Simple Summary
Avipoxvirus, the causative agent of avian pox disease, can infect more than 278 species of wild and domestic birds. It causes multiple negative consequences for both the economy and the ecosystem. These include reduced egg production, reduced mating success, growth retardation, and occasionally death. Several diagnostic methods are currently available, and prevention is mainly performed by implementing sanitary measures and vaccination. In order to evaluate and characterize the Avipoxviruses circulating in Portugal between 2017 and 2023, ten samples positive for this virus were analyzed. They were then compared with other isolates from other hosts, countries, and years of collection. As a result, it was possible to understand that certain variants of the virus continued to circulate over the years despite the introduction of new viruses.
Abstract
Avipoxvirus (APV), a linear dsDNA virus belonging to the subfamily Chordopoxvirinae of the family Poxviridae, infects more than 278 species of domestic and wild birds. It is responsible for causing avian pox disease, characterized by its cutaneous and diphtheric forms. With a high transmission capacity, it can cause high economic losses and damage to the ecosystem. Several diagnostic methods are available, and bird vaccination can be an effective preventive measure. Ten APV-positive samples were analyzed to update the molecular characterization and phylogenetic analysis of viruses isolated in Portugal between 2017 and 2023. A P4b gene fragment was amplified using a PCR, and the nucleotide sequence of the amplicons was determined using Sanger sequencing. The sequences obtained were aligned using ClustalW, and a maximum likelihood phylogenetic tree was constructed. With this study, it was possible to verify that the analyzed sequences are distributed in subclades A1, A2, B1, and B3. Since some of them are quite similar to others from different countries and obtained in different years, it is possible to conclude that there have been several viral introductions in Portugal. Finally, it was possible to successfully update the data on Avipoxviruses in Portugal.
1. Introduction
The family Poxviridae is divided into subfamilies Chordopoxvirinae and Entomopoxvirinae, responsible for infecting vertebrates and insects, respectively [1]. The genus Avipoxvirus is included in the subfamily Chordopoxvirinae, along with seventeen other genera [2]. Regarding the different bird species that the virus infects, there are twelve species of Avipoxviruses (APVs) [3].
According to Manarolla et al. [4], the nomenclature given to APVs is based on the type of species present in each clade, with five clades already identified, A to E. Clade A is associated with fowlpox, clade B with canarypox, and clade C with psittacinepox viruses [5]. To date, seven subclades within clade A and four subclades within clade B have been described. Clade D includes a unique strain, QP-241, which was isolated from a quail in Italy [6]. Clade E contains sequences isolated from APVs from chickens in Brazil [7] and Mozambique [8] and also from a turkey in Hungary [9]. These studies of phylogenetic classification were conducted with fragments of the genes encoding the 4b core-like protein (P4b) and the DNA polymerase, both highly conserved among poxviruses [10].
Avipoxviruses are very large viruses [11] that can be either oval or brick-shaped [12]. The dsDNA genome is in a linear configuration [13] and has a very low GC content [14]. Of all the poxviruses, Avipoxviruses have the largest genome, approximately 288–300 kbp long [15], and encode more than 320 putative genes [14]. The replicative cycle, unlike other virus species, is characterized by transcription carried out in the cytoplasm [1].
APV infection can lead to the development of avian pox disease, which can present in its cutaneous or diphtheric forms [16]. These are caused by different routes of infection. The cutaneous form is caused by mechanical trauma [17] and is responsible for nodular lesions in featherless areas, being associated with very low mortality rates [18]. The diphtheric form, on the other hand, occurs after inhalation or ingestion of the virus [17] and is characterized by the formation of proliferative nodular lesions in the mucous membranes of the digestive and respiratory tracts [19]. As the nodular lesions can cause breathing and eating difficulties, the mortality rate is higher in this form of the disease [20]. The severity of the clinical symptoms depends on the initial virulence and pathogenicity of the virus strain [21]. Nevertheless, the avian species, age [22], and the presence of secondary viral or bacterial infections are also important factors [21]. Transmission can occur via mechanical vectors such as arthropods, aerosols released by sick birds, direct contact with lesions, or ingestion of contaminated food or drink [22]. Wild bird behavior, such as migration, introduction of new species, and habitat change, can also lead to transmission [23]. However, the disease can be prevented by implementing sanitary measures to avoid mechanical vectors and contaminated sources [21] or vaccinating birds [24].
Infection leads to a number of negative consequences, both economic and ecological. A decrease in egg production and immunity is common, resulting in a reduced ability to survive secondary infections. Reduced mating success, growth retardation, blindness, feeding difficulties, and death may also occur [15,23,25]. As a result, poultry farmers suffer significant economic losses due to the need to replace livestock, lost sales, and sanitation costs [21,26]. At the ecosystem level, the birds are more vulnerable, leading to increased predation and reduced mating, resulting in population decline [27].
In this study, the molecular characterization and phylogenetic analysis of Avipoxviruses isolated from ten samples detected in Portugal between 2017 and 2023 are performed in order to infer the possible origin of the viruses circulating in the country.
2. Materials and Methods
2.1. Samples
Several samples arrive each year at INIAV for screening for Avipoxvirus infection. This screening is performed using the method referred to by Huw Lee and Hwa Lee [28] and described in Section 2.3. Between 2017 and 2023, ten positive samples were obtained (Table 1) and preserved at −80 °C. The positive results obtained were only communicated to the respective customers and were not published anywhere.

Table 1.
Avipoxvirus-positive samples were diagnosed at INIAV since 2017 and used in this study.
2.2. Nucleic Acids Extraction
The viral DNA extraction was performed in a nucleic acid extraction workstation, Kingfisher Flex (Thermo Fisher Scientific, Waltham, MA, USA), using the IndiMag Kit (Indical Bioscience, Leipzig, Germany), following the manufacturer’s instructions. After extraction, the DNA was stored at 4 °C.
2.3. PCR Amplification of the P4b Gene Fragment
The amplification of the P4b gene fragment was performed using conventional PCR with primers described by Huw Lee and Hwa Lee [28], using NZYTaq II 2× Green Master Mix (0.2 U/μL) (NZYTech, Lisboa, Portugal). The reaction contained 0.5 μL of each primer (50 pmol/μL) and 5 μL of DNA, for a total of 25 μL. The PCR program was executed in a UNO II thermocycler thermoblock (Biometra, Göttingen, Germany) and consisted of an initial denaturation at 95 °C for 2 min, followed by 50 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 40 s, and extension at 72 °C for 1 min. A final extension was performed at 72 °C for 10 min. The PCR product was separated with a 1% agarose gel electrophoresis stained with GreenSafe Premium (NZYTech, Lisboa, Portugal). The expected fragments of 578 bp, corresponding to the P4b gene amplified fragment, were excised and purified using the NZYGelpure Kit (NZYTech, Lisboa, Portugal), according to the manufacturer’s instructions.
2.4. Sanger Sequencing of the P4b Gene Fragment
The P4b gene fragments were sequenced using the Sanger method, using the BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, MA, USA), according to the manufacturer’s guidelines. This fragment was chosen since it was mostly used in the phylogenetic analysis of these viruses, and the majority of the nucleotide sequences available in the database belong to this fragment. The primers used in the sequencing reaction are listed in Table 2. To improve the sequencing quality, different sample volumes were used in each reaction depending on the DNA concentration. Samples were sequenced in a 3130 Genetic Analyzer (Applied Biosystems, Waltham, MA, USA). Sequence alignment was performed with SeqScape Software from Applied Biosystems. Using Clustal Omega by EMBL-EBI (https://www.ebi.ac.uk/Tools/msa/clustalo, accessed on 9 March 2023), percent identity matrices were also generated between the sequences of subclades A1, A2, B1, and B3 and the sequences isolated and sequenced in this work. The GenBank accession numbers of the sequences obtained in this study are shown in Table 1.

Table 2.
Details of APV sequences published in GenBank and used in this study.
2.5. Phylogenetic Analysis
To study the phylogenetic origin of the Avipoxviruses detected and their phylogenetic relationship with the sequences published in GenBank (Table 2), a multiple alignment was performed between them using the ClustalW (https://www.genome.jp/tools-bin/clustalw, accessed on 10 March 2023) method. Sequences from different countries, years, and clades were chosen in order to obtain a reliable analysis. The sequences used for phylogenetic analysis were shortened in order to have the same length as those from the literature.
A phylogenetic tree was generated using the maximum likelihood method, using MEGA 11 with the Tamura 3-parameter G + I [29], determined as the best fit model also by MEGA 11, and 1000 bootstrap replicates [30]. Neighbor-joining and Bayesian methods were also used to generate the phylogenetic trees.
3. Results and Discussion
3.1. PCR Amplification of the P4b Gene Fragment
Amplification of the 578 bp P4b gene fragment was confirmed via 1% agarose gel electrophoresis. The results obtained are shown in Supplementary Materials Figure S1. The presence of the expected fragment can be observed in all lanes except lanes 3, 4, and 5, corresponding to samples 11612-19 (puffin), 37026-19 (canary), and 03779-20 (canary), respectively. Amplification of DNA from samples 37026-19 and 03779-20 obtained in a previous extraction procedure was achieved (lanes 11 and 12) (Supplementary Materials Figure S1a). A new PCR reaction resulted in the amplification of the sample 11612-19 (Supplementary Materials Figure S1b). The P4b gene fragment from samples P-09292-22 and 00917-23 was previously obtained.
3.2. Sequencing of the P4b Gene Fragment
Alignment of the forward and reverse nucleotide sequences yielded a nucleotide sequence of 538 bp for all samples, except for sample 03779-20 (canary), after removal of the primer sequences. Due to incomplete sequencing of the 3′ end, only 513 bp were obtained for this sample. This result can be explained by DNA damage, incorrect amplification, or insufficient purification.
The alignment of the sequences studied is presented in Figure 1. Figure 1A presents the nucleotide alignment, while the alignment of the deduced amino acid sequences is presented in Figure 1B. The analysis of the alignments is carried out in the following section, together with the phylogenetic analysis.
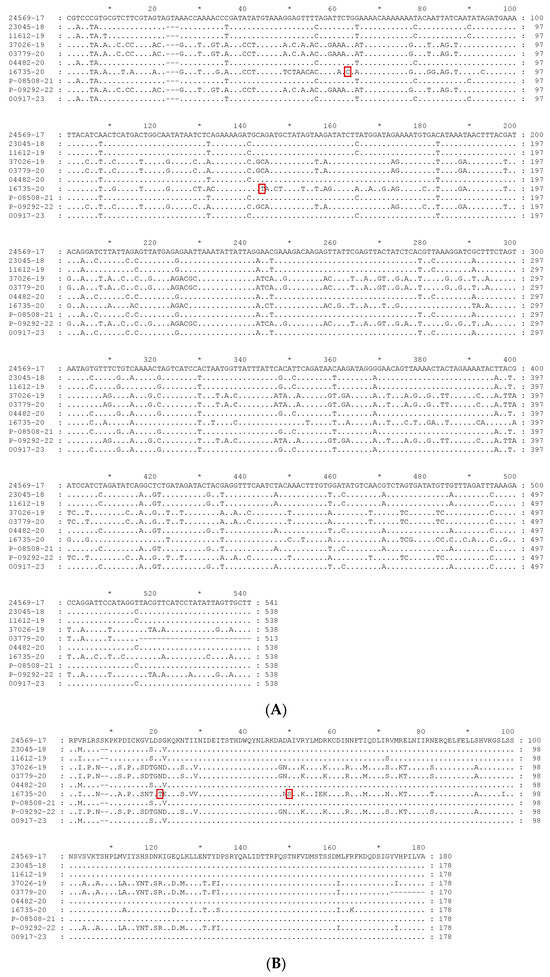
Figure 1.
Alignment of the sequences obtained in this study. (A) Nucleotide sequences alignment; (B) deduced amino acid sequences alignment. Unique mutations observed in sequence 16735-20, when compared with all the sequences present in the phylogenetic tree, are heightened by a red square in both alignments.
3.3. Phylogenetic Analysis
The Avipoxvirus nucleotide sequences obtained were phylogenetically analyzed using different algorithms, namely maximum likelihood, neighbor-joining, and Bayesian analysis. The three algorithms produced phylogenetic trees with similar topologies. However, the maximum likelihood tree gave a better resolution of the sequences and was therefore chosen (Figure 2). The phylogenetic tree obtained, including representatives of all subclades, shows that clades C and D share a common most recent ancestor, as observed in previous studies [21,22].

Figure 2.
The evolutionary history was inferred using the maximum likelihood method and the Tamura 3-parameter model [29]. The tree with the highest log likelihood (−3244.07) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Tamura 3 parameter model and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories (+G, parameter = 0.8553)). The rate variation model allowed some sites to be evolutionarily invariable ([+I], 23.87% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 63 nucleotide sequences. There was a total of 463 positions in the final dataset. Evolutionary analyses were conducted in MEGA11 [30]. Star pentagrams indicate the strains of this study. A, B, C, D and E characters indicate groups of strains belonging to clades A, B, C, D and E, respectively.
The phylogenetic analysis shows that the samples from Portugal are well distributed, with five samples belonging to subclade A1, one to subclade A2, three to subclade B1, and one to subclade B3. Moreover, their distribution is quite consistent with the nomenclature given since samples from Gallus gallus belong to the fowlpox clade (clade A), whereas samples from Turdus merula and Serinus canaria, both Passeriformes, belong to the canarypox clade (clade B). Phoenicopterus ruber, Fratercula, and Spheniscidae are neither fowl nor Passeriforme’s representatives. The viruses detected in the first two hosts belong to the fowlpox clade, whereas the penguinpox virus belongs to the canarypox clade.
Comparing the alignment (Figure 1) and the percentage identity matrix of the sequences obtained in this study (Table 3), several conclusions can be drawn. Sample 16735-20 (blackbird) is the most distinct, as it has several different nucleotides along the entire sequence. Therefore, it has a very low percentage of identity compared to all the other sequences, with the highest percentage of identity being only 81.60%. The nucleotide sequence of sample 24569-17 (Flamingo) is the second most distinct sequence, with a maximum identity percentage of 90.52%. It also contains an additional valine codon at position 23.

Table 3.
Percent identity matrix between APV sequences isolated in this study.
By comparing the alignment of all the sequences present in the phylogenetic tree, it is observed that sample 16735-20 (Blackbird) has two unique mutations. It contains a cytosine instead of a thymine in position nt 63 and a thymine instead of a guanine or an adenine at position nt 148. These mutations originate amino acid changes, namely for a threonine (T) and a serine (S), as referred to below.
Interestingly, when analyzing the percentage identity matrix between the sequences of the A1 subclade, it can be observed that the puffin (Fratercula) isolate sequence is not completely identical to any other sequence, but the percentages of identity are quite high, showing that the sequences are very similar.
On the other hand, all the chicken (Gallus gallus) isolates have an identity percentage of 100%, except for sample sequence 04482-20. However, the identity percentage is still very high, 99.63%, with only two nucleotides differing. It can also be observed that the sequences of the samples isolated from turkeys (Meleagris, Germany, 2001) and wild turkeys (Meleagris gallopavo, Iran, 2015) are also similar to those isolated from chickens. By analyzing the position of the remaining sequences in the phylogenetic tree, it is possible to verify the presence of samples isolated from chickens in subclades C and E. When comparing those of subclade A1 with these, it is found that the percentage of identity is quite low, around 74%, confirming the classification in different clades.
Regarding the sequences of subclade A2, it can be observed that the sequence of the sample isolated from the flamingo (Phoenicopterus ruber) is different from all the other sequences. However, the percentages are quite high, varying between 92.15% (pigeon) (Columbia livia, USA, 1995) and 99.63% (pigeon) (Columbidae, Tanzania, 2013). Furthermore, it can be seen that the two APVs isolated from flamingos in Portugal belong to different clades: the one from 2010 belongs to B2 and the one from 2017 to A2. In fact, the percentage of identity between them is only 75.84%. When they are aligned, it can be seen that the 2010 sequence has an extra isoleucine codon at position nt 24, while the 2017 sequence has an extra phenylalanine codon at position nt 60.
The analysis of the sequences of subclade B1 shows that the two canary (Serinus canaria) isolates from Portugal have the same nucleotide sequence. The canary’s isolate from Austria is not 100% identical to these canary sequences, but it is very similar, with a 99.81% identity percentage and only one nucleotide difference. The sequence of the penguin (Spheniscidae) isolate is 100% identical to the sequences from three other isolates: the golden eagle isolate (Aquila chrysaetos, Spain, 2000), the stone-curlew isolate (Burhinidae, United Arab Emirates, 1998), and the crossbill isolate (Loxia curvirostra, Spain, 1930). A comparison of the sequence of the Avipoxvirus isolated from the penguin in Portugal with that from Argentina shows that they belong to different subclades, B1 and A3, respectively. These two sequences show several mutations between them, with a percentage of identity of only 72.68%.
Comparing the sequences of subclade B3, it can be seen that the sequence isolated from the blackbird (Turdus merula) is not completely identical to any other sequence. However, the percentage of identity is quite high among all sequences (97.03% to 97.58%).
Concerning the deduced amino acid sequences, it is possible to distinguish two major groups, one including sequences from clade A (24569-17, 23045-18, 11612-19, 04482-20, P-08508-21, and 00917-23), and another including sequences from clade B (37026-19, 03779-20, 16735-20, and P-09292-22), as expected. The most divergent sequence is that from strain 16735-20 from subclade B3, as occurred already with the nucleotide sequences. Amino acid sequences from strains 37026-19, 03779-20, and P-09292-22, all from the subclade B1, are almost identical. Regarding sequences from clade A, the same is observed, with that from subclade A2 showing more amino acid differences than those from A1, which are almost similar. When comparing the amino acid alignment of all the sequences present in the phylogenetic tree, it can be seen that sample 16735-20 (Blackbird) also has two unique mutations: a threonine (T) at site 21 and a serine (S) at site 50, as a result of the unique nucleotide mutations already referred.
The results of this study indicate that the APVs circulating in Portugal in chickens and canaries are quite similar since the nucleotide sequences of the isolates have not undergone significant changes. However, the isolates from flamingos show the opposite situation, as the virus circulating in 2010 is very different from the one circulating in 2017. This suggests the presence of a second virus introduction in Portugal. Finally, since the nucleotide sequence of the isolate from penguins is very similar to that of the isolates from golden eagles, stone curlews, and crossbills, it is possible to hypothesize that there has been a third viral introduction in Portugal.
4. Conclusions
This study has updated the molecular and taxonomic data of Avipoxviruses in Portugal based on ten viruses obtained between 2017 and 2023.
In conclusion, it was possible to verify that the samples studied were widely distributed in the phylogenetic tree, with representatives in subclades A1, A2, B1, and B3. It was also observed that some isolates, such as those from chickens and canaries, suggest that the APV circulating in Portugal originates from the same virus. Other isolates, such as those from flamingo and penguin isolates, suggest new virus introductions since they are similar to other strains from different years and geographical origins. These data suggest that the viruses circulating in Portugal have distinct origins. Interestingly, the sequence from the blackbird isolate has two unique mutations compared with all the sequences used in the phylogenetic analysis.
The major drawback of this study is the small sampling size used. However, there was no access to other sequences since only ten positive samples arrived at the laboratory between 2017 and 2023. It would be very interesting to analyze positive sequences that will arrive in the future to go deeper into the molecular characterization and phylogenetic analysis of Avipoxviruses circulating in Portugal.
Supplementary Materials
The following supporting informations can be downloaded at: https://www.mdpi.com/article/10.3390/vetsci10120693/s1, Figure S1. A 1% agarose gel electrophoresis of PCR amplification of the 578 bp P4b gene fragment. (a) Lane 1 corresponds to the molecular weight marker, NZYDNA Ladder V (NZYTech, Portugal). Lanes 2 to 9 correspond to the amplification of samples 24569-17 (flamingo), 11612-19 (puffin), 37026-19 (canary), 03779-20 (canary), 04482-20 (chicken), 16735-20 (blackbird), P-08508-21 (chicken), and 23049-18 (chicken), respectively. Lane 10 is the negative control. Lanes 11 and 12 correspond to the amplification of samples 37026-19 (canary) and 03779-20 (canary) obtained from DNA previously extracted. (b) Lane 1 corresponds to the molecular weight marker, NZYDNA Ladder V. Lanes 2 and 3 correspond to sample 11612-19 (puffin) from different extraction reactions. Lanes 4 and 5 are the negative and positive controls, respectively.
Author Contributions
Conceptualization, S.C.B. and A.M.H.; methodology, D.S., T.F. and F.R.; software, A.D.; investigation, T.L.; writing—original draft preparation, D.S.; writing—review and editing, M.D.D. and A.M.H.; supervision, A.M.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available upon request.
Acknowledgments
The authors thank Maria João Teixeira, Fátima Cordeiro, Rosário Ferreira, and Sandra Nunes for excellent technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Murphy, F.A.; Gibbs, E.P.J.; Horzinek, M.C.; Studdert, M.J. Veterinary Virology, 3rd ed.; Academic Press: San Diego, CA, USA, 1999; ISBN 978-0-12-511340-3. [Google Scholar]
- Brennan, G.; Stoian, A.M.M.; Yu, H.; Rahman, M.J.; Banerjee, S.; Stroup, J.N.; Park, C.; Tazi, L.; Rothenburg, S. Molecular Mechanisms of Poxvirus Evolution. mBio 2023, 14, e01526-22. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S.; Sutherland, M. Molecular Characterisation of a Novel Pathogenic Avipoxvirus from an Australian Little Crow (Corvus Bennetti) Directly from the Clinical Sample. Sci. Rep. 2022, 12, 15053. [Google Scholar] [CrossRef] [PubMed]
- Manarolla, G.; Pisoni, G.; Sironi, G.; Rampin, T. Molecular Biological Characterization of Avian Poxvirus Strains Isolated from Different Avian Species. Vet. Microbiol. 2010, 140, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lecis, R.; Secci, F.; Antuofermo, E.; Nuvoli, S.; Cacciotto, C.; Pittau, M.; Alberti, A. Detection and Characterization of an Avipoxvirus in a Common Buzzard (Buteo Buteo) in Italy Using a Multiple Gene Approach. J. Wildl. Dis. 2019, 55, 142. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Godoy, F.; Ledesma, I.; De Oca, V.; Cervantes, R.; Chávez-Maya, F.; Garcí-Espinosa, G. First Report of Psittacid Avipoxvirus in Agapornis in Mexico. Braz. J. Vet. Pathol. 2023, 16, 64–70. [Google Scholar] [CrossRef]
- Asif, K.; O’Rourke, D.; Legione, A.R.; Shil, P.; Marenda, M.S.; Noormohammadi, A.H. Whole-Genome Based Strain Identification of Fowlpox Virus Directly from Cutaneous Tissue and Propagated Virus. PLoS ONE 2021, 16, e0261122. [Google Scholar] [CrossRef] [PubMed]
- Mapaco, L.P.; Lacerda, Z.; Monjane, I.V.A.; Gelaye, E.; Sussuro, A.H.; Viljoen, G.J.; Dundon, W.G.; Achá, S.J. Identification of Clade E Avipoxvirus, Mozambique, 2016. Emerg. Infect. Dis. 2017, 23, 1602–1604. [Google Scholar] [CrossRef]
- Bányai, K.; Palya, V.; Dénes, B.; Glávits, R.; Ivanics, É.; Horváth, B.; Farkas, S.L.; Marton, S.; Bálint, Á.; Gyuranecz, M.; et al. Unique Genomic Organization of a Novel Avipoxvirus Detected in Turkey (Meleagris Gallopavo). Infect. Genet. Evol. 2015, 35, 221–229. [Google Scholar] [CrossRef]
- Carulei, O.; Douglass, N.; Williamson, A.-L. Comparative Analysis of Avian Poxvirus Genomes, Including a Novel Poxvirus from Lesser Flamingos (Phoenicopterus Minor), Highlights the Lack of Conservation of the Central Region. BMC Genom. 2017, 18, 947. [Google Scholar] [CrossRef]
- Malik, Y.S. , Singh, R.K., Yadav, M.P. (Eds.). Recent Advances in Animal Virology; Springer: Singapore, 2019; ISBN 9789811390722. [Google Scholar]
- Bolte, A.L.; Meurer, J.; Kaleta, E.F. Avian Host Spectrum of Avipoxviruses. Avian Pathol. 1999, 28, 415–432. [Google Scholar] [CrossRef]
- Moss, B. Poxvirus DNA Replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010199. [Google Scholar] [CrossRef] [PubMed]
- Henriques, A.M.; Fagulha, T.; Duarte, M.; Ramos, F.; Barros, S.C.; Luís, T.; Bernardino, R.; Fernandes, T.L.; Lapão, N.; Da Silva, J.F.; et al. Avian Poxvirus Infection in a Flamingo (Phoenicopterus Ruber) of the Lisbon Zoo. J. Zoo Wildl. Med. 2016, 47, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Lebdah, M.; Ali, A.M.; Ali, A.A.; Hassanin, O. Insights into Pathological and Molecular Characterization of Avipoxviruses Circulating in Egypt. Br. Poult. Sci. 2019, 60, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S.; Athukorala, A.; Bowden, T.R.; Boyle, D.B. Genomic Characterisation of a Novel Avipoxvirus Isolated from an Endangered Yellow-Eyed Penguin (Megadyptes Antipodes). Viruses 2021, 13, 194. [Google Scholar] [CrossRef] [PubMed]
- Jarmin, S.; Manvell, R.; Gough, R.E.; Laidlaw, S.M.; Skinner, M.A. Avipoxvirus Phylogenetics: Identification of a PCR Length Polymorphism That Discriminates between the Two Major Clades. J. Gen. Virol. 2006, 87, 2191–2201. [Google Scholar] [CrossRef] [PubMed]
- Bertelloni, F.; Ceccherelli, R.; Marzoni, M.; Poli, A.; Ebani, V.V. Molecular Detection of Avipoxvirus in Wild Birds in Central Italy. Animals 2022, 12, 338. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Nashiruddullah, N.; Bhat, M.A.; Taku, A.; Roychoudhury, P.; Ahmed, J.A.; Sood, S.; Mehmood, S. Occurrence and Phylogenetic Analysis of Avipoxvirus Isolated from Birds around Jammu. Virusdisease 2019, 30, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Kirubaharan, J.J.; Rajasekaran, R.; Shilpa, P.; Vidhya, M.; Rajalakshmi, S. Isolation, Molecular Detection and Phylogenetic Analysis of Avipox Virus Obtained from Pigeon. Indian J. Vet. Sci. Biotechnol. 2018, 14, 40–43. [Google Scholar] [CrossRef]
- Chacón, R.D.; Astolfi-Ferreira, C.S.; Pereira, P.C.; Assayag, M.S.; Campos-Salazar, A.B.; De La Torre, D.; Sá, L.R.M.D.; Almeida, S.R.Y.D.; Rici, R.E.G.; Ferreira, A.J.P. Outbreaks of Avipoxvirus Clade E in Vaccinated Broiler Breeders with Exacerbated Beak Injuries and Sex Differences in Severity. Viruses 2022, 14, 773. [Google Scholar] [CrossRef]
- Van Der Meer, C.S.; Paulino, P.G.; Jardim, T.H.A.; Senne, N.A.; Araujo, T.R.; Dos Santos Juliano, D.; Massard, C.L.; Peixoto, M.P.; Da Costa Angelo, I.; Santos, H.A. Detection and Molecular Characterization of Avipoxvirus in Culex Spp. (Culicidae) Captured in Domestic Areas in Rio de Janeiro, Brazil. Sci. Rep. 2022, 12, 13496. [Google Scholar] [CrossRef]
- Sarker, S.; Athukorala, A.; Raidal, S.R. Molecular Characterisation of a Novel Pathogenic Avipoxvirus from an Australian Passerine Bird, Mudlark (Grallina Cyanoleuca). Virology 2021, 554, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Boyle, D.B. Genus Avipoxvirus. In Poxviruses; Mercer, A.A., Schmidt, A., Weber, O., Eds.; Birkhäuser Basel: Basel, Switzerland, 2007; pp. 217–251. ISBN 978-3-7643-7556-0. [Google Scholar]
- Ghalyanchilangeroudi, A.; Hosseini, H.; Morshed, R. Molecular Characterization and Phylogenetic Analysis of Avian Pox Virus Isolated from Pet Birds and Commercial Flocks, in Iran. Slov. Vet. Res. 2018, 55, 213–218. [Google Scholar] [CrossRef]
- Deng, L.; Liu, C.; Li, L.; Hao, P.; Wang, M.; Jin, N.; Yin, R.; Du, S.; Li, C. Genomic Characteristics of an Avipoxvirus 282E4 Strain. Virus Res. 2023, 336, 199218. [Google Scholar] [CrossRef] [PubMed]
- Loc, G.L.; Bertagnoli, S.; Ducatez, M.; Camus-Bouclainville, C.; Guérin, J.-L. Diversity of Avipoxviruses in Captive-Bred Houbara Bustard. Vet. Res. 2014, 45, 1–10. [Google Scholar] [CrossRef][Green Version]
- Lee, L.H.; Lee, K.H. Application of the Polymerase Chain Reaction for the Diagnosis of Fowl Poxvirus Infection. J. Virol. Methods 1997, 63, 113–119. [Google Scholar] [CrossRef]
- Tamura, K. Estimation of the Number of Nucleotide Substitutions When There Are Strong Transition-Transversion and G + C-Content Biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).