 | 99.2 & | 259 | 100 & |
| LA + SV265/96 + PG1779/03 + EVI88/95 + B6 | 257 | 99.2 & | 258 | 99.6 & |
| LA + PG1779/03 + EVI88/95 + A663 + ISO97/95 | 257 | 99.2 & | 257 | 99.2 & |
| LA + PG1779/03 + EVI88/95 + A663 + B6 | 257 | 99.2 & | 251 | 96.9 |
| LA + EVI88/95 + A663 + ISO97/95 + B6 | 259 | 100 & | 256 | 98.8 & |
| SV265/96 + PG1779/03 + EVI88/95 + A663 + ISO97/95 | 254 | 98.1 & | 259 | 100 & |
| SV265/96 + PG1779/03 + EVI88/95 + A663 + B6 | 254 | 98.1 & | 259 | 100 & |
| PG1779/03 + EVI88/95 + A663 + ISO97/95 + B6 | 256 | 98.8 & | 258 | 99.6 & |
| LA + SV265/96 + PG1779/03 + EVI88/95 + A663 + SO97/95 | 257 | 99.2 & | 259 | 100 & |
| LA + SV265/96 + PG1779/03 + EVI88/95 + A663 + B6 | 257 | 99.2 & | 259 | 100 & |
| SV265/96 + PG1779/03 + EVI88/95 + A663 + ISO97/95 + B6 | 256 | 98.8 & | 259 | 100 & |
| LA + SV265/96 + PG1779/03 + EVI88/95 + A663 + ISO97/95 + B6 | 259 | 100.0 | 259 | 100 & |
| SN | sAgELISA | TOTAL | |
|---|---|---|---|
| POSITIVE | NEGATIVE | ||
| POSITIVE | 250 (a) | 9 (b) | 259 (a + b) |
| NEGATIVE | 13 (c) | 328 (d) | 341 (c + d) |
| TOTAL | 263 (a + c) | 337 (b + d) | 600 (a + b + c + d) |
| Sensitivity (a/a + c) × 100 = 96.56%; Specificity (d/d + b) × 100 = 96.1%; Positive predictive value (PPV) (a/a + b) × 100 = 95%; Negative predictive value (NPV) (d/c + d) × 100 = 97.3%; Kappa correlation: | |||
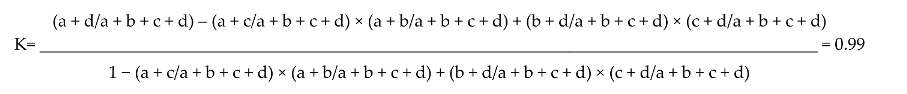 McNemar’s = [(a − d)/−1]2/a + d = significant at p > 0.05. | |||
| sAgELISA * | mAgELISA | TOTAL | |
|---|---|---|---|
| POSITIVE | NEGATIVE | ||
| POSITIVE | 262 | 1 | 263 |
| NEGATIVE | 1 | 336 | 337 |
| TOTAL | 263 | 337 | 600 |
| SN | mAgELISA | TOTAL | |
|---|---|---|---|
| POSITIVE | NEGATIVE | ||
| POSITIVE | 250 | 9 | 259 |
| NEGATIVE | 13 | 328 | 341 |
| TOTAL | 263 | 337 | 600 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scheffer, C.M.; Petzhold, S.A.; Varela, A.P.M.; Paim, W.P.; Duarte, P.M.; Loiko, M.R.; Cerva, C.; Schmidt, C.; Wendlant, A.; Cibulski, S.P.; et al. An ELISA to Detect Antibodies to Bovine Alphaherpesviruses 1 and 5 and Bubaline Alphaherpesvirus 1 in Cattle Sera. Vet. Sci. 2023, 10, 110. https://doi.org/10.3390/vetsci10020110
Scheffer CM, Petzhold SA, Varela APM, Paim WP, Duarte PM, Loiko MR, Cerva C, Schmidt C, Wendlant A, Cibulski SP, et al. An ELISA to Detect Antibodies to Bovine Alphaherpesviruses 1 and 5 and Bubaline Alphaherpesvirus 1 in Cattle Sera. Veterinary Sciences. 2023; 10(2):110. https://doi.org/10.3390/vetsci10020110
Chicago/Turabian StyleScheffer, Camila Mengue, Sylio Alfredo Petzhold, Ana Paula Muterle Varela, Willian Pinto Paim, Phelipe Magalhães Duarte, Márcia Regina Loiko, Cristine Cerva, Candice Schmidt, Adrieli Wendlant, Samuel Paulo Cibulski, and et al. 2023. "An ELISA to Detect Antibodies to Bovine Alphaherpesviruses 1 and 5 and Bubaline Alphaherpesvirus 1 in Cattle Sera" Veterinary Sciences 10, no. 2: 110. https://doi.org/10.3390/vetsci10020110
APA StyleScheffer, C. M., Petzhold, S. A., Varela, A. P. M., Paim, W. P., Duarte, P. M., Loiko, M. R., Cerva, C., Schmidt, C., Wendlant, A., Cibulski, S. P., Lima, D. A. d., Tochetto, C., Santos, A. C. R. d., Herpich, J. I., Teixeira, T. F., Santos, H. F. d., Campos, F. S., Franco, A. C., & Roehe, P. M. (2023). An ELISA to Detect Antibodies to Bovine Alphaherpesviruses 1 and 5 and Bubaline Alphaherpesvirus 1 in Cattle Sera. Veterinary Sciences, 10(2), 110. https://doi.org/10.3390/vetsci10020110











