Simple Summary
Livestock guardian and herding shepherd dogs are morphologically and behaviourally different, due to the long selection for different tasks made by farmers and breeders. This study aimed to identify genomic regions that best distinguish and characterise four livestock guardian and five herding Italian dog breeds. Genomic SNP data of 158 dogs were compared using two analyses, allowing for the identification of regions harbouring 29 genes. Sixteen runs of homozygosity islands were found in livestock guardians, four of which were partially shared with the fifteen found in herding shepherd dogs. The identified genes were related to dog domestication and behaviour, including herding behaviour, body size and muscle development, the prick or drop ear phenotype, and eye development and functionality. These results contribute to a better understanding of how human selection shaped the genome of dogs selected for different tasks, even considering a limited geographic area.
Abstract
Livestock guardian (LGD) and herding shepherd (HSD) dogs have distinct morphological and behavioural characteristics, long selected by farmers and breeders, to accomplish different tasks. This study aimed to find the genomic regions that best differentiate and characterise Italian LGD and HSD. Genomic data of 158 dogs of four LGD and five HSD breeds, obtained with the 170K canine SNPchip, were collected. The two groups were compared using FST and XP-EHH analyses, identifying regions containing 29 genes. Moreover, 16 islands of runs of homozygosity were found in LGD, and 15 in HSD; 4 of them were partially shared. Among the genes found that better differentiated HSD and LGD, several were associated with dog domestication and behavioural aspects; particularly, MSRB3 and LLPH were linked to herding behaviour in previous studies. Others, DYSK, MAP2K5, and RYR, were related to body size and muscle development. Prick ears prevailed in sampled HSD, and drop ears in LGD; this explains the identification of WIF1 and MSRB3 genes. Unexpectedly, a number of genes were also associated with eye development and functionality. These results shed further light on the differences that human selection introduced in dogs aimed at different duties, even in a limited geographic area such as Italy.
1. Introduction
Dogs were domesticated 12,000 to 31,000 years ago, probably as help for hunters to find and capture large prey [1]; however, after the ascent of an agricultural and farming society (11,000–7,000 BC), dogs assumed a new role, becoming fundamental for the management of livestock [2]. Ancient writings suggest that the first shepherd dogs worked primarily as guardians, rather than herders, but it is likely that this distinction was not as clear as today [3]. In fact, at present, shepherd dogs can be distinguished into two main categories: livestock guardian (LGD) and herding shepherd (HSD) dogs.
Livestock guardians are meant to constantly watch and protect the herd from wild predators as well as rustlers. They are usually large, but at the same time fast and strong, in order to deal with large animals such as bears and wolves. To handle the harsh climates that they can live in, they are provided with a thick coat [1,4]. Their attention, trustworthiness, and protectiveness are the result of early socialization with the herd rather than from actual training: around eight weeks of age, they are introduced to the herd, and will live with it full time, sometimes even being breastfed by sheep or goats, thus creating such a strong social bond that they are considered to be part of the flock [1,3,5,6]. Their main characteristic is that they never display predatory behaviour toward the livestock, probably as a consequence of a selection to mature at an early ontogenetic stage, before the emergence of predatory sequences [3,7].
On the other hand, herding shepherd dogs work actively with shepherds, helping them to conduct the livestock and keep animals in a group. To do this, they have to show a hunting motor pattern that is interrupted before the crush-bite-kill sequence [3].
The selection of shepherd dogs has been typically based on their attitude to work: shepherds chose dogs that displayed the best behavioural characteristics, and natural and artificial selection led to the reproductive success of the healthiest and best-adapted dogs [5,7]. Even so, various breeds originated over the decades, and differences also arose among dogs of the same breed, according to the topography of the geographic area they lived and worked in.
Italy counts 17 officially recognized dog breeds, including the Bergamasco shepherd dog and the Maremma and the Abruzzi sheepdog. The first is a herding shepherd dog, with a typical felted coat. Unfortunately, after World War II this breed declined, following a marked decrease in sheep and goat breeding in the Alpine region; thanks to the work of a small group of enthusiasts, the Bergamasco shepherds survived, even though only a small number of dogs are registered every year and they are not diffusely used to work in farms (www.enci.it accessed on 10 December 2022). On the other hand, the Maremma and the Abruzzi sheepdog are livestock guardians most widely used to protect flocks from wild predators. Indeed, several national and international projects provided these dogs to farmers to limit the damage caused by the increasing population of wolves. Furthermore, Italy counts many other local breeds which are in the process of being recognised and should be valorised as well. Among these, we can find several shepherd breeds, such as the Pastore della Sila, the Pastore d’Oropa, the Pastore Apuano, the Pastore della Lessinia e del Lagorai, and the Lupino del Gigante, belonging to Group 1—Sheepdogs and Cattledogs (except Swiss Cattledogs), and the Mannara dog and Fonni’s dog, which are instead placed in Group 2—Pinscher and Schnauzer—Molossoid and Swiss Mountain and Cattledogs.
In the literature, only a few genomic studies focused on the Italian shepherd dog breeds, mostly based on microsatellites [8,9,10,11,12,13] or on the identification of genes specifically related to sheepdogs [14,15]. Therefore, the aim of the present study was to investigate the genomic regions that best differentiate and characterise Italian herding and livestock guardian shepherd dogs genotyped with a high-density SNP chip.
2. Materials and Methods
2.1. Animals and Genotyping
The present study included 158 dogs belonging to the following breeds: Maremma and the Abruzzi sheepdog (MARM, n. 20), Mannara dog (MANN, n. 12), Pastore della Sila (SILA, n. 14), and Fonni’s dog (FONN, n. 30), which comprised the livestock guardian group (LGD); Pastore d’Oropa (DORO, n. 15), Pastore Apuano (APUA, n. 19), Bergamasco shepherd dog (BERG, n. 15), Pastore della Lessinia e del Lagorai (PALA, n. 10), and Lupino del Gigante (LUGI, n. 23), which represented the herding shepherd dog group (HSD) (Table S1).
Blood samples from 26 of the FONN and of all the APUA were collected according to the Ethics Committee’s statement of the University of Messina number 040/2020bis. DNA was extracted according to the recommended manufacturer’s protocol using the DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany), and genotyped in outsourcing with a 170K canine SNPchip (CanineHD BeadChip, Illumina, San Diego, CA, USA). Genomic data of all the other dogs came from previous studies [9,16].
2.2. Quality Control
Raw genotype data underwent a quality control, performed with PLINK 1.9 software [17]: only those individuals were retained with a call rate ≥ 95% and not directly related to each other (Table S1), and SNPs with call rates ≥ 95%, with a minor allele frequency (MAF) > 1%, and located on autosomes. The software BEAGLE 4.1 was used for phasing genotype data.
2.3. Genomic Analyses
The population structure was assessed through a multidimensional scaling analysis (MDS) of the identity-by-state (IBS) distances using PLINK 1.9 [17]. The genomic backgrounds of all the dogs included in the study were assessed using ADMIXTURE v1.3.0 [18], with a number of clusters (K) ranging from two to eleven: the best-fitting model was identified as the one with the lowest cross-validation value (CV), and individuals’ probabilities of assignment to each K group (Q-value) were analysed.
In order to have homogeneous breed groups and avoid possible bias due to an over-representation of single breeds within a group, before looking for the selection signatures we performed a sample size reduction in the FONN breed in order to have a number of individuals more similar to the other breeds of the LGD group (Table S1).
The identification of selection signatures in each group of breeds (LGD and HSD) was performed by investigating the runs of homozygosity (ROH), according to the fact that in genomic regions undergoing selection, nucleotide diversity decreases while homozygosity increases around the selected locus [19]. ROH were calculated by applying a sliding window method in PLINK. The sliding window was 50 SNPs long, and no heterozygous SNPs were admitted; ROH was called if a selection (i) consisted of ≥ 50 consecutive homozygous SNPs; (ii) was ≥ 1 Mb long; (iii) had a density ≥ 1 SNP per 50 kb; and (iv) had gaps between two consecutive SNPs that were ≤ 100 kb long. A homozygosity score (H-score) was estimated for each SNP maker as the ratio between the number of its appearances in ROH and the number of the individuals in each group. Only the markers with the top 1% H-scores were considered to identify ROH islands, and were further investigated for the presence of annotated genes in the reference genome CanFam3.1.
Lastly, we investigated the selection signatures that emerged in comparing the two groups, LGD and HSD, using two different and complementary approaches: Wright’s fixation index (FST), using PLINK 1.9 [17], and single SNP cross-population extended haplotype homozygosity (XP-EHH), using SELSCAN 1.1.0 software [20]. While ROH analysis investigated intra-population selection sweep, FST and XP-EHH are inter-population analyses based on the degree of differentiation between groups due to locus-specific allele frequencies at a single-site (FST) or haplotype (XP-EHH) level. In particular, FST depends on the proportion of genetic diversity in terms of allele frequency between two populations, thus, detecting the genetic variances that actually underwent divergent selection in the two groups [21], whereas XP-EHH compares the haplotype lengths at each marker between two populations [22], identifying alleles nearly fixated in only one of them. XP-EHH is more powerful in detecting hard sweeps and polygenic selection [23].
In order to minimize the impact of outlier values, the FST of each SNP was averaged with those of the five adjacent SNPs at both the flanking regions [24]. All of the markers within the top 1% of the empirical distribution both of FST and normalized XP-EHH values were retained and mapped to CanFam3.1 genome assembly. All of the relevant genes were further investigated in terms of function and related pathways.
3. Results
3.1. Population Structure
After the procedures of quality control, the dataset consisted of 72 HSD (18 APUA, 11 BERG, 15 DORO, 18 LUGI, and 10 PALA) and 72 LGD (30 FONN, 12 MANN, 16 MARM, and 14 SILA). A total of 120,568 SNPs were retained.
The graphical representation of the first two principal components of the MDS analysis is shown in Figure 1. The two groups are well-separated along the first component (horizontal axis), with LGD on the right and HSD on the left. The second component, instead, isolates DORO from the other dogs. The single breeds are mostly identifiable too, despite partial superimposition of close breeds.
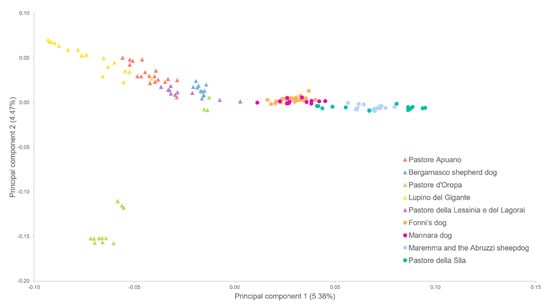
Figure 1.
Multidimensional scaling plot representing the first and second principal components. Herding shepherd dogs are represented with diamonds, whereas livestock guardians are represented with circles; each colour indicates a different breed.
An admixture analysis was performed, in order to explore the genetic background of all the dogs enrolled in the study. It is interesting to notice that when considering only two structural divisions (K = 2, Figure 2A), all of the LGD showed the prevalence of one cluster (orange, 69 ± 1.4%), while the HSD showed the other cluster (light blue, 79 ± 1.4%); the Q-values for the two clusters were significantly different between the two groups (p < 0.0001). The best-fitting model obtained included seven different clusters (K = 7, Figure 2B). A specific cluster (i.e., a prevailing unique colour) was clearly evident for APUA, BERG, DOPO, FONN, LUGI, MARM, and SILA. However, within almost all of the breeds, single dogs showed variable Q-scores for their respective cluster (Table S2); the introgression of other clusters may reflect historical phylogenetic relationships among the breeds, as well as the effect of unplanned matings that may occur during the traditional transhumance, when these kinds of dogs are not surveilled. Instead, for K = 7, MANN and PALA did not have a unique specific cluster, being the result of a mixture of the other clusters, with MARM prevailing in the MANN and LUGI in the PALA. However, it should be reported that the eighth cluster distinguished MANN. The introgression of MARM could be observed in almost all of the livestock guardians and, to a lesser extent, in PALA.
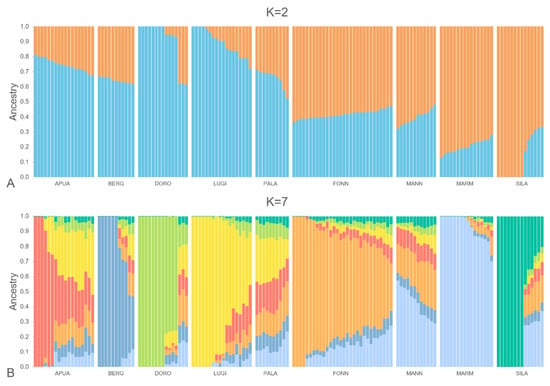
Figure 2.
Admixture analysis of all the enrolled individuals, considering a number of clusters (K) equal to two (A) and seven (B), which resulted in the best fitting model. Each colour represents a different cluster. Herding dog breeds: Pastore Apuano (APUA), Bergamasco shepherd dog (BERG), Pastore d’Oropa (DORO), Lupino del Gigante (LUGI), and Pastore della Lessinia e del Lagorai (PALA); livestock guardian breeds: Fonni’s dog (FONN), Mannara dog (MANN), Maremma and the Abruzzi sheepdog (MARM), and Pastore della Sila (SILA).
3.2. Genomic Regions Differentiating Livestock Guardian and Herding Shepherd Dogs
The sample size reduction procedure, applied to make the sizes of the compared breeds uniform, excluded six FONNs. Therefore, the HSD and LGD groups used for selection signature analysis consisted of 72 and 66 dogs, respectively.
The comparison between HSD and LGD led to the identification of 488 SNPs, mapping regions that contained 179 different genes characterised by the top 1% FST values (0.13−0.31, Figure 3A) and of 393 SNPs, mapping 137 different genes with the top 1% XP-EHH values (2.7–5.1, Figure 3B).
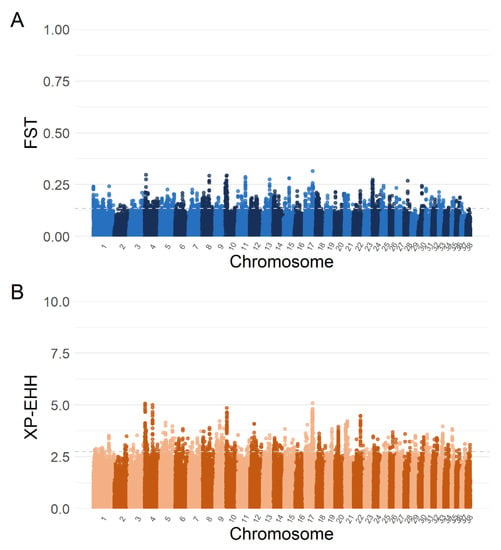
Figure 3.
Manhattan plots of Wright’s fixation index (FST) (A) and single SNP cross-population extended haplotype homozygosity (XP-EHH) (B) analyses comparing livestock guardian and herding shepherd dog breeds. Top 1% FST absolute values ranged from 0.13 to 0.31, and top 1% log (XP-EHH) from 2.7 to 5.1, as indicated by the grey dashed lines.
In cross-referencing the results of both analyses, we found 48 SNPs that mapped onto regions containing 29 different genes that were shared between the two (Table 1).

Table 1.
List of genes shared between Wright’s fixation index (FST) and single SNP cross-population extended haplotype homozygosity (XP-EHH) analyses comparing livestock guardian and herding shepherd dogs.
3.3. ROH Analysis
The top 1% H-score comprised SNPs located on 16 ROH islands on 11 chromosomes in LGD (H-score: 0.22−0.68), and 15 ROH islands on 12 chromosomes in HSD (H-score: 0.26−0.51); 4 ROH islands, located on chromosomes 1, 13, 25, and 30, were partially shared between the two groups (Table 2). We mapped the SNPs on 43 genes that were harboured in ROH identified in both the groups, 141 in LGD only, and 98 in HSD only (Table S3).

Table 2.
Runs of homozygosity islands found in livestock guardian and herding shepherd dogs.
4. Discussion
Livestock guardian and herding shepherd dogs present several differences in physical appearance and behaviour. For this reason, as it is well known by the shepherds, a dog cannot meet both requirements. In light of this, the main aim of the present study was to determine if these dogs are different in their genome as well and, if so, to identify the regions that diverge the most between them.
The MDS plot and admixture analyses confirmed that these dogs can also be distinguished from a genetic perspective. This is consistent with Talenti et al. (2018) [9], who investigated the phylogenetic relationship and breed status of several Italian dog populations, finding that Italian livestock guardian and herding shepherd dogs belonged to two different clades; the similarity that we observed between Pastore Apuano, whose SNP data are presented here for the first time, and Lupino del Gigante supports the notion that the first may be allocated in the herding shepherd dog clade as well. It must be said that the geographic localization of these dogs may also have influenced these results, enhancing the differentiation between the Italian herding shepherd dogs, whose cradle is in northern Italy, and the guardian livestock breeds, which instead originated for the most part in central-southern Italy and isles, as already seen in Italian sheep and goats [25,26].
In light of these considerations, our aim was to identify those SNPs and related genes that could better distinguish these two groups, or that may have been specifically selected in one of them. Even though it is for certain that we do not know the amount of variation in specific phenotypes that is explained by most of these markers, it is noteworthy that there is evidence from the literature that some of them are related to behavioural and morphological traits that distinguish HSD and LGD.
Several of the genes we identified are associated with dog domestication and behavioural or cognitive aspects. For example, in herding shepherd dogs we found ROH on CFA 4 (RYR2, MTR, and ACTN2 genes) and 10 (LLPH gene), and a large region on CFA 17 that were identified by Kukekova et al. (2018) [27] as differentiating tame and aggressive fox populations developed in the famous Russian farm-fox experiment. Among these genes, four (ACTN2, CCSER1, DYSF, and NID1) were also identified as differentiating LGD and HSD in the present study. Moreover, with regards to aggression, mice deficient of HSF1, a gene that comprised ROH found in LGD, showed increased offensive aggression towards intruders [28], whereas in dogs, EXOC4 was associated with the tendency of aggression during a state of nervousness [29], and MAP2K5 was associated with friendliness toward conspecifics [29]; both genes could distinguish HSD and LGD in our study.
Of particular interest are two genes found in HSD ROH only, namely MSRB3 and LLPH, which have been strongly related to dog herding behaviour in another study [30]. Moreover, some genes that we found on a homozygous region on CFA 22 were highly represented in both LGD and HSD, and harboured RCBTB1, PHF11, SETDB2, CDADC1, CYSLTR2, and RCBTB2 genes, which have been already reported by other authors to distinguish hunting and herding dogs [31,32].
Guardian and herding purposes can require specific cognitive abilities. For example, herding shepherd dogs strongly rely on communication with the shepherds during their work, while livestock guardians are more independent. In this regard, we found two genes, NFIA and VPS13B, that have been previously related to communication in dogs [33], and seem to be differentially selected in the two groups or highly homozygous in HSD, respectively. Moreover, the AGBL4 gene, identified by FST and XP-EHH analyses, was associated with an ease for dogs to become provoked by uncomfortable or frightening stimuli [29].
Furthermore, other genes differentiating HSD and LGD are related to neurological functionality. Specifically, DLG2 encodes an excitatory postsynaptic protein involved in the development of striatal connectivity; this gene has been related to autism spectrum disorders in humans, and mice with a deficiency in DLG2 activity showed decreased sociability and increased stereotypies [34]. Instead, the RELN gene encodes reelin, a protein controlling neuronal migration during brain development, and is known to be associated with a number of neurological disturbances in humans; furthermore, a study showed that in mutant zebrafish, the knockout of RELN led to a selective reduction in preference for social novelty, and increased serotonin signaling [35].
As previously mentioned, despite considerable breed variability, LGD and HSD differ in their appearance: LGD are usually larger, with a protective double coat and floppy ears; on the other hand, most of HSD are medium-sized and display erect, pricked, or folded-over ears [3,36]. The GABRB1, HELB, and RELN genes, which differentiated the two groups in our study, have been previously associated with dog size [29]; moreover, other genes are related with human body height and body mass index: XPO4, SSBP2, SH3RF3, RGL1, RELN, PEAK1, MAP2K5, KIZ, HELB, DYSF, EPB41L1, AGBL4, and DLG2 (www.genecard.org accessed on 10 December 2022). Moreover, we identified some genes playing a role in muscle development and functionality: ACTN2 encodes α-actinin-2, which anchors actin filaments at the Z-lines in both skeletal and cardiac muscles [37], and is considered to be a candidate gene associated with physical activity [38]; DYSF and MAP2K5, instead, encode proteins involved in muscle cell contraction and differentiation. A gene that was frequently contained in ROH of both dog groups was RYR, which is related to muscle development in sporting dogs, and was also identified as an adaptation to altitude in the Tibetan Mastiff [39,40,41,42]. Interestingly, AGBL4, which differentiated the two groups, was associated to altitude adaptation as well, but in this case, in yak species [43]. Moreover, CHRM5, identified in LGD only, is related to the development of muscle mass in dogs [41].
Interestingly, several HSD dogs had ROH on the region of the WIF1 and MSRB3 genes, which are related to the pricked/drop-ear phenotype in dogs [29,44,45]. It should be noted that all of the guardian dogs included in this study had drop ears, while all of the herding shepherd dogs had upright ears, with the exception of the semi-drop ears of the Bergamasco shepherd. One of the reasons why the drop ear phenotype tends to be preferred in livestock guardian dogs is that it confers them a more inoffensive appearance [46]. Moreover, it is interesting to note that the so-called “domestication syndrome” is thought to have influenced the rates of ontogenetic processes in dogs, leading to a prolonged duration of the sensitive period of socialization, which is also one of the mechanisms accounting for the suppression of predatory behaviour in LGD, and to the retention of juvenile physical features, including floppy ears [47].
Unexpectedly, several of the genes that differentiated the two dog groups are associated with eye development and functionality: KIZ, NFIA, AGBL4, DLG2, DYNC2H1, and NID1 (www.genecards.org accessed on 10 December 2022). Particularly, mutations in three of them, AGBL4, DYNC2H1, and KIZ, cause retinal pathologies in humans [48,49,50,51,52], while a NID1 mutation was found to be the cause of the development of recessive cataracts in Romagnola cattle [53]. These breeds are not commonly screened for eye pathologies. Only one study analysed the epidemiology of inherited ocular disorders in the Maremma sheepdog, finding that over onethird of the dogs were affected by at least one oculopathy; specifically, the most common problems were cataracts, entropion, corneal dystrophy, and retinal dysplasia [54]. It is our opinion that screening for hereditary pathologies should be performed on these breeds; in this way, possible disease predispositions could be identified and controlled, thanks to cautious and targeted selection.
5. Conclusions
From a genetic perspective, Italian livestock guardian and herding shepherd dog breeds appear to be well-differentiated. Particularly, some of the most differentiating genes confirm findings of previous studies that compared dogs belonging to different functional groups. Moreover, several genes are associated with behavioural traits, cognitive abilities, communication, and/or neurologic development. Other regions, instead, have been previously associated with body and muscular development, ear shape, or eye functionality. These results shed further light on the differences that human selection introduced in the genome of dogs aimed at different tasks, even within a rather limited geographic area, such as Italy.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/vetsci10010003/s1, Table S1: Dog breeds included in the present study. Table S2: Mean Q-scores of each of the seven clusters identified in the admixture analysis. Table S3: Genes located within runs of homozygosity islands found in livestock guardian and herding shepherd dogs (assembly: CanFam3.1).
Author Contributions
Conceptualization, P.C. and L.L.; methodology, A.B., M.C., L.L. and P.C.; formal analysis, M.C.; investigation, A.B. and M.C.; resources, P.C., L.L. and D.B.; data curation, A.B. and M.C.; writing—original draft preparation, A.B., M.C. and P.C.; writing—review and editing, A.B., M.C., P.C., L.L. and D.B.; visualization, A.B. and M.C.; supervision, P.C. and L.L.; project administration, P.C.; funding acquisition, L.L. and V.C. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was supported by the Consortium of Research for Meat Chain and Agrifood (CoRFilCarni) through the BIOSAVE project, “Use of phenotypic and genomic descriptors for the recovery, definition of genetic originality, and zootechnical management of Sicilian endangered local breeds”, financed by the PSR Sicilia 2014–2020—Sub-measure 10.2b “support for the conservation of genetic resources in agriculture and forestry”; the Department of Agriculture, Rural Development and Mediterranean Fisheries—Sicilian Region (Dipartimento Regionale Agricoltura—Assessorato Regionale dell’Agricoltura, dello Sviluppo Rurale e della Pesca Mediterranea—Regione Siciliana) Italy, n. G49J21006760009, prot. 0012062 (principal investigator: Vincenzo Chiofalo).
Institutional Review Board Statement
The animal study protocol was approved by the Ethics Committee of the University of Messina (protocol code number 040/2020bis).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Genomic data can be downloaded from the Gene Expression Omnibus database under accession number GSE121027 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE121027, accessed on 10 December 2022).
Acknowledgments
The authors thanks all those collaborators who supplied the samples and genomic data, including Elaine Ostrander and Dayna Dreger, all the breeders, and the Italian dog breed associations, particularly Cristian Ielli of the Cane Lupino del Gigante and Gabriele Pucciarelli of the Cane da Pastore Apuano associations.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Huson, H.J. Genetic Aspects of Performance in Working Dogs. In The Genetics of the Dog; Ostrander, E., Ruvinsky, A., Eds.; CAB International: Croydon, UK, 2012; pp. 477–495. ISBN 9781845939403. [Google Scholar]
- Olsen, S.J. Origins of the Domestic Dog: The Fossil Record; University of Arizona Press: Tucson, AZ, USA, 1985. [Google Scholar]
- Coppinger, L.; Coppinger, R. Dogs for Herding and Guarding Livestock. In Livestock Handling and Transport; Grandin, T., Ed.; CAB International: Croydon, UK, 2007; ISBN 9781845932190. [Google Scholar]
- Rigg, R. Livestock Guarding Dogs: Their Current Use World Wide; Canid Specialist Group: Oxford, UK, 2001. [Google Scholar]
- Grandin, T.; Deesing, M.J. Genetics and the Behavior of Domestic Animals; Academic Press: Cambridge, MA, USA, 2013; ISBN 9780123945860. [Google Scholar]
- Urbigkit, C. Brave and Loyal; Skyhorse Publishing: New York, NY, USA, 2016; ISBN 978150709112. [Google Scholar]
- Serpell, J. The Domestic Dog, 2nd ed.; Cambridge University Press: New York, NY, USA, 2016; ISBN 9781107024144. [Google Scholar]
- Sechi, S.; Polli, M.; Marelli, S.P.; Talenti, A.; Crepaldi, P.; Fiore, F.; Spissu, N.; Dreger, D.L.; Zedda, M.; Dimauro, C.; et al. Fonni’s dog: Morphological and genetic characteristics for a breed standard definition. Ital. J. Anim. Sci. 2017, 16, 22–30. [Google Scholar] [CrossRef]
- Talenti, A.; Dreger, D.L.; Frattini, S.; Polli, M.; Marelli, S.P.; Harris, A.C.; Liotta, L.; Cocco, R.; Hogan, A.N.; Bigi, D.; et al. Studies of modern Italian dog populations reveal multiple patterns for domestic breed evolution. Ecol. Evol. 2018, 8, 2911–2925. [Google Scholar] [CrossRef] [PubMed]
- Liotta, L.; Bionda, A.; Cortellari, M.; Negro, A. From phenotypical to genomic characterisation of the mannara dog: An italian shepherd canine resource. Ital. J. Anim. Sci. 2021, 20, 1431–1443. [Google Scholar] [CrossRef]
- Dreger, D.L.; Davis, B.W.; Cocco, R.; Sechi, S.; Di Cerbo, A.; Parker, H.G.; Polli, M.; Marelli, S.P.; Crepaldi, P.; Ostrander, E.A. Commonalities in development of pure breeds and population isolates revealed in the genome of the Sardinian Fonni’s dog. Genetics 2016, 204, 737–755. [Google Scholar] [CrossRef] [PubMed]
- Bigi, D.; Marelli, S.P.; Randi, E.; Polli, M. Genetic characterization of four native Italian shepherd dog breeds and analysis of their relationship to cosmopolitan dog breeds using microsatellite markers. Animal 2015, 9, 1921–1928. [Google Scholar] [CrossRef]
- Bigi, D.; Marelli, S.P.; Liotta, L.; Frattini, S.; Talenti, A.; Pagnacco, G.; Polli, M.; Crepaldi, P. Investigating the population structure and genetic differentiation of livestock guard dog breeds. Animal 2018, 12, 2009–2016. [Google Scholar] [CrossRef]
- Jones, P.; Chase, K.; Martin, A.; Davern, P.; Ostrander, E.A.; Lark, K.G. Single-nucleotide-polymorphism-based association mapping of dog stereotypes. Genetics 2008, 179, 1033–1044. [Google Scholar] [CrossRef]
- Chase, K.; Jones, P.; Martin, A.; Ostrander, E.A.; Lark, K.G. Genetic mapping of fixed phenotypes: Disease frequency as a breed characteristic. J. Hered. 2009, 100, S37–S41. [Google Scholar] [CrossRef]
- Parker, H.G.; Shearin, A.L.; Ostrander, E.A. Man’s best friend becomes biology’s best in show: Genome analyses in the domestic dog. Annu. Rev. Genet. 2010, 44, 309–336. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef]
- Pemberton, T.J.; Absher, D.; Feldman, M.W.; Myers, R.M.; Rosenberg, N.A.; Li, J.Z. Genomic Patterns of Homozygosity in Worldwide Human Populations. Am. J. Hum. Genet. 2012, 91, 275–292. [Google Scholar] [CrossRef]
- Szpiech, Z.A.; Hernandez, R.D. selscan: An Efficient Multithreaded Program to Perform EHH-Based Scans for Positive Selection. Mol. Biol. Evol. 2014, 31, 2824–2827. [Google Scholar] [CrossRef]
- Holsinger, K.E.; Weir, B.S. Genetics in geographically structured populations: Defining, estimating and interpreting FST. Nat. Rev. Genet. 2009, 10, 639–650. [Google Scholar] [CrossRef]
- Vitti, J.J.; Grossman, S.R.; Sabeti, P.C. Detecting Natural Selection in Genomic Data. Annu. Rev. Genet. 2013, 47, 97–120. [Google Scholar] [CrossRef]
- Saravanan, K.A.; Panigrahi, M.; Kumar, H.; Bhushan, B.; Dutt, T.; Mishra, B.P. Selection signatures in livestock genome: A review of concepts, approaches and applications. Livest. Sci. 2020, 241, 104257. [Google Scholar] [CrossRef]
- Onzima, R.B.; Upadhyay, M.R.; Doekes, H.P.; Brito, L.F.; Bosse, M.; Kanis, E.; Groenen, M.A.M.; Crooijmans, R.P.M.A. Genome-Wide Characterization of Selection Signatures and Runs of Homozygosity in Ugandan Goat Breeds. Front. Genet. 2018, 9, 318. [Google Scholar] [CrossRef]
- Cortellari, M.; Barbato, M.; Talenti, A.; Bionda, A.; Carta, A.; Ciampolini, R.; Ciani, E.; Crisà, A.; Frattini, S.; Lasagna, E.; et al. The climatic and genetic heritage of Italian goat breeds with genomic SNP data. Sci. Rep. 2021, 11, 10986. [Google Scholar] [CrossRef]
- Ciani, E.; Crepaldi, P.; Nicoloso, L.; Lasagna, E.; Sarti, F.M.; Moioli, B.; Napolitano, F.; Carta, A.; Usai, G.; D’Andrea, M.; et al. Genome-wide analysis of Italian sheep diversity reveals a strong geographic pattern and cryptic relationships between breeds. Anim. Genet. 2014, 45, 256–266. [Google Scholar] [CrossRef]
- Kukekova, A.V.; Johnson, J.L.; Xiang, X.; Feng, S.; Liu, S.; Rando, H.M.; Kharlamova, A.V.; Herbeck, Y.; Serdyukova, N.A.; Xiong, Z.; et al. Red fox genome assembly identifies genomic regions associated with tame and aggressive behaviours. Nat. Ecol. Evol. 2018, 2, 1479–1491. [Google Scholar] [CrossRef]
- Uchida, S.; Hara, K.; Kobayashi, A.; Fujimoto, M.; Otsuki, K.; Yamagata, H.; Hobara, T.; Abe, N.; Higuchi, F.; Shibata, T.; et al. Impaired hippocampal spinogenesis and neurogenesis and altered affective behavior in mice lacking heat shock factor 1. Proc. Natl. Acad. Sci. USA 2011, 108, 1681–1686. [Google Scholar] [CrossRef] [PubMed]
- Morrill, K.; Hekman, J.; Li, X.; McClure, J.; Logan, B.; Goodman, L.; Gao, M.; Dong, Y.; Alonso, M.; Carmichael, E.; et al. Ancestry-inclusive dog genomics challenges popular breed stereotypes. Science 2022, 376, eabk0639. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.; Xu, F.; Brenig, B. Genome-Wide Association Studies Reveal Neurological Genes for Dog Herding, Predation, Temperament, and Trainability Traits. Front. Vet. Sci. 2021, 8, 693290. [Google Scholar] [CrossRef] [PubMed]
- Ostrander, E.A.; Wayne, R.K.; Freedman, A.H.; Davis, B.W. Demographic history, selection and functional diversity of the canine genome. Nat. Rev. Genet. 2017, 18, 705–720. [Google Scholar] [CrossRef] [PubMed]
- Akkad, D.A.; Gerding, W.M.; Gasser, R.B.; Epplen, J.T. Homozygosity mapping and sequencing identify two genes that might contribute to pointing behavior in hunting dogs. Canine Genet. Epidemiol. 2015, 2, 5. [Google Scholar] [CrossRef]
- Gnanadesikan, G.E.; Hare, B.; Snyder-Mackler, N.; Call, J.; Kaminski, J.; Miklósi, Á.; MacLean, E.L. Breed Differences in Dog Cognition Associated with Brain-Expressed Genes and Neurological Functions. Integr. Comp. Biol. 2020, 60, 976–990. [Google Scholar] [CrossRef]
- Yoo, T.; Kim, S.-G.; Yang, S.H.; Kim, H.; Kim, E.; Kim, S.Y. A DLG2 deficiency in mice leads to reduced sociability and increased repetitive behavior accompanied by aberrant synaptic transmission in the dorsal striatum. Mol. Autism 2020, 11, 19. [Google Scholar] [CrossRef]
- Dalla Vecchia, E.; Di Donato, V.; Young, A.M.J.; Del Bene, F.; Norton, W.H.J. Reelin Signaling Controls the Preference for Social Novelty in Zebrafish. Front. Behav. Neurosci. 2019, 13, 214. [Google Scholar] [CrossRef]
- Vorwald Dohner, J. Farm Dogs; Storey Puglishing: North Adams, MA, USA, 2016. [Google Scholar]
- Oikonomou, K.G.; Zachou, K.; Dalekos, G.N. Alpha-actinin: A multidisciplinary protein with important role in B-cell driven autoimmunity. Autoimmun. Rev. 2011, 10, 389–396. [Google Scholar] [CrossRef]
- Lightfoot, J.T. Can You Be Born a Couch Potato? The Genomic Regulation of Physical Activity. In Exercise Genomics; Coleman, W.B., Tsongalis, G.J., Eds.; Humana Press: Totowa, NJ, USA, 2011; pp. 45–72. [Google Scholar]
- Kim, J.; Williams, F.J.; Dreger, D.L.; Plassais, J.; Davis, B.W.; Parker, H.G.; Ostrander, E.A. Genetic selection of athletic success in sport-hunting dogs. Proc. Natl. Acad. Sci. USA 2018, 115, E7212–E7221. [Google Scholar] [CrossRef]
- Li, Y.; Wu, D.-D.; Boyko, A.R.; Wang, G.-D.; Wu, S.-F.; Irwin, D.M.; Zhang, Y.-P. Population Variation Revealed High-Altitude Adaptation of Tibetan Mastiffs. Mol. Biol. Evol. 2014, 31, 1200–1205. [Google Scholar] [CrossRef]
- Mosher, D.S.; Quignon, P.; Bustamante, C.D.; Sutter, N.B.; Mellersh, C.S.; Parker, H.G.; Ostrander, E.A. A Mutation in the Myostatin Gene Increases Muscle Mass and Enhances Racing Performance in Heterozygote Dogs. PLoS Genet. 2007, 3, e79. [Google Scholar] [CrossRef]
- vonHoldt, B.M.; Pollinger, J.P.; Lohmueller, K.E.; Han, E.; Parker, H.G.; Quignon, P.; Degenhardt, J.D.; Boyko, A.R.; Earl, D.A.; Auton, A.; et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 2010, 464, 898–902. [Google Scholar] [CrossRef]
- Guang-Xin, E.; Basang, W.-D.; Zhu, Y.-B. Whole-genome analysis identifying candidate genes of altitude adaptive ecological thresholds in yak populations. J. Anim. Breed. Genet. 2019, 136, 371–377. [Google Scholar] [CrossRef]
- Plassais, J.; Kim, J.; Davis, B.W.; Karyadi, D.M.; Hogan, A.N.; Harris, A.C.; Decker, B.; Parker, H.G.; Ostrander, E.A. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat. Commun. 2019, 10, 1489. [Google Scholar] [CrossRef]
- Vaysse, A.; Ratnakumar, A.; Derrien, T.; Axelsson, E.; Rosengren Pielberg, G.; Sigurdsson, S.; Fall, T.; Seppälä, E.H.; Hansen, M.S.T.; Lawley, C.T.; et al. Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 2011, 7, e1002316. [Google Scholar] [CrossRef]
- Van Bommel, L.; Johnson, C.N. Where do livestock guardian dogs go? Movement patterns of free-ranging Maremma sheepdogs. PLoS ONE 2014, 9, e111444. [Google Scholar] [CrossRef]
- Trut, L.N.; Oskina, I.N.; Kharlamova, A. V Experimental Studies of Early Canid Domestication. In The Genetics of the Dog, 2nd ed.; Ostrander, E.A., Ruvinsky, A., Eds.; CPI Group (UK) Ltd.: Croydon, UK, 2012; pp. 12–37. ISBN 978-1-84593-940-3. [Google Scholar]
- Gustafson, K.; Duncan, J.; Biswas, P.; Soto-Hermida, A.; Matsui, H.; Jakubosky, D.; Suk, J.; Telenti, A.; Frazer, K.; Ayyagari, R. Whole Genome Sequencing Revealed Mutations in Two Independent Genes as the Underlying Cause of Retinal Degeneration in an Ashkenazi Jewish Pedigree. Genes 2017, 8, 210. [Google Scholar] [CrossRef]
- Zhao, Y.; Coussa, R.G.; DeBenedictis, M.J.M.; Traboulsi, E.I. Retinal dystrophy associated with a Kizuna KIZ mutation and a predominantly macular phenotype. Ophthalmic Genet. 2019, 40, 455–460. [Google Scholar] [CrossRef]
- El Shamieh, S.; Neuillé, M.; Terray, A.; Orhan, E.; Condroyer, C.; Démontant, V.; Michiels, C.; Antonio, A.; Boyard, F.; Lancelot, M.-E.; et al. Whole-exome sequencing identifies KIZ as a ciliary gene associated with autosomal-recessive rod-cone dystrophy. Am. J. Hum. Genet. 2014, 94, 625–633. [Google Scholar] [CrossRef]
- Vig, A.; Poulter, J.A.; Ottaviani, D.; Tavares, E.; Toropova, K.; Tracewska, A.M.; Mollica, A.; Kang, J.; Kehelwathugoda, O.; Paton, T.; et al. DYNC2H1 hypomorphic or retina-predominant variants cause nonsyndromic retinal degeneration. Genet. Med. 2020, 22, 2041–2051. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, W.; Li, J.-K.; Wang, Z.-S.; Bai, J.-Y.; Xu, L.; Xing, B.; Yang, W.; Wang, Z.-W.; Wang, L.-S.; et al. Genetic and clinical findings of panel-based targeted exome sequencing in a northeast Chinese cohort with retinitis pigmentosa. Mol. Genet. Genom. Med. 2020, 8, e1184. [Google Scholar] [CrossRef] [PubMed]
- Murgiano, L.; Jagannathan, V.; Calderoni, V.; Joechler, M.; Gentile, A.; Drögemüller, C. Looking the Cow in the Eye: Deletion in the NID1 Gene Is Associated with Recessive Inherited Cataract in Romagnola Cattle. PLoS ONE 2014, 9, e110628. [Google Scholar] [CrossRef] [PubMed]
- Guandalini, A.; Di Girolamo, N.; Santillo, D.; Andreani, V.; Corvi, R.; Bandini, M.; Peruccio, C. Epidemiology of ocular disorders presumed to be inherited in three large Italian dog breeds in Italy. Vet. Ophthalmol. 2017, 20, 420–426. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).