Molecular Survey of Hepatozoon canis Infection in Domestic Dogs from Sardinia, Italy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
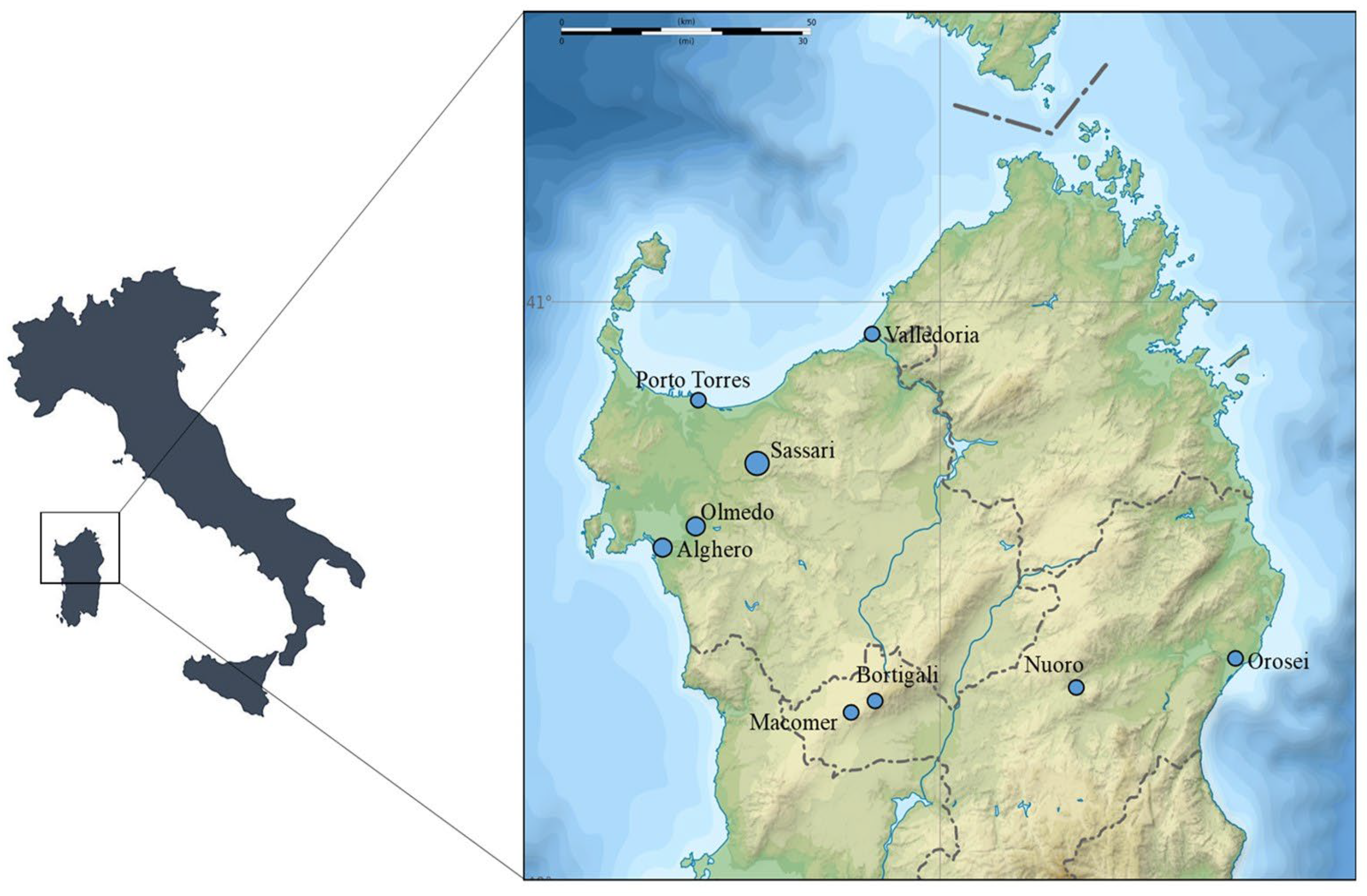
2.1. Collection of Samples
2.2. DNA Extraction and PCR Analysis
2.3. DNA Purification, Sequencing, and Phylogenetic Analyses
3. Results
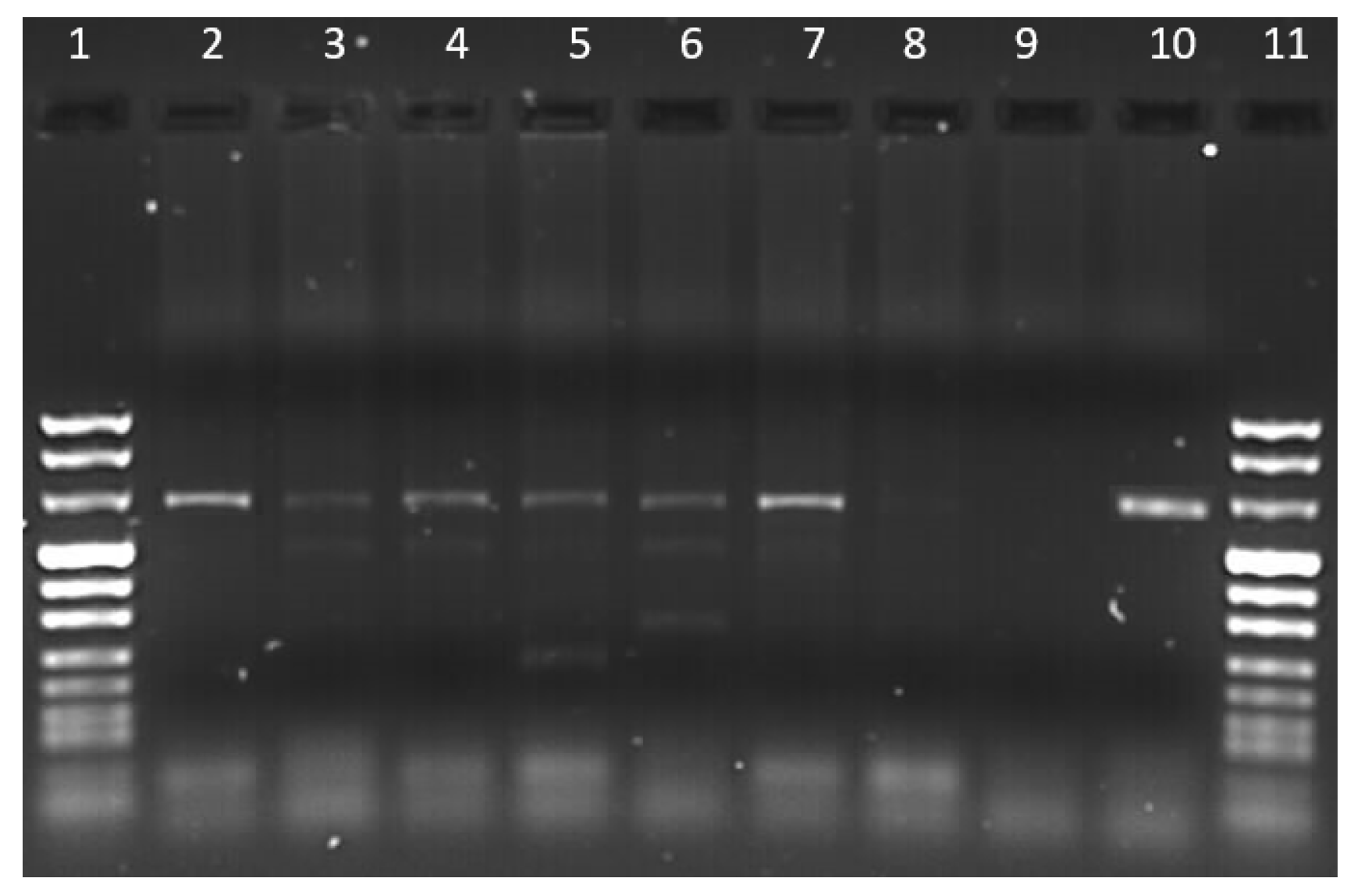
3.1. Collection of Samples and Detection of Hepathozoon spp. by PCR
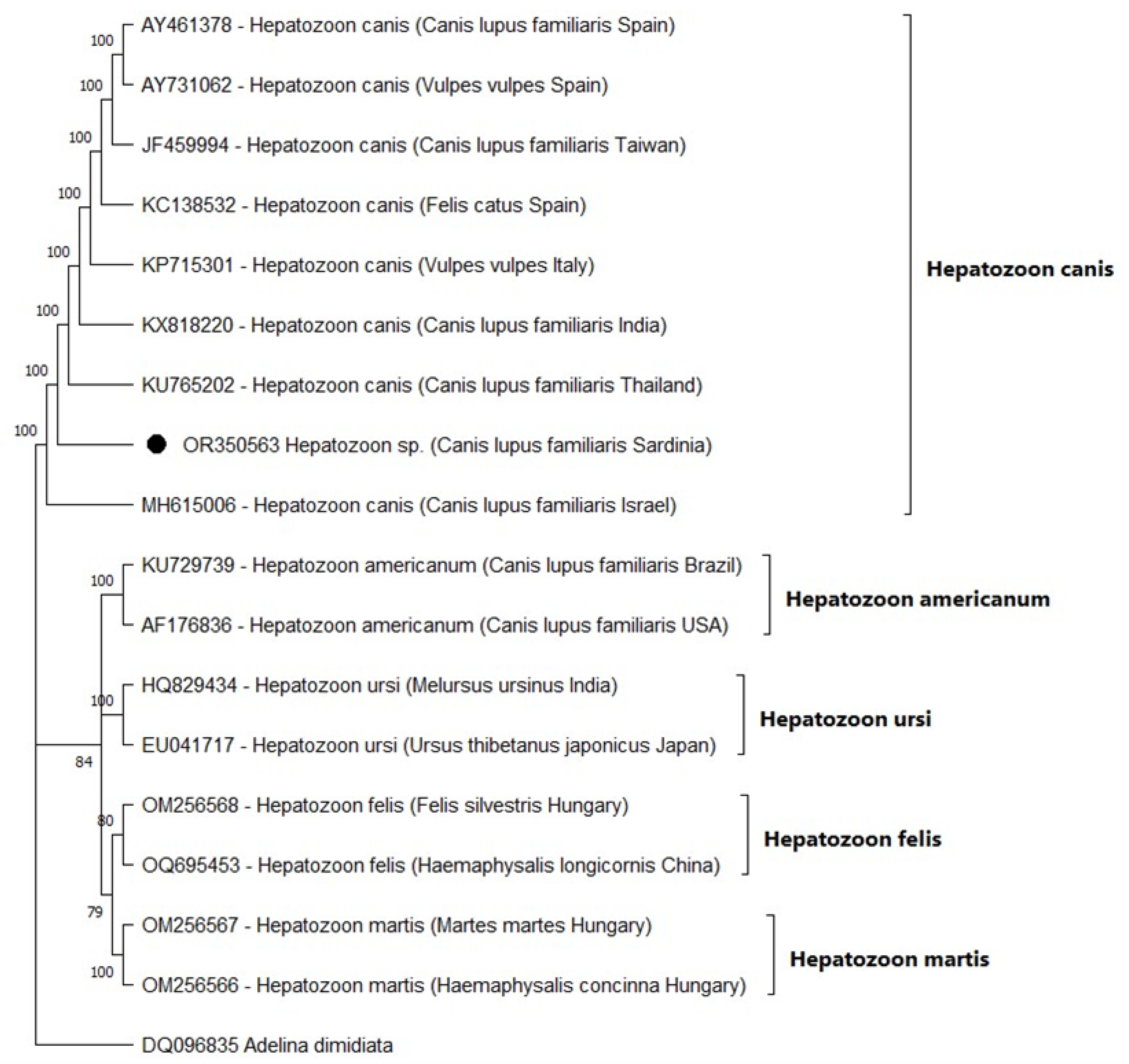
3.2. Nucleotide Sequence and Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Al-Qurarishy, S.; Abdel-Ghaffar, F.; Dkhil, M.A.; Abdel-Gaber, R. Haemogregarines and criteria for identification. Animals 2021, 11, 170. [Google Scholar] [CrossRef] [PubMed]
- Morrissette, N.S.; Sibley, L.D. Cytoskeleton of Apicomplexan Parasites. Microbiol. Mol. Biol. Rev. 2002, 66, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Liberato, G.A.; Lotta-Arévalo, I.A.; Rodríguez-Almonacid, C.C.; Vargas-Ramírez, M.; Matta, N.E. Molecular and morphological description of the first Hepatozoon (Apicomplexa: Hepatozoidae) species infecting a neotropical turtle, with an approach to its phylogenetic relationships. Parasitology 2021, 148, 747–759. [Google Scholar] [CrossRef]
- Merino, S.; Martínez, J.; Masello, J.F.; Bedolla, Y.; Quillfeldt, P. First molecular characterization of a Hepatozoon species (Apicomplexa: Hepatozoidae) infecting birds and description of a new species infecting storm petrels (Aves: Hydrobatidae). J. Parasitol. 2014, 100, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, W.A.; Perles, L.; Picelli, A.M.; Correa, J.K.C.; André, M.R.; Viana, L.A. Hepatozoon parasites (Apicomplexa: Hepatozoidae) in fish Hoplias aimara (Characiformes, Erythrinidae) from the Eastern Amazon, Brazil. Parasitol. Res. 2022, 121, 1041–1046. [Google Scholar] [CrossRef] [PubMed]
- Baneth, G.; Aroch, I.; Tal, N.; Harrus, S. Hepatozoon species infection in domestic cats: A retrospective study. Vet. Parasitol. 1998, 79, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.G. The genus Hepatozoon (Apicomplexa: Adeleina). J. Parasitol. 1996, 82, 565–585. [Google Scholar] [CrossRef]
- Ewing, S.A.; Panciera, R.J. American canine hepatozoonosis. Clin. Microbiol. Rev. 2003, 16, 688–697. [Google Scholar] [CrossRef]
- Baneth, G.; Samish, M.; Shkap, V. Life cycle of Hepatozoon canis (Apicomplexa: Adeleorina: Hepatozoidae) in the tick Rhipicephalus sanguineus and domestic dog (Canis familiaris). J. Parasitol. 2007, 93, 283–299. [Google Scholar] [CrossRef]
- Cardoso, L.; Cortes, H.C.; Eyal, O.; Reis, A.; Lopes, A.P.; Vila-Viçosa, M.J.; Rodrigues, P.A.; Baneth, G. Molecular and histopathological detection of Hepatozoon canis in red foxes (Vulpes vulpes) from Portugal. Parasites Vectors 2014, 7, 113. [Google Scholar] [CrossRef]
- Murata, T.; Inoue, M.; Tateyama, S.; Taura, Y.; Nakama, S. Vertical transmission of Hepatozoon canis in dogs. J. Vet. Med. Sci. 1993, 55, 867–868. [Google Scholar] [CrossRef] [PubMed]
- Uiterwijk, M.; Vojta, L.; Šprem, N.; Beck, A.; Jurković, D.; Kik, M.; Duscher, G.G.; Hodžić, A.; Reljić, S.; Sprong, H.; et al. Diversity of Hepatozoon species in wild mammals and ticks in Europe. Parasites Vectors 2023, 16, 27. [Google Scholar] [CrossRef] [PubMed]
- Baneth, G.; Shkap, V. Monozoic cysts of Hepatozoon canis. J. Parasitol. 2003, 89, 379–381. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, I.; Müller, E.; Nijhof, A.M.; Aupperle-Lellbach, H.; Loesenbeck, G.; Cramer, S.; Naucke, T.J. First evidence of vertical Hepatoozoon canis transmission in dogs in Europe. Parasites Vectors 2022, 15, 296. [Google Scholar] [CrossRef] [PubMed]
- Otranto, D.; Dantas-Torres, F.; Weigl, S.; Latrofa, M.S.; Stanneck, D.; Decaprariis, D.; Capelli, G.; Baneth, G. Diagnosis of Hepatozoon canis in young dogs by cytology and PCR. Parasites Vectors 2011, 4, 55. [Google Scholar] [CrossRef] [PubMed]
- Baneth, G.; Mathew, J.S.; Shkap, V.; Macintire, D.K.; Barta, J.R.; Ewing, S.A. Canine hepatozoonosis: Two disease syndromes caused by separate Hepatozoon spp. Trends Parasitol. 2003, 19, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Tołkacz, K.; Kretschmer, M.; Nowak, S.; Mysłajek, R.W.; Alsarraf, M.; Wężyk, D.; Bajer, A. The first report on Hepatozoon canis in dogs and wolves in Poland: Clinical and epidemiological features. Parasites Vectors 2023, 16, 313. [Google Scholar] [CrossRef]
- Baneth, G.; Bourdeau, P.; Bourdoiseau, G.; Bowman, D.; Breitschwerdt, E.; Capelli, G.; Dantas-Torres, F.; Day, M.; Dedet, J.-P.; Dobler, G.; et al. Vector-borne diseases–constant challenge for practicing veterinarians: Recommendations from the CVBD World Forum. Parasit Vectors 2012, 5, 55. [Google Scholar] [CrossRef]
- Baneth, G. Perspectives on canine and feline hepatozoonosis. Vet. Parasitol. 2011, 181, 3–11. [Google Scholar] [CrossRef]
- de Miranda, R.L.; de Castro, J.R.; Olegário, M.M.; Beletti, M.E.; Mundim, A.V.; O’Dwyer, L.H.; Eyal, O.; Talmi-Frank, D.; Cury, M.C.; Baneth, G. Oocysts of Hepatozoon canis in Rhipicephalus (Boophilus) microplus collected from a naturally infected dog. Vet. Parasitol. 2011, 177, 392–396. [Google Scholar] [CrossRef]
- Orkun, O.; Nalbantoğlu, S. Hepatozoon canis in Turkish red foxes and their ticks. Vet. Parasitol. Reg. Stud. Rep. 2018, 13, 35–37. [Google Scholar] [CrossRef] [PubMed]
- Léveillé, A.N.; Baneth, G.; Barta, J.R. Next generation sequencing from Hepatozoon canis (Apicomplexa: Coccidia: Adeleorina): Complete apicoplast genome and multiple mitochondrion-associated sequences. Int. J. Parasitol. 2019, 49, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Chisu, V.; Foxi, C.; Mannu, R.; Satta, G.; Masala, G. A five-year survey of tick species and identification of tick-borne bacteria in Sardinia, Italy. Ticks Tick Borne Dis. 2018, 9, 678–681. [Google Scholar] [CrossRef] [PubMed]
- Chisu, V.; Serra, E.; Foxi, C.; Chessa, G.; Masala, G. Molecular Investigation of Theileria and Babesia Species in Domestic Mammals from Sardinia, Italy. Vet. Sci. 2023, 10, 59. [Google Scholar] [CrossRef] [PubMed]
- Inokuma, H.; Okuda, M.; Ohno, K.; Shimoda, K.; Onishi, T. Analysis of the 18S rRNA gene sequence of a Hepatozoon detected in two Japanese dogs. Vet. Parasitol. 2002, 106, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Pacifico, L.; Braff, J.; Buono, F.; Beall, M.; Neola, B.; Buch, J.; Sgroi, G.; Piantedosi, D.; Santoro, M.; Tyrrell, P.; et al. Hepatozoon canis in hunting dogs from Southern Italy: Distribution and risk factors. Parasitol. Res. 2020, 119, 3023–3031. [Google Scholar] [CrossRef]
- Ebani, V.V.; Nardoni, S.; Mancianti, F. Arthropod-Borne Pathogens in Wild Canids. Vet. Sci. 2023, 10, 165. [Google Scholar] [CrossRef]
- André, M.R.; Adania, C.H.; Teixeira, R.H.; Vargas, G.H.; Falcade, M.; Sousa, L.; Salles., A.R.; Allegretti., S.M.; Felippe., P.A.; Machado., R.Z. Molecular detection of Hepatozoon spp. in Brazilian and exotic wild carnivores. Vet. Parasitol. 2010, 173, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Baneth, G.; Shkap, V.; Samish, M.; Pipano, E.; Savitsky, I. Antibody response to Hepatozoon canis in experimentally infected dogs. Vet. Parasitol. 1998, 74, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Ebani, V.V.; Bertelloni, F.; Turchi, B.; Filogari, D.; Cerri, D. Molecular survey of tick-borne pathogens in Ixodid ticks collected from hunted wild animals in Tuscany, Italy. Asian Pac. J. Trop. Med. 2015, 8, 714–717. [Google Scholar]
- Cassini, R.; Zanutto, S.; Frangipane di Regalbono, A.; Gabrielli, S.; Calderini, P.; Moretti, A.; Tampieri, M.P.; Pietrobelli, M. Canine piroplasmosis in Italy: Epidemiological aspects in vertebrate and invertebrate hosts. Vet. Parasitol. 2009, 165, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Maurelli, M.P.; Pepe, P.; Colombo, L.; Armstrong, R.; Battisti, E.; Morgoglione, M.E.; Counturis, D.; Rinaldi, L.; Cringoli, G.; Ferroglio, E.; et al. A national survey of Ixodidae ticks on privately owned dogs in Italy. Parasites Vectors 2018, 11, 420. [Google Scholar] [CrossRef] [PubMed]
- Bhusri, B.; Lekcharoen, P.; Changbunjong, T. First detection and molecular identification of Babesia gibsoni and Hepatozoon canis in an Asiatic wild dog (Cuon alpinus) from Thailand. Int. J. Parasitol. Parasites Wildl. 2022, 17, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Latrofa, M.S.; Dantas-Torres, F.; Giannelli, A.; Otranto, D. Molecular detection of tick-borne pathogens in Rhipicephalus sanguineus group ticks. Ticks Tick Borne Dis. 2014, 5, 943–946. [Google Scholar] [CrossRef]
- Giannelli, A.; Lia, R.P.; Annoscia, G.; Buonavoglia, C.; Lorusso, E.; Dantas-Torres, F.; Baneth, G.; Otranto, D. Rhipicephalus turanicus, a new vector of Hepatozoon canis. Parasitology 2017, 144, 730–737. [Google Scholar] [CrossRef]
- Chisu, V.; Loi, F.; Foxi, C.; Chessa, G.; Masu, G.; Rolesu, S.; Masala, G. Coexistence of tick-borne pathogens in ticks collected from their hosts in Sardinia: An update. Acta Parasitol. 2020, 65, 999–1004. [Google Scholar] [CrossRef]
- Lauzi, S.; Maia, J.P.; Epis, S.; Marcos, R.; Pereira, C.; Luzzago, C.; Santos, M.; Puente-Payo, P.; Giordano, A.; Pajoro, M.; et al. Molecular detection of Anaplasma platys, Ehrlichia canis, Hepatozoon canis and Rickettsia monacensis in dogs from Maio Island of Cape Verde archipelago. Ticks Tick Borne Dis. 2016, 7, 964–969. [Google Scholar] [CrossRef]
- Rojas, A.; Rojas, D.; Montenegro, V.; Gutiérrez, R.; Yasur-Landau, D.; Baneth, G. Vector-borne pathogens in dogs from Costa Rica: First molecular description of Babesia vogeli and Hepatozoon canis infections with a high prevalence of monocytic ehrlichiosis and the manifestations of co-infection. Vet. Parasitol. 2014, 199, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Aktas, M.; Özübek, S.; Altay, K.; Balkaya, İ.; Utuk, A.E.; Kırbas, A.; Şimsek, S.; Dumanlı, N.A. A molecular and parasitological survey of Hepatozoon canis in domestic dogs in Turkey. Vet. Parasitol. 2015, 209, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Gomes, P.V.; Mundim, M.J.S.; Mundim, A.V.; Ávila, D.F.; Guimarães, E.C.; Cury, M.C. Occurrence of Hepatozoon sp. in dogs in the urban area originating from a municipality in southeastern Brazil. Vet. Parasitol. 2010, 174, 155–161. [Google Scholar] [CrossRef] [PubMed]
- De Bonis, A.; Colombo, M.; Terragni, R.; Bacci, B.; Morelli, S.; Grillini, M.; Vignoli, M. Potential role of Hepatozoon canis in a fatal systemic disease in a puppy. Pathogens 2021, 10, 1193. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.S.; Mylonakis, M.E.; Garg, R.; Vatsya, S.; Yadav, C.L. Concurrent hepatozoonosis, monocytic and granulocytic ehrlichiosis in a dog. J. Vet. Parasitol. 2008, 22, 9–11. [Google Scholar]
- Sasanelli, M.; Paradies, P.; Greco, B.; Eyal, O.; Zaza, V.; Baneth, G. Failure of imidocarb dipropionate to eliminate Hepatozoon canis in naturally infected dogs based on parasitological and molecular evaluation methods. Vet. Parasitol. 2010, 171, 194–199. [Google Scholar] [CrossRef]
| Parameters | Variables | Sample Analyzed | Positive Dogs (%) | 95% CI |
|---|---|---|---|---|
| Gender | Male | 20 | 2 (10) | 0–23 |
| Female | 31 | 7 (22.6) | 8–38 | |
| Age (years) | <1 | 2 | 0 | 0 |
| 1–5 | 25 | 5 (20) | 4–36 | |
| 5–10 | 14 | 2 (14.3) | 0–32 | |
| >10 | 7 | 2 (28.6) | 0–63 | |
| Not determined | 3 | 0 | 0 | |
| Ectoparasiticide treatments | Yes | 46 | 8 (17.4) | 6–28 |
| No | 5 | 1 (20) | 0–55 | |
| Environment | Urban | 19 | 6 (32) | 11–43 |
| Rural | 32 | 3 (9.4) | 0–19 | |
| Geographical location | Sassari | 48 | 9 (18.8) | 8–30 |
| Nuoro | 3 | 0 | 0 | |
| Clinical signs | Lymphadenomegaly | 7 | 1 (14) | 0–40 |
| Fever | 3 | 1 (33) | 0–86 | |
| Anorexia | 2 | 0 | 0 | |
| Muscular pain | 11 | 4 (36) | 8–64 | |
| No clinical symptoms | 28 | 3 (11) | 0–23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chisu, V.; Giua, L.; Bianco, P.; Masala, G.; Sechi, S.; Cocco, R.; Piredda, I. Molecular Survey of Hepatozoon canis Infection in Domestic Dogs from Sardinia, Italy. Vet. Sci. 2023, 10, 640. https://doi.org/10.3390/vetsci10110640
Chisu V, Giua L, Bianco P, Masala G, Sechi S, Cocco R, Piredda I. Molecular Survey of Hepatozoon canis Infection in Domestic Dogs from Sardinia, Italy. Veterinary Sciences. 2023; 10(11):640. https://doi.org/10.3390/vetsci10110640
Chicago/Turabian StyleChisu, Valentina, Laura Giua, Piera Bianco, Giovanna Masala, Sara Sechi, Raffaella Cocco, and Ivana Piredda. 2023. "Molecular Survey of Hepatozoon canis Infection in Domestic Dogs from Sardinia, Italy" Veterinary Sciences 10, no. 11: 640. https://doi.org/10.3390/vetsci10110640
APA StyleChisu, V., Giua, L., Bianco, P., Masala, G., Sechi, S., Cocco, R., & Piredda, I. (2023). Molecular Survey of Hepatozoon canis Infection in Domestic Dogs from Sardinia, Italy. Veterinary Sciences, 10(11), 640. https://doi.org/10.3390/vetsci10110640






