Long Non-Coding RNA as a Potential Biomarker for Canine Tumors
Abstract
:Simple Summary
Abstract
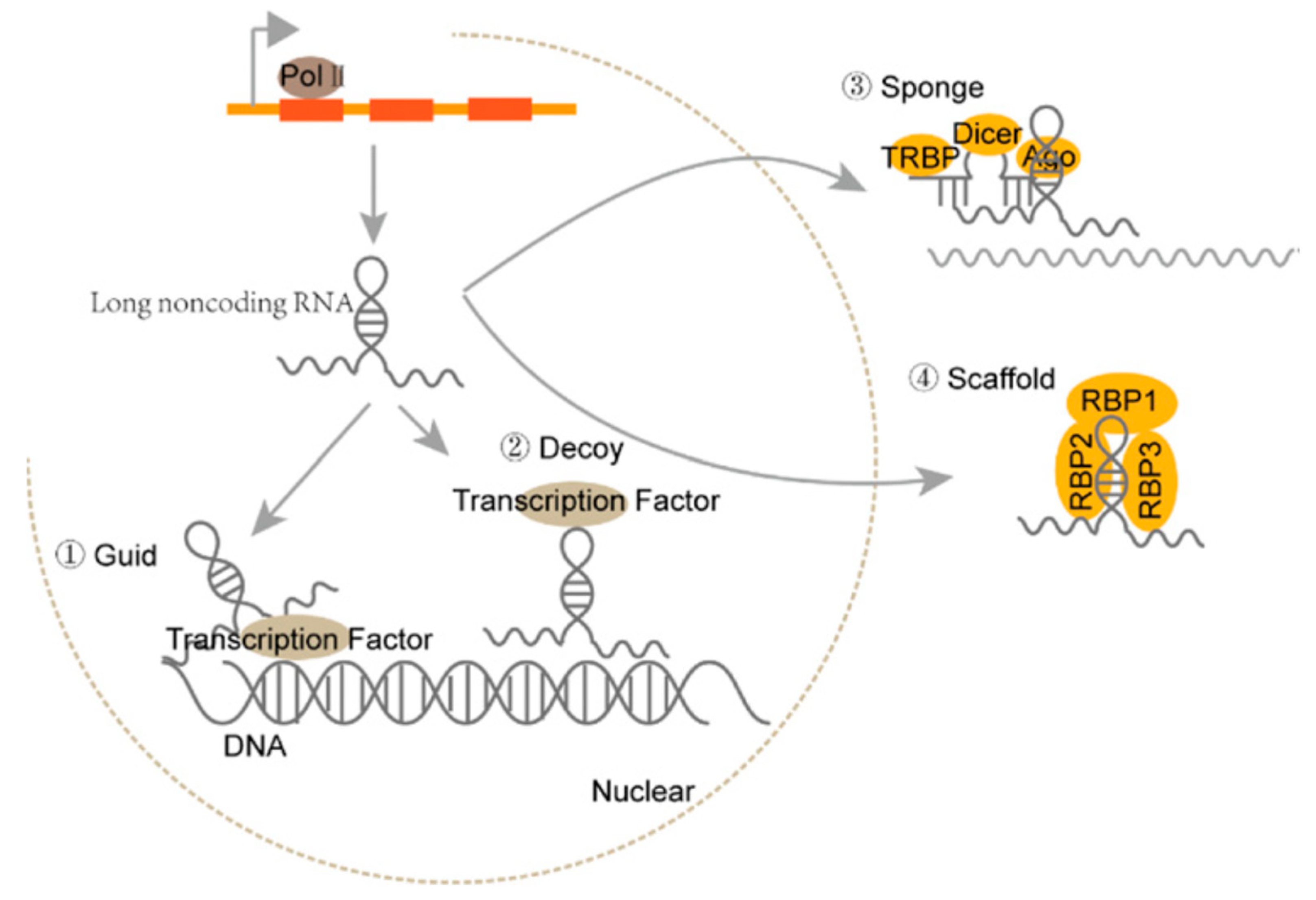
1. Introduction
2. The Molecular Function and Clinical Use of lncRNA as Canine Mammary Tumor Biomarkers
3. The Molecular Function and Clinical Use of lncRNA as Canine Melanoma Biomarkers
4. The Molecular Function and Clinical use of lncRNA as Canine Lymphoma Biomarkers
5. The Molecular Function and Clinical Use of lncRNA in Other Canine Tumors
6. Summary and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Vail, D.M.; Thamm, D.H.; Liptak, J.M. Withrow and MacEwen’s Small Animal Clinical Oncology; Elsevier: Amsterdam, The Netherlands, 2020; p. xix. [Google Scholar]
- Davis, B.W.; Ostrander, E.A. Domestic Dogs and Cancer Research: A Breed-Based Genomics Approach. ILAR J. 2014, 55, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S. The Emperor of All Maladies: A biography of Cancer; Scribner: New York, NY, USA, 2011. [Google Scholar]
- Oh, J.H.; Cho, J.-Y. Comparative oncology: Overcoming human cancer through companion animal studies. Exp. Mol. Med. 2023, 55, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.Y.; Moskwa, N.; Kang, W.; Fan, T.M.; Lee, C. Canine as a Comparative and Translational Model for Human Mammary Tumor. J. Breast Cancer 2023, 26, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Diao, H.; Zhao, Y.; Xu, H.; Pei, S.; Gao, J.; Wang, J.; Hussain, T.; Zhao, D.; Zhou, X.; et al. Overexpression of matrix metalloproteinase-9 in breast cancer cell lines remarkably increases the cell malignancy largely via activation of transforming growth factor beta/SMAD signalling. Cell Prolif. 2019, 52, e12633. [Google Scholar] [CrossRef] [PubMed]
- Leon-Ferre, R.A.; Goetz, M.P. Advances in systemic therapies for triple negative breast cancer. BMJ 2023, 381, e071674. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Nelson, B.R.; Makarewich, C.A.; Anderson, D.M.; Winders, B.R.; Troupes, C.D.; Wu, F.; Reese, A.L.; McAnally, J.R.; Chen, X.; Kavalali, E.T.; et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016, 351, 271–275. [Google Scholar] [CrossRef]
- Gibb, E.A.; Brown, C.J.; Lam, W.L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer 2011, 10, 38. [Google Scholar] [CrossRef]
- Postepska-Igielska, A.; Giwojna, A.; Gasri-Plotnitsky, L.; Schmitt, N.; Dold, A.; Ginsberg, D.; Grummt, I. LncRNA Khps1 Regulates Expression of the Proto-oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure. Mol. Cell 2015, 60, 626–636. [Google Scholar] [CrossRef]
- Grelet, S.; Link, L.A.; Howley, B.; Obellianne, C.; Palanisamy, V.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nature 2017, 19, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Souquere, S.; Chujo, T.; Kobelke, S.; Chong, Y.S.; Fox, A.H.; Bond, C.S.; Nakagawa, S.; Pierron, G.; Hirose, T. Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol. Cell 2018, 70, 1038–1053.e7. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.; Weidmann, C.A.; Hilimire, T.A.; Yee, E.; Hatfield, B.M.; Schneekloth, J.S.; Weeks, K.M.; Novina, C.D. Targeting the Oncogenic Long Non-coding RNA SLNCR1 by Blocking Its Sequence-Specific Binding to the Androgen Receptor. Cell Rep. 2020, 30, 541–554.e5. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Bu, P. Non-coding RNA in cancer. Essays Biochem. 2021, 65, 625–639. [Google Scholar] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Hoeppner, M.P.; Lundquist, A.; Pirun, M.; Meadows, J.R.; Zamani, N.; Johnson, J.; Sundström, G.; Cook, A.; FitzGerald, M.G.; Swofford, R.; et al. An improved canine genome and a comprehensive catalogue of coding genes and non-coding transcripts. PLoS ONE 2014, 9, e91172. [Google Scholar] [CrossRef]
- Wucher, V.; Legeai, F.; Hédan, B.; Rizk, G.; Lagoutte, L.; Leeb, T.; Jagannathan, V.; Cadieu, E.; David, A.; Lohi, H.; et al. FEELnc: A tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 2017, 45, e57. [Google Scholar] [CrossRef] [PubMed]
- Le Béguec, C.; Wucher, V.; Lagoutte, L.; Cadieu, E.; Botherel, N.; Hédan, B.; De Brito, C.; Guillory, A.-S.; André, C.; Derrien, T.; et al. Characterisation and functional predictions of canine long non-coding RNAs. Sci. Rep. 2018, 8, 13444. [Google Scholar] [CrossRef]
- Benavente, M.A.; Bianchi, C.P.; Aba, M.A. Canine Mammary Tumors: Risk Factors, Prognosis and Treatments. J. Veter Adv. 2016, 6, 1291–1300. [Google Scholar] [CrossRef]
- Stratmann, N.; Failing, K.; Richter, A.; Wehrend, A. Mammary Tumor Recurrence in Bitches After Regional Mastectomy. Veter Surg. 2008, 37, 82–86. [Google Scholar] [CrossRef]
- Azari, M.; Bahreini, F.; Uversky, V.N.; Rezaei, N. Current therapeutic approaches and promising perspectives of using bioengineered peptides in fighting chemoresistance in triple-negative breast cancer. Biochem. Pharmacol. 2023, 210, 115459. [Google Scholar] [CrossRef]
- Kaszak, I.; Ruszczak, A.; Kanafa, S.; Kacprzak, K.; Król, M.; Jurka, P. Current biomarkers of canine mammary tumors. Acta Vet. Scand. 2018, 60, 66. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Wu, J.; Chen, H.; Li, S.; Jia, K. LncRNA Expression Profiles in Canine Mammary Tumors Identify lnc34977 as a Promoter of Proliferation, Migration and Invasion of Canine Mammary Tumor Cells. Vet. Sci. 2022, 9, 82. [Google Scholar] [CrossRef]
- Lu, B.; Zhu, Y.; Wu, J.; Qiu, H.; Wang, J.; Ma, Z.; Jia, K. LncRNA34977 promotes the proliferation, migration, and invasion and inhibits the apoptosis of canine mammary tumors by regulating the expression of miR-8881/ELAVL4. Funct. Integr. Genom. 2023, 23, 31. [Google Scholar] [CrossRef] [PubMed]
- Xu, E.; Hu, M.; Ge, R.; Tong, D.; Fan, Y.; Ren, X.; Liu, Y. LncRNA-42060 Regulates Tamoxifen Sensitivity and Tumor Development via Regulating the miR-204-5p/SOX4 Axis in Canine Mammary Gland Tumor Cells. Front. Vet. Sci. 2021, 8, 654694. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Willenbrock, S.; Nolte, I.; Escobar, H.M. Comparison of non-coding RNAs in human and canine cancer. Front. Genet. 2013, 4, 46. [Google Scholar] [CrossRef] [PubMed]
- Palma, S.D.; McConnell, A.; Verganti, S.; Starkey, M. Review on Canine Oral Melanoma: An Undervalued Authentic Genetic Model of Human Oral Melanoma? Vet. Pathol. 2021, 58, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Prouteau, A.; André, C. Canine Melanomas as Models for Human Melanomas: Clinical, Histological, and Genetic Comparison. Genes 2019, 10, 501. [Google Scholar] [CrossRef]
- Brockley, L.; Cooper, M.; Bennett, P. Malignant melanoma in 63 dogs (2001–2011): The effect of carboplatin chemotherapy on survival. N. Z. Vet. J. 2013, 61, 25–31. [Google Scholar] [CrossRef]
- Ramos-Vara, J.A.; Beissenherz, M.E.; Miller, M.A.; Johnson, G.C.; Pace, L.W.; Fard, A.; Kottler, S.J. Retrospective Study of 338 Canine Oral Melanomas with Clinical, Histologic, and Immunohistochemical Review of 129 Cases. Vet. Pathol. 2000, 37, 597–608. [Google Scholar] [CrossRef]
- Gillard, M.; Cadieu, E.; De Brito, C.; Abadie, J.; Vergier, B.; Devauchelle, P.; Degorce, F.; Dréano, S.; Primot, A.; Dorso, L.; et al. Naturally occurring melanomas in dogs as models for non-UV pathways of human melanomas. Pigment. Cell Melanoma Res. 2013, 27, 90–102. [Google Scholar] [CrossRef]
- Hitte, C.; Le Béguec, C.; Cadieu, E.; Wucher, V.; Primot, A.; Prouteau, A.; Botherel, N.; Hédan, B.; Lindblad-Toh, K.; André, C.; et al. Genome-Wide Analysis of Long Non-Coding RNA Profiles in Canine Oral Melanomas. Genes 2019, 10, 477. [Google Scholar] [CrossRef]
- Husna, A.A.; Rahman, M.; Chen, H.; Hasan, N.; Nakagawa, T.; Miura, N. Long non-coding RNA and transfer RNA-derived small fragments in exosomes are potential biomarkers for canine oral melanoma. Vet. Comp. Oncol. 2022, 20, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Van Rooyen, L.J.; Hooijberg, E.; Reyers, F. Breed prevalence of canine lymphoma in South Africa. J. S. Afr. Vet. Assoc. 2018, 89, e1–e11. [Google Scholar] [CrossRef] [PubMed]
- Modiano, J.F.; Breen, M.; Burnett, R.C.; Parker, H.G.; Inusah, S.; Thomas, R.; Avery, P.R.; Lindblad-Toh, K.; Ostrander, E.A.; Cutter, G.C.; et al. Distinct B-Cell and T-Cell Lymphoproliferative Disease Prevalence among Dog Breeds Indicates Heritable Risk. Cancer Res. 2005, 65, 5654–5661. [Google Scholar] [CrossRef] [PubMed]
- Comazzi, S.; Marelli, S.; Cozzi, M.; Rizzi, R.; Finotello, R.; Henriques, J.; Pastor, J.; Ponce, F.; Rohrer-Bley, C.; Rütgen, B.C.; et al. Breed-associated risks for developing canine lymphoma differ among countries: An European canine lymphoma network study. BMC Vet. Res. 2018, 14, 232. [Google Scholar] [CrossRef] [PubMed]
- Kluin, P.; Schuuring, E. Molecular cytogenetics of lymphoma: Where do we stand in 2010? Histopathology 2011, 58, 128–144. [Google Scholar] [CrossRef]
- Montaner-Angoiti, E.; Marín-García, P.J.; Llobat, L. Epigenetic Alterations in Canine Malignant Lymphoma: Future and Clinical Outcomes. Animals 2023, 13, 468. [Google Scholar] [CrossRef] [PubMed]
- Thamm, D.H. Novel Treatments for Lymphoma. Vet. Clin. N. Am. Small Anim. Pract. 2019, 49, 903–915. [Google Scholar] [CrossRef]
- Aresu, L.; Ferraresso, S.; Marconato, L.; Cascione, L.; Napoli, S.; Gaudio, E.; Kwee, I.; Tarantelli, C.; Testa, A.; Maniaci, C.; et al. New molecular and therapeutic insights into canine diffuse large B-cell lymphoma elucidates the role of the dog as a model for human disease. Haematologica 2019, 104, e256–e259. [Google Scholar] [CrossRef]
- Verma, A.; Jiang, Y.; Du, W.; Fairchild, L.; Melnick, A.; Elemento, O. Transcriptome sequencing reveals thousands of novel long non-coding RNAs in B cell lymphoma. Genome Med. 2015, 7, 110. [Google Scholar] [CrossRef]
- Cascione, L.; Giudice, L.; Ferraresso, S.; Marconato, L.; Giannuzzi, D.; Napoli, S.; Bertoni, F.; Giugno, R.; Aresu, L. Long Non-Coding RNAs as Molecular Signatures for Canine B-Cell Lymphoma Characterization. Non-Coding RNA 2019, 5, 47. [Google Scholar] [CrossRef] [PubMed]
- Kreilmeier, T.; Mejri, D.; Hauck, M.; Kleiter, M.; Holzmann, K. Telomere Transcripts Target Telomerase in Human Cancer Cells. Genes 2016, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- Leitner, N.; Ertl, R.; Gabner, S.; Fuchs-Baumgartinger, A.; Walter, I.; Hlavaty, J. Isolation and Characterization of Novel Canine Osteosarcoma Cell Lines from Chemotherapy-Naïve Patients. Cells 2023, 12, 1026. [Google Scholar] [CrossRef] [PubMed]
- Farzaneh, M.; Najafi, S.; Anbiyaee, O.; Azizidoost, S.; Khoshnam, S.E. LncRNA MALAT1-related signaling pathways in osteosarcoma. Clin. Transl. Oncol. 2022, 25, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Zhang, T.; Chen, B. Long Non-Coding RNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1) Promotes Proliferation and Metastasis of Osteosarcoma Cells by Targeting c-Met and SOX4 via miR-34a/c-5p and miR-449a/b. Med. Sci. Monit. 2019, 25, 1410–1422. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Piao, C.-D.; Ding, J.; Li, Z.-W. LncRNA MALAT1 facilitates lung metastasis of osteosarcomas through miR-202 sponging. Sci. Rep. 2020, 10, 12757. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Z.; Yang, D.; Liu, L.; Liu, Z.; Wang, J.; He, D.; Wu, H.; Wang, J.; Ma, Z. Genome-wide identification and characterization of long non-coding RNAs in MDCK cell lines with high and low tumorigenicities. Genomics 2020, 112, 1077–1086. [Google Scholar] [CrossRef]
- Li, Y.; Shan, Z.; Yang, B.; Yang, D.; Men, C.; Cui, Y.; Wu, J. LncRNA HULC promotes epithelial and smooth-muscle-like differentiation of adipose-derived stem cells by upregulation of BMP9. Die Pharm. 2018, 73, 49–55. [Google Scholar] [CrossRef]
- Hédan, B.; Cadieu, É.; Rimbault, M.; Vaysse, A.; de Citres, C.D.; Devauchelle, P.; Botherel, N.; Abadie, J.; Quignon, P.; Derrien, T.; et al. Identification of common predisposing loci to hematopoietic cancers in four dog breeds. PLoS Genet. 2021, 17, e1009395. [Google Scholar] [CrossRef]
| Cancer Types | lncRNA | Function | Reference |
|---|---|---|---|
| Mammary tumor | lncRNA 40589 | inhibits cell proliferation, migration, and invasion | [24] |
| lncRNA 34977 | promotes the development of tumor cells and suppresses apoptosis | [24] | |
| lncRNA 42060 | inhibiting tumor cell progression | [26] | |
| Melanoma | lncRNA SOX21-AS | regulatory elements in the cell cycle and carbohydrate metabolism | [33] |
| lncRNA ZEB2-AS | regulatory elements in the cell cycle and carbohydrate metabolism | [33] | |
| lncRNA CASC15 | regulatory elements in the cell cycle and carbohydrate metabolism | [33] | |
| lncRNAs ENSCAFT00000069599.1 | correlation with the patient’s ROC | [34] | |
| lncRNAs ENSCAFT00000090032.1 | NA | [34] | |
| Lymphoma | A group of lncRNAs | a non-negligible fraction of human DLBCL | [42] |
| A group of lncRNAs | differentiates B-cell lymphoma subtypes and indicates mortality rates | [43] | |
| Soft tissue sarcoma | lncRNA TERRA | regulates the cell growth | [44] |
| Osteosarcoma | lncRNA MALAT1 | regulates the cell growth | [45] |
| MDCK tumorigenicity | lncRNA MSTRG.1056.2 | contributes to tumorigenesis | [49] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Wu, M.; Zhou, J.; Diao, H. Long Non-Coding RNA as a Potential Biomarker for Canine Tumors. Vet. Sci. 2023, 10, 637. https://doi.org/10.3390/vetsci10110637
Zhang Y, Wu M, Zhou J, Diao H. Long Non-Coding RNA as a Potential Biomarker for Canine Tumors. Veterinary Sciences. 2023; 10(11):637. https://doi.org/10.3390/vetsci10110637
Chicago/Turabian StyleZhang, Yan, Meijin Wu, Jiahao Zhou, and Hongxiu Diao. 2023. "Long Non-Coding RNA as a Potential Biomarker for Canine Tumors" Veterinary Sciences 10, no. 11: 637. https://doi.org/10.3390/vetsci10110637
APA StyleZhang, Y., Wu, M., Zhou, J., & Diao, H. (2023). Long Non-Coding RNA as a Potential Biomarker for Canine Tumors. Veterinary Sciences, 10(11), 637. https://doi.org/10.3390/vetsci10110637






