Abstract
Traumatic brain injuries may cause intracranial hemorrhages (ICH). ICH could lead to disability or death if it is not accurately diagnosed and treated in a time-sensitive procedure. The current clinical protocol to diagnose ICH is examining Computerized Tomography (CT) scans by radiologists to detect ICH and localize its regions. However, this process relies heavily on the availability of an experienced radiologist. In this paper, we designed a study protocol to collect a dataset of 82 CT scans of subjects with a traumatic brain injury. Next, the ICH regions were manually delineated in each slice by a consensus decision of two radiologists. The dataset is publicly available online at the PhysioNet repository for future analysis and comparisons. In addition to publishing the dataset, which is the main purpose of this manuscript, we implemented a deep Fully Convolutional Networks (FCNs), known as U-Net, to segment the ICH regions from the CT scans in a fully-automated manner. The method as a proof of concept achieved a Dice coefficient of 0.31 for the ICH segmentation based on 5-fold cross-validation.
Dataset: https://physionet.org/content/ct-ich/1.3.0/, doi:10.13026/w8q8-ky94.
Dataset License: Creative Commons Attribution 4.0 International Public License.
1. Introduction
Traumatic brain injury (TBI) is a major cause of death and disability in the United States. It contributed to about 30% of all injury deaths in 2013 [1]. After accidents with TBI, extra-axial intracranial lesions, such as intracranial hemorrhage (ICH), may occur. ICH is a critical medical lesion that results in a high rate of mortality [2]. It is considered to be clinically dangerous because of its high risk for turning into a secondary brain injury that may lead to paralysis and even death if it is not treated in a time-sensitive procedure. Depending on its location in the brain, ICH is divided into five sub-types: Intraventricular (IVH), Intraparenchymal (IPH), Subarachnoid (SAH), Epidural (EDH) and Subdural (SDH). In addition, the ICH that occurs within the brain tissue is called Intracerabral hemorrhage.
The Computerized Tomography (CT) scan is commonly used in the emergency evaluation of subjects with TBI for ICH [3]. The availability of the CT scan and its rapid acquisition time makes it a preferred diagnostic tool over Magnetic Resonance Imaging for the initial assessment of ICH. CT scans generate a sequence of images using X-ray beams where brain tissues are captured with different intensities depending on the amount of tissue X-ray absorbency (Hounsfield units (HU)). CT scans are displayed using a windowing method. This method transforms the HU numbers into grayscale values ([0, 255]) according to the window level and width parameters. By selecting different window parameters, different features of the brain tissues are displayed in the grayscale image (e.g., brain window, stroke window, and bone window) [4]. In the CT scan images, which are displayed using the brain window, the ICH regions appear as hyperdense regions with a relatively undefined structure. These CT images are examined by an expert radiologist to determine whether ICH has occurred and if so, detect its type and region. However, this diagnosis process relies on the availability of a subspecialty-trained neuroradiologist, and as a result, could be time inefficient and even inaccurate, especially in remote areas where specialized care is scarce.
Recent advances in convolutional neural networks (CNNs) have demonstrated that the method has an excellent performance in automating multiple image classification and segmentation tasks [5]. Hence, we hypothesized that deep learning algorithms have the potential to automate the procedure of the ICH detection and segmentation. We implemented a fully convolutional network (FCN), known as U-Net [6], to segment the ICH regions in each CT slice. An automated tool for ICH detection and classification can be used to assist junior radiology trainees when experts are not immediately available in the emergency rooms, especially in developing countries or remote areas. Such a tool can help reduce the time and error in the ICH diagnosis significantly.
Furthermore, there are two publicly available datasets for the ICH classification, and no publicly available dataset for the ICH segmentation. The first public dataset is called CQ500 that consists of 491 head CT scans [7], and the second one was published in September 2019 for the RSNA challenge at Kaggle that consists of over 25k CT scans. Arbabshirani et al. and Lee et al. indicated the availability of their data upon request [8,9]. Many papers proposed ICH segmentation approaches in addition to the ICH detection and classification. However, many of these approaches were not validated due to the lack of public or private datasets with ICH masks [10,11,12,13,14], and the other approaches were validated on private datasets that have different characteristics such as the number of CT scans and the diagnosed ICH types [9,15,16,17,18,19,20,21,22,23,24,25]. With these differences, an objective comparison between the different approaches is not feasible. Hence, there is a need for a dataset which can help to benchmark and extend the work in ICH segmentation. Therefore, the main focus of this work was collecting head CT scans with ICH segmentation and making them publicly available. We also performed a comprehensive literature review in the area of ICH detection and segmentation. Our contributions to fill the gap in knowledge are listed in Table 1.

Table 1.
Contributions of this paper.
The paper is organized as follows. Section 2 provides a review of the ICH detection and segmentation methods proposed in the literature. Section 3 describes the study used to collect a dataset of CT scans, and the deep learning method used to perform the ICH segmentation. Section 4 describes three experiments that were performed using the proposed method and provides the results. We discuss the results in Section 5. The paper is concluded in Section 6.
2. Related Work
Much interesting work has been performed for automating the process of ICH diagnosis. The majority of this work has focused either on a two-class detection problem where the method detects the presence of an ICH [7,8,9,10,11,12,14,15,18,21,24,28,29,30] or as a multi-class classification problem, where the goal is to detect the ICH sub-types [7,9,10,14,18,21,24]. Some researchers have extended the scope and performed the ICH segmentation to identify the region of ICH [9,15,16,17,18,19,20,21,22,23,24,25]. Most researchers tested their algorithms using small datasets [9,10,11,12,15,16,17,18,19,22,23,25,28,29], while a few used large datasets for the validation and testing [7,8,14,20,21,24,30]. This section provides a comprehensive review of the published papers for the ICH detection, classification, and segmentation (Figure 1).
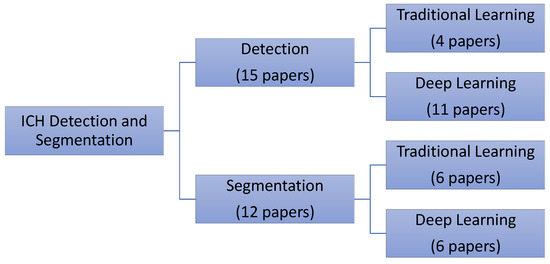
Figure 1.
The distribution of the reviewed papers for intracranial hemorrhages (ICH) detection and segmentation.
2.1. Intracranial Hemorrhage Detection and Classification
Several traditional and deep learning approaches were developed in the literature. Regarding the traditional machine learning methods, Yuh et al. developed a threshold-based algorithm to detect ICH. The method detected the ICH sub-types based on their location, shape, and volume [10]. The authors optimized the value of the threshold using the retrospective samples of 33 CT scans and evaluated their model on 210 CT scans of subjects with suspected TBI. Their algorithm achieved 98% sensitivity and 59% specificity for the ICH detection and an intermediate accuracy in detecting the ICH sub-types. In another work, Li et al. proposed two methods to segment the SAH space and then used the segmented regions to detect the SAH hemorrhage [11,12]. One method used elastic registration with the SAH space atlas, whereas the other method extracted distance transform features and trained a Bayesian decision method to perform the delineation. After the SAH space segmentation, mean gray value, variance, entropy, and energy were extracted and used to train a support vector machine classifier for the SAH hemorrhage detection. They used 60 CT scans (30 with SAH hemorrhage) to train the algorithm and tested the model on 69 CT scans (30 with SAH hemorrhage). The best performance was reported using the Bayesian decision method with 100% testing sensitivity, 92% specificity, and 91% accuracy [12].
Regarding the deep learning approaches, all the methods were based on CNNs and its variants except for the approaches in [20,23,24], which were based on an FCN model. In these approaches, the spatial dependence between the adjacent slices was considered using a second model such as random forest [7] or RNN [14,29]. Some authors also modified CNNs to process some part or the entire CT scan [21,30] or used an interpolation layer [9]. Other approaches were 1-stage, meaning that they did not consider the spatial dependency between the slices [20,24,28]. Prevedello et al. proposed two algorithms based on CNNs [28]. One of their algorithms was focused on detecting ICH, mass effect, and hydrocephalus at the scan level, while their other algorithm was developed to detect the suspected acute infarcts. A total of 246 CT scans were used for training and validation (100 hydrocephalus, 22 suspected acute infarct, and 124 noncritical findings), and a total of 100 CT scans were used for testing (50 hydrocephalus, 15 SAI, and 35 noncritical findings). The testing predictions were validated with the final radiology report or with the neuroradiologist’ review for the equivocal findings. The hydrocephalus detection algorithm yielded 90% sensitivity, 85% specificity, and an area under the curve (AUC) of 0.91. The suspected acute infarct detection algorithm resulted in a lower specificity and an AUC of 0.81.
Chilamkurthy et al. proposed four algorithms to detect the sub-types of ICH, calvarial fractures, midline shift, and mass effect [7]. They trained and validated their algorithms on a large dataset with 290k and 21k CT scans, respectively. Two datasets were used for testing. One of them had 491 scans and it was made publicly available (called CQ500). Clinical radiology reports were used as the gold-standard to label the training and validation of CT scans. These reports were used to label each scan, utilizing a natural language processing algorithm. The testing scans were then annotated by the majority votes of the ICH sub-types reported by three expert radiologists. Different deep models were developed for each of the four detection categories. ResNet18 was trained with five parallel fully connected layers as the output layers. The results of these output layers for each slice were fed to a random forest algorithm to predict the scan-level confidence for the presence of an ICH. They reported an average AUC of 0.93 for the ICH sub-type classification on both datasets. Considering the high sensitivity operating point, the average sensitivity was 92%, which was similar to that of the radiologists. However, the average specificity was 70%, which was significantly lower than the gold-standard. It also varied for different ICH sub-types. The lowest specificity of 68% was for the SDH detection.
Two approaches based on CNN with RNN were proposed to detect ICH [14,29]. Grewal et al. [29] proposed a 40-layer CNN, called DenseNet, with a bidirectional long short-term memory (LSTM) layer for the ICH detection. They also introduced three auxiliary tasks after each dense convolutional block to compute the binary segmentation of the ICH regions. Each of these tasks consisted of one convolutional layer followed by a deconvolution layer in order to upsample the feature maps to the original image size. The LSTM layer was added to incorporate the inter-slice dependencies of the CT scans of each subject. They considered 185 CT scans for the training, 67 for the validation, and 77 for the testing. The training data was augmented by rotation and horizontal flipping to balance the number of scans for each of the two classes. The network detection of the test data was evaluated against the annotation of three expert radiologists for each CT slice. They reported 81% accuracy, 88% recall (sensitivity), 81% precision, and 84% F1 score. The model F1 score was higher than the F1 scores of two of the three radiologists. Adding attention layers provided a significant increase in the model sensitivity. In [14], Ye et al. presented a 3D joint convolutional and recurrent neural network (CNN-RNN) to detect and classify ICH regions. The overall architecture of this model was similar to the model proposed by Grewal et al. [29]. VGG-16 was used as the CNN model, and the bidirectional Gated Recurrent Unit (GRU) was used as the RNN model. The RNN layer had the same functionality of the slice interpolation technique proposed by Lee et al. [9], but it was more flexible with respect to the number of adjacent slices included in the classification. The algorithm was trained and validated on 2,537 CT scans and tested on 299 CT scans. They reported a precise slice-level ICH detection with 99% for both the sensitivity and specificity and an AUC of 1. However, for the classification of the ICH sub-types, they reported a lower performance with 80% average sensitivity, 93.2% average specificity, and an AUC of 0.93. The lowest sensitivity was reported for SAH and EDH, which was 69% for both sub-types.
In three approaches, the CNN model was modified to process a number of CT slices at once [8,21,30]. Jnawalia et al. [30] proposed an ensemble of three different CNN models to perform the ICH detection. The CNN models were based on the architectures of AlexNet and GoogleNet that were extended to a 3D model by taking all the slices for each CT scan. They also have a lower number of parameters by reducing the number of layers and filter specifications. They trained, validated, and tested their model on a large dataset with 40,000 CT scans. About 34,000 CT scans were used for training (26,000 normal scans). The method that was used to label the CT scans was not reported. The positive slices were oversampled and augmented to make a balanced training dataset. About 2000 and 4000 scans were used for validation and testing, respectively. The AUC of the ensemble of the CNN models was 87% with a precision of 80%, a recall of 77%, and an F1-score of 78%. Chang et al. also developed a deep learning algorithm to detect ICH and its sub-types (except for IVH) with an ability to segment the ICH regions and quantify the ICH volume [21]. Their deep model is based on a region-of-interest CNN that estimates regions that contain ICHs for each five CT slices and then generates segmentation masks for these regions. The authors trained their algorithm on a dataset with 10k CT scans and tested it on a prospective dataset of 862 CT scans. They reported a sensitivity of 95%, and a specificity of 97% and an AUC of 0.97 for the classification of ICH sub-types and an average Dice score of 0.85 for the ICH segmentation. The lowest detection sensitivity of 90% and Dice score of 0.77 were reported for SAH. In the work of Arbabshirani et al. [8], an ensemble of four 3D CNN models with an input shape of was implemented and evaluated using 9499 retrospective and 347 prospective CT scans. An AUC of 0.846 was achieved on the retrospective study, and an average sensitivity of 71.5% and specificity of 83.5% were obtained on both testing datasets.
Similar to the work of Jnawalia et al. [30], Lee et al. used the transfer learning on an ensemble of four well-known CNN models to detect the ICH sub-types and bleeding points [9]. The four models were VGG-16, ResNet-50, Inception-v3, and Inception-ResNet-v2. The spatial dependency between the adjacent slices was taken into consideration by introducing a slice interpolation technique. This ensemble model was trained and validated using a dataset with 904 CT scans, and tested using a retrospective dataset with 200 CT scans and a prospective dataset with 237 scans. On average, the ICH detection algorithm resulted a testing AUC of 0.98 with 95% sensitivity and specificity. However, the algorithm resulted in a significantly lower sensitivity for the classification of the ICH sub-types with 78.3% sensitivity and 92.9% specificity. The lowest sensitivity of 58.3% was reported for the EDH slices in the retrospective test set and 68.8% for the IPH slices in the prospective test set. The overall localization accuracy of the attention maps was 78.1% between the model segmentation and the radiologists’ maps of bleeding points.
2.2. Intracranial Hemorrhage Segmentation
It is essential to localize and find the ICH volume to decide on the appropriate medical and surgical intervention [31]. Several methods were proposed to automate the process of the ICH segmentation [9,15,16,17,18,19,20,21,22,23,24,25]. Similar to the ICH detection, the ICH delineation approaches can be divided into traditional [15,16,17,18,19,25] and deep learning methods [9,20,21,22,23,24].
The traditional methods usually require preprocessing of the CT scans to remove the skull and noise. They also require us to register the segmented brains and extract some complicated engineered features. Many of these methods are based on unsupervised clustering to segment the ICH regions [16,17,18,25]. The methods proposed by Prakash et al. [16] and Shahangian et al. [18] use the Distance Regularized Level Set Evolution (DRLSE) method to fit active contours on ICH regions. Prakash et al. modified DRLSE for the segmentation of the IVH and IPH regions after preprocessing the CT scans for the skull removal and noise filtering [16]. Validating the method on 50 test CT scans resulted in an average Dice coefficient of 0.88, a sensitivity of 79.6%, and a specificity of 99.9%. Shahangian et al. used DRLSE for the segmentation of the EDH, IPH and SDH regions and also proposed a supervised method based on support vector machine for the classification of the ICH slices [18]. The first step in their method was segmenting the brain by removing the skull and brain ventricles. Next, they performed the ICH segmentation based on DRLSE. Then, they extracted the shape and texture features of the ICH regions, and finally, they performed the ICH detection. This method resulted in an average Dice coefficient of 58.5, a sensitivity of 82.5%, and a specificity of 90.5% on 627 CT slices. The other traditional unsupervised studies [17,25] used a fuzzy c-means clustering approach for the ICH segmentation. In [17], Bhadauria et al. proposed a method based on a spatial fuzzy c-means clustering and region-based active contour model. A retrospective set of 20 CT scans with an ICH was used. The authors reported a sensitivity of 79%, and a specificity of 99%, and an average Jaccard index of 0.78. Similarly, Gautam et al. proposed a method based on the white matter fuzzy c-means clustering followed by a wavelet-based thresholding technique [25]. They evaluated their method on 20 CT scans with an ICH and reported a Dice coefficient of 0.82.
Unlike the unsupervised methods, the traditional supervised approaches [15,19] use labeled slices to train the classifiers. Chan et al. [15] proposed a semi-automatic ICH segmentation method where the brain in each CT slice was first segmented and aligned. Then, the candidate ICH regions were selected using the top-hat transformation and extraction of the asymmetrical high intensity regions. Finally, the candidate regions were fed to a knowledge-based classifier for the ICH detection. This method resulted in 100% slice-level sensitivity, 84.1% slice-level specificity, and 82.6% lesion-level sensitivity. In another work, Muschelli et al. [19] proposed a fully-automatic method. They compared multiple traditional supervised methods for the segmentation of intracerebral hemorrhage [19]. For this purpose, the brain was first extracted from the CT scan and registered using a CT brain-extracted template. Next, multiple features were extracted from each scan. The features consisted of a threshold-based information of the CT voxel intensity, local moment information, such as the mean and standard deviation (STD), within-plane standard scores, initial segmentation using an unsupervised model, contralateral difference images, distance to the brain center, and the standardized-to-template intensity that contrasts a given CT scan with an averaged CT scans from healthy subjects. The classification models considered in this study were a logistic regression, a generalized additive model, and a random forest. These models were trained on 10 CT scans and tested on 102 CT scans. The random forest resulted in the highest Dice coefficient of 0.899.
The deep learning approaches for the ICH segmentation were either based on CNNs [9,21,22] or the FCNs design [20,23,24]. In the previous section, two methods for the ICH segmentation based on CNNs were reviewed [9,21]. Another work was developed by Nag et al. where the authors first selected the CT slices with ICHs using a trained autoencoder and then segmented the ICH areas using the active contour Chan-Vese model [22]. A dataset with 48 CT scans was used to evaluate the method. The autoencoder was trained on half of the data and all the data was used to test the algorithm. This work reported a sensitivity of 71%, a positive predictive value of 73%, and a Jaccard index of 0.55.
FCNs provides an ability to predict the presence of ICH at the pixel level. This ability of FCNs can also be used for the ICH segmentation. Several architectures of FCNs were used for the ICH segmentation as follows: dilated residual net (DRN) [20], modified VGG16 [24], and U-Net [23]. Kuo et al. [20] proposed a cost-sensitive active learning system. The system consisted of an ensemble of patch-based FCNs (PatchFCN). After the PatchFCN, the uncertainty score was calculated for each patch, and the sum of these patches’ scores was maximized under the estimated labeling time constraint. The authors used 934 CT scans for training and validating purposes and 313 retrospective scans and 100 prospective scans for testing purposes. They reported 92.85% average precision for the ICH detection at the scan level using both test sets and 77.9% average precision for the segmentation. The application of the cost-sensitive active learning technique improved the model performance on the prospective test set by annotating the new CT scans and increasing the size of the training data/scans. In the work of Cho et al. [24], the CNN cascade model was used for the ICH detection and the dual FCN models was used for the ICH segmentation. The CNN cascade model was based on the GoogLeNet network, and the dual FCN model was based on a pre-trained VGG16 network that was modified and fine-tuned using CT slices in the brain and stroke windows. The methods were evaluated using 5-fold cross-validation of about 6k CT scans. The authors reported a sensitivity and specificity of about 98% for the ICH detection and an accuracy ranging from 70% to 90% for the ICH sub-type classification. The lowest accuracy was reported for the EDH detection. For the ICH segmentation, they reported 80.19% precision and 82.15% recall. Kuang et al. proposed a semi-automatic method to segment the regions of intracerebral hemorrhage in addition to the ischemic infarct segmentation [23]. The method consisted of U-Net models for the ICH and infarct segmentation that was fed beside a user initialization of the ICH and infarct regions to a multi-region contour evolution. A set of hand-crafted features based on the bilateral density difference between the symmetric brain regions in the CT scan was introduced into the U-Net. The authors weighted the U-Net cross-entropy loss by the Euclidean distance between a given pixel and the boundaries of the true masks. The proposed semi-automatic method with the weighted loss outperformed the traditional U-net where it achieved a Dice similarity coefficient of 0.72.
Table 2 and Table 3 summarize the methods for the ICH detection and segmentation. As expected, a high testing sensitivity and specificity were reported on large datasets, and the performance of the ICH detection algorithms was equivalent to the results from the senior expert radiologists [7,9,14,21]. However, the sensitivity of the detection of some ICH sub-types was equivalent to the results from the junior radiology trainees [14]. SAH and EDH were the most difficult ICH sub-types to be classified by all the machine learning models [9,14,21,23]. It is interesting to note that SAH is also reported to be the most miss-classified sub-type by radiology residents [32]. For the ICH segmentation, the machine learning methods achieved a relatively high-performance [16,17,19,20,21,23,24,25]. However, there is still a need for a method that can precisely delineate the regions of all ICH sub-types.

Table 2.
Review of the methods proposed for the ICH detection and segmentation. Some papers used retrospective and prospective sets to test their models (i.e., retrospective + prospective), so the reported results are the average of both sets.

Table 3.
Review of the methods proposed for the ICH segmentation only.
3. Methods
This section first describes the dataset collection and annotation protocol, then it describes the FCN implemented in this work.
3.1. Dataset
A retrospective study was designed to collect head CT scans of subjects with TBI. The study was approved by the research and ethics board in the Iraqi ministry of health-Babil Office. The CT scans were collected between February and August 2018 from Al Hilla Teaching Hospital-Iraq. The CT scanner was Siemens/ SOMATOM Definition AS which had an isotropic resolution of 0.33 mm, 100 kV, and a slice thickness of 5mm. The information of each subject was anonymized. A total of 82 subjects (46 male) with an average age of 27.8 ± 19.5 years were included in this study (refer to Table 4 for the subject demographics). Each CT scan includes about 34 slices on average. Two radiologists annotated the non-contrast CT scans and recorded the ICH sub-types if an ICH was diagnosed. The two expert radiologists reviewed the non-contrast CT scans together and at the same time to reduce the effort and time in the ICH segmentation process. Once they reached a consensus on the ICH diagnosis, which consisted of the presence of ICH as well as its shape and location, the delineation of the ICH regions was performed. The radiologists did not have access to the clinical history of the subjects.

Table 4.
Subject demographics.
During the data collection process, Syngo by Siemens Medical Solutions was first used to read the CT DICOM files and save two videos (AVI format), one after windowing using the brain window (level = 40, width = 120) and one using the bone window (level = 700, width = 3200). Second, a custom tool was implemented in Matlab and used to record the radiologist annotations and delineating the ICH regions. The gray-scale 650 × 650 images (JPG format) for each CT slice were also saved for both brain and bone windows (Figure S1). The raw CT scans in DICOM format were anonymized and transferred directly to NIfTI using NiBabel library in Python. Likewise, the segmentation masks were saved as NIfTI files.
Out of the 82 subjects, 36 were diagnosed with an ICH with the following types: IVH, IPH, SAH, EDH, and SDH. See Figure 2 for some examples. One of the CT scans had a chronic ICH, and it was excluded from this study. Table 5 shows the number of slices with and without an ICH as well as the numbers with different ICH sub-types. It is important to note that the number of the CT slices for each ICH sub-type in this dataset is not balanced as the majority of the CT slices do not have an ICH. Besides that, the IVH was only diagnosed in five subjects and the SDH hemorrhage in only four subjects. Some slices were annotated with two or more ICH sub-types.
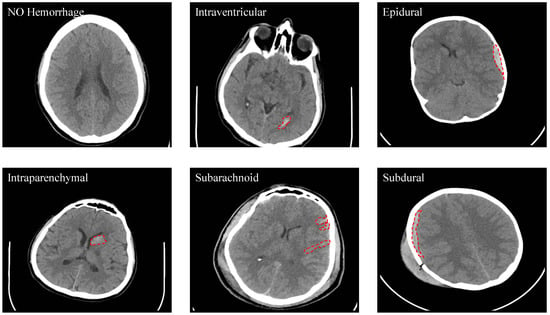
Figure 2.
Samples from the dataset that show the different types of ICH (Intraventricular (IVH), Intraparenchymal (IPH), Subarachnoid (SAH), Epidural (EDH) and Subdural (SDH)).

Table 5.
The number of slices with and without an ICH as well as different ICH sub-types.
As shown in Table 3, only an average of about 60 CT scans were used to test the ICH segmentation methods in the literature, so 82 CT scans in our dataset have a comparable (and even larger) size. However, more CT scans with ICH masks are still preferable. The dataset, including both the CT scans and the ICH masks, was released in JPG and NIfTI formats at PhysioNet (https://physionet.org/content/ct-ich/1.3.0/), which is a repository of freely-available medical research data [26,27]. The license is Creative Commons Attribution 4.0 International Public License.
3.2. ICH Segmentation Using U-Net
A Fully Convolutional Network (FCN) is an end-to-end or 1-stage algorithm used for semantic segmentation. Recently, FCN is the state-of-art performance in many applications involving the delineation of the objects. U-Net was developed by Ronneberger et al. as a type of FCNs [6]. For biomedical image segmentation, U-Net was shown to be effective on small training datasets [6], which motivated us to use it for the ICH segmentation in our study. In this work, we investigated the first application of U-Net for the ICH segmentation. The architecture of U-Net is illustrated in Figure 3.
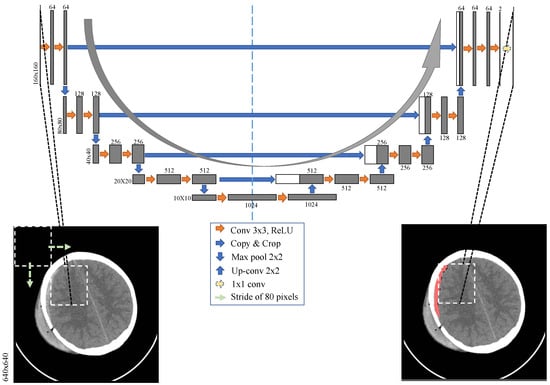
Figure 3.
The U-Net architecture implemented in this study. A sliding window of size with a stride of 80 pixels was used to divide each CT slice into 49 windows before feeding them to the U-Net for the ICH segmentation.
The architecture is symmetrical because it builds upon two paths: a contracting path and an expansive path. In the contracting path, four blocks of typical components of a convolutional network are used. Each block is constructed by two convolutional filtering layers along with padding, which is followed by a rectified linear unit (ReLU) and then by a max-pooling layer. In the expansive path, four blocks are also built that consist of two convolutional filtering layers followed by ReLU layers. Each block is preceded by upsampling the feature maps followed by a convolution (up-convolution), which are then concatenated with the corresponding cropped feature map from the contracting path. The skip connections between the two paths are intended to provide the local or the fine-grained spatial information to the global information while upsampling for the precise localization. After the last block in the expansive path, the feature maps are first filtered using two convolutional filters to produce two images; one is for the ICH regions and one for the background. The final stage is a convolutional filter with a sigmoid activation layer to produce the ICH probability in each pixel. In summary, the network has 24 convolutional layers, four max-pooling layers, four upsampling layers, and four concatenations. No dense layer is used in this architecture, in order to reduce the number of parameters and the computation time.
4. Results
We did not perform any preprocessing on the original CT slices, except for removing 5 pixels from the image borders which were only the black regions with no important information. This process resulted in CT slices. We performed three experiments to validate the performance of U-Net and compare it with a simple threshold-based method.
In the first experiment, a grid search was implemented to select the lower and upper thresholds of the ICH regions. The thresholds that resulted in the highest Jaccard index on the training data were selected and used in the testing procedure. It is expected that using U-Net with the full CT slices creates a bias of the model to the negative class (i.e., non-ICH class) because only a small number of pixels would belong to the positive class (i.e., ICH class) in each CT scan. In the second experiment, we investigated this effect by training and testing U-Net using the full CT slices. For the same reason, Kuo et al. [20] used windows instead of the entire CT slice and achieved a more precise model. This approach can also balance the training data by undersampling the negative windows. Therefore, in the third experiment, each slice from the CT scan was first divided using window with a stride of 80 pixels. This process resulted in 49 overlapped windows of size , which were then passed through U-Net for the ICH segmentation. Next, the masks of each CT scan were mapped to their original spatial positions on the original CT slice. The overlapped masks were then averaged to produce the full 640 × 640 ICH masks. This process resulted in four predictions for every pixel in the CT slice except for the ones in the edges and corners, where two and one predictions were made, respectively. The average of all the predictions at every pixel provided the final predictions. Finally, two consecutive morphological operations were performed on the ICH masks, which were closing and opening. The closing operation was performed to fill in the gaps in the ICH regions and the opening operation was performed to remove outliers and non-ICH regions.
For the evaluation, we used slice-level Jaccard index (Equation (1)) and Dice similarity coefficient (Equation (2)) to quantify how well the model segmentation on each CT slice fits the gold-standard segmentation. In Equations (1) and (2), refers to the neurologists’ segmentation and to the segmentation that resulted from U-Net.
Subject-based, 5-fold cross-validation (at the patient level) was used to train, validate, and test the developed model for all the experiments. For the first experiment, a grid search was implemented to select a lower threshold in the 100 to 210 range and an upper threshold in the 210 to 255 range. This process resulted in thresholds of 140 and 230 with a testing Jaccard index of 0.08 and Dice coefficient of 0.135.
During the second and third experiments, we implemented the U-Net architecture, illustrated in Figure 3, in the Python environment using Keras library with TensorFlow as backend [33]. The shape of the input image was in the second experiment and in the third experiment. The CT slices or the windows and their corresponding segmentation masks were used to train the network in each experiment. In our dataset, 36 out of 82 subjects were diagnosed with ICHs, resulting in only 318 ICH slices out of 2491 (i.e., less than 10% of the images). In order to avoid any class-imbalance issues between data with and without ICH, we applied a random undersampling to the training data and reduced the number of non-ICH data to the same level as the data with ICH. At every cross-validation iteration, one fold of the CT scans was left out as a held-out set for testing, one fold was used for validation, and three folds were used for training. U-Net was trained for 150 epochs on the CT slices or windows and their corresponding segmentation windows. For our implementation purposes, we used GeForce RTX2080 GPU with 11 GB of memory. The training took approximately 5 hours in each cross-validation iteration. During the training and at each iteration, random slices were selected from the training data, and a data augmentation was performed randomly from the following linear transformations:
- Rotation with a maximum of 20 degrees
- Width shift with a maximum of 10% of the image width
- Height shift with a maximum of 10% of the image height
- Shear with a maximum intensity of 0.1
- Zoom with a maximum range of 0.2
The dataset includes subjects with a wide age-range, which implies the presence of a wide range of head shapes and sizes in this dataset. To account for such a variability, we used zooming and shearing transformations for the data augmentation purposes. The head orientation can be different from subject to subject. Hence, rotation, as well as width and height shifts, were applied to increase the model generalizability. These linear transformations will yield valid CT slices as would present in real CT data. It is worth mentioning that the non-linear deformations may provide slices that would not be seen in real CT data. As a result, we only used linear transformations in our analysis. In addition, all the subjects entered the CT scanner with their heads facing to the same direction. So, the horizontal flipping will lead to CT slices that will not be generated in the data acquisition process. Therefore, it was not used as an augmentation method.
Adam optimizer with cross-entropy loss and learning rate was used. A mini-batch of size 2 was used for the second experiment and 32 in the third experiment. The trained model was validated after each epoch. The best-trained model with the lowest validation Jaccard index was saved and used for the testing. The training evaluation metric was the average cross-entropy loss.
During the second experiment with the full CT slices, U-Net failed to detect any ICH regions and resulted in only black masks (i.e., negative class with no ICH). Although we used only the CT slices with an ICH, there were only a few number of ICH pixels in the dataset, which caused the model to be biased towards the negative class with no ICH. Windowing the CT slices and undersampling the negative windows in the third experiment improved the biasing issue. The 5-fold cross-validation of the developed U-Net resulted in a better performance for the third experiment as shown in Table 6. The testing Jaccard index was 0.21 and the Dice coefficient was 0.31. The slice-level sensitivity was 97.2% and the slice-level specificity was 50.4%. Increasing the threshold on the predicted probability masks yielded a better testing specificity at the expense of the testing sensitivity as shown in Table 7. Figure 4 provides the segmentation result of the trained U-Net on some test windows along with the radiologist delineation of the ICHs. The boundary effect of each predicted mask was minimal. The boundaries show low probabilities for the non-ICH regions instead of zero, and they were zeroed out after thresholding and performing the morphological operations. The final segmented ICH regions after combining the windows, thresholding, and performing the morphological operations for some CT slices are shown in Figure 5. As shown in this figure, the model matched the radiologist ICH segmentation perfectly in the slices shown on the left side, but there were some false-positive ICH regions in the right-side slices. Note that the CT slice in Figure 5, bottom right panel, shows the ending of an EDH region where it is partially segmented by the model.

Table 6.
The testing results of the U-Net model trained on windows and used for the ICH segmentation.

Table 7.
The testing slice-level results of the U-Net model trained on windows using different thresholds.
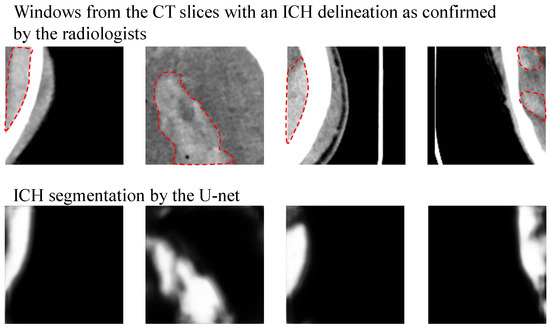
Figure 4.
Samples from the windows of the testing CT slices are shown on the top. The mask or delineation of the ICH is shown with a red dotted line. The output of U-Net before thresholding and applying the morphological operations is shown on the bottom.
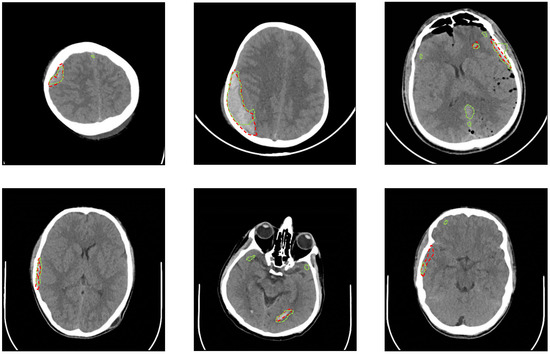
Figure 5.
Samples from the testing CT slices along with the radiologist delineation of the ICH (red dotted lines) and the U-Net segmentation (green dotted lines) are provided. A precise match of the U-Net segmentation is shown in the slices on the left side. There are some false-positive regions in the slices on the right side.
The results based on the ICH sub-type showed that the U-Net performed the best with a Dice coefficient of 0.52 for the SDH segmentation. The average Dice scores for the segmentation of EDH, IVH, IPH and SAH were 0.35, 0.3, 0.28 and 0.23, respectively. The minimum Dice coefficient and Jaccrad index in Table 6 was zero when the U-Net failed to localize the ICH regions in the CT scans of two subjects. One of the subjects had only a small IPH region in one CT slice, and the other subject had only a small IPH region in two CT slices. The width and height of the IPH regions for these subjects were less than 10mm, which sets the lower limit of the ICH segmentation by the proposed U-Net architecture. The results based on the subjects’ age show that the Dice coefficient of the subjects younger than 18 years old is 0.321, and it is 0.309 for the subjects older than 18. This analysis confirms that there is no significant difference between the method’s performance for the subjects younger and older than 18 years old.
5. Discussion
The U-Net model based on the windows of the CT slices resulted in a Dice coefficient of 0.31 for the ICH segmentation and a high sensitivity in detecting the ICH regions to be considered as the baseline for this dataset. This performance is comparable to the deep learning methods in the literature that were trained on small datasets [22,23]. Kuang et al. reported a Dice coefficient of 0.65 when a semi-automatic method based on U-Net and a contour evolution were used for the ICH segmentation. They reported a Dice coefficient of 0.35 when only U-Net was used [23]. The performance of the U-Net trained in our study is comparable to their results considering that we used a smaller dataset that had all the ICH sub-types and not only intracerebral hemorrhage. Nag et al. [22] tested autoencoder and active contour Chan-Vese model on a dataset that did not contain any SDH cases and reported an average Jaccard index of 0.55. The autoencoder was trained on half of the dataset, and then the entire dataset was used for testing, which could boost the average Jaccard index. The other deep learning-based models in [9,20,21,24] were trained and tested on larger datasets and achieved higher performance for the ICH segmentation. Chang et al. [21] reported an average Dice coefficient of 0.85, Lee et al. [9] reported a 78% overlap between the attention maps of their CNN model and the gold-standard bleeding points, Kuo et al. [20] reported 78% average precision, and Cho et al. [24] reported 80.19% precision and 82.15% recall. In addition to the deep learning methods, in the study of Shahangian et al. [18], DRLSE was used for the segmentation of EDH, IPH, and SDH, and Dice coefficients of 0.75, 0.62 and 0.37 were reported for each sub-type, respectively. Our method achieved a higher Dice coefficient of 0.52 in segmenting SDH. Some traditional methods reported better dice coefficient (0.87 [17], 0.89 [19], and 0.82 [25]) for the ICH segmentation when a small dataset was used.
Regarding the ICH detection, U-Net achieved a slice-level sensitivity of 97.2% and specificity of 50.4%, which is comparable to the results reported by Yuh et al. [10] when 0.5 threshold was used. Increasing the threshold to 0.8 resulted in 73.7% sensitivity, 82.4% specificity, and 82.5% accuracy, which is comparable to some methods in the literature that were trained on large datasets [8,29]. In the work of Arbabshirani et al. [8], an ensemble of four 3D CNN models was trained on 10k CT scans and yielded 71.5% sensitivity and 83.5% specificity. In the work of Grewal et al. [29], a deep model based on DenseNet and RNN achieved 81% accuracy.
One limitation of U-Net was the false-positive segmentation as shown in Figure 5, which was the main reason for the method’s low Dice coefficient. The false-positive segmentation was more prevalent near the bones, where the intensity in the grayscale image is similar to the intensity of the ICH regions. Another limitation is that the developed U-Net model failed to localize the ICH regions in the CT scans of two subjects who had small IPH regions. Hence, the current method as stands can be used as an assistive software to the radiologists for the ICH segmentation but is not yet at a precision that can be used as a standalone segmentation method. The future work can include collecting further CT scans and also enhancing U-Net with a recurrent neural network such as LSTM networks to consider the relationship between the adjacent slices when segmenting the ICH regions. Furthermore, we plan to improve the accuracy of our method by utilizing the transfer learning. The publicly available datasets for ICH detection and classification can be used for the transfer learning.
6. Conclusions
ICH is a critical medical lesion that requires immediate medical attention, or it may turn into a secondary brain injury, which can lead to paralysis or even death. The contribution of this paper is two-fold. First, a new dataset with 82 CT scans was collected. The dataset is made publicly available online at the PhysioNet to address the need for more publicly available benchmark datasets toward developing reliable techniques for ICH segmentation. Second, a deep learning method for the ICH segmentation was developed as a proof-of-concept. The developed method was assessed on the collected CT scans with 5-fold cross-validation. It resulted in a Dice coefficient of 0.31, which has a comparative performance for deep learning methods reported in the literature, and trained using small datasets. The U-Net model developed in this manuscript can be used as add-on software to process the CT scans. The processed CT scans with the potential ICH areas can then be reviewed by the radiologists. This preprocessing can help the radiologists to perform the final segmentation more effectively (with more accuracy) and efficiently (in a shorter time). Moreover, the paper provides a detailed review of the methods proposed for the ICH detection and classification as well as segmentation.
Supplementary Materials
The following are available at https://www.mdpi.com/2306-5729/5/1/14/s1, Figure S1: Windowing of the CT scans.
Author Contributions
Conceptualization, M.D.H., M.S.C., A.D.S., H.F.A.-k., and B.G.; data curation, M.D.H., and Z.A.Y.; formal analysis, M.H.; investigation, H.F.A.-k., and B.G.; methodology, M.D.H., M.S.C., A.D.S., H.F.A.-k., and B.G.; resources, H.F.A.-k., Z.A.Y., and B.G.; software, M.D.H.; validation, M.D.H.; writing—original draft, M.D.H. and B.G.; writing—review and editing, M.D.H., M.S.C., A.D.S. and B.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
Thanks for Mohammed Ali for the clinical support and all the patients participated in the data collection.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| CT | Computerized Tomography |
| TBI | Traumatic brain injury |
| ICH | Intracranial hemorrhage |
| IVH | Intraventricular hemorrhage |
| IPH | Intraparenchymal hemorrhage |
| SAH | Subarachnoid hemorrhage |
| EDH | Epidural hemorrhage |
| SDH | Subdural hemorrhage |
| CNN | Convolutional neural networks |
| RNN | Recurrent neural network |
| FCN | Fully convolutional networks |
| LSTM | Long short-term memory network |
| AUC | Area under the ROC curve |
References
- Taylor, C.A.; Bell, J.M.; Breiding, M.J.; Xu, L. Traumatic brain injury-related emergency department visits, hospitalizations, and deaths-United States, 2007 and 2013. Morb. Mortal. Wkly. Rep. Surveill. Summ. 2017, 66, 1–16. [Google Scholar] [CrossRef] [PubMed]
- van Asch, C.J.; Luitse, M.J.; Rinkel, G.J.; van der Tweel, I.; Algra, A.; Klijn, C.J. Incidence, case fatality, and functional outcome of intracerebral haemorrhage over time, according to age, sex, and ethnic origin: A systematic review and meta-analysis. Lancet Neurol. 2010, 9, 167–176. [Google Scholar] [CrossRef]
- Currie, S.; Saleem, N.; Straiton, J.A.; Macmullen-Price, J.; Warren, D.J.; Craven, I.J. Imaging assessment of traumatic brain injury. Postgrad. Med. 2016, 92, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Antani, S.; Long, L.R.; Demner-Fushman, D.; Thoma, G.R. Window classification of brain CT images in biomedical articles. In AMIA Annual Symposium Proceedings; American Medical Informatics Association: Bethesda, MD, USA, 2012; Volume 2012, p. 1023. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Chilamkurthy, S.; Ghosh, R.; Tanamala, S.; Biviji, M.; Campeau, N.G.; Venugopal, V.K.; Mahajan, V.; Rao, P.; Warier, P. Deep learning algorithms for detection of critical findings in head CT scans: A retrospective study. Lancet 2018, 392, 2388–2396. [Google Scholar] [CrossRef]
- Arbabshirani, M.R.; Fornwalt, B.K.; Mongelluzzo, G.J.; Suever, J.D.; Geise, B.D.; Patel, A.A.; Moore, G.J. Advanced machine learning in action: Identification of intracranial hemorrhage on computed tomography scans of the head with clinical workflow integration. NPJ Digit. Med. 2018, 1, 9. [Google Scholar] [CrossRef]
- Lee, H.; Yune, S.; Mansouri, M.; Kim, M.; Tajmir, S.H.; Guerrier, C.E.; Ebert, S.A.; Pomerantz, S.R.; Romero, J.M.; Kamalian, S.; et al. An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat. Biomed. Eng. 2019, 3, 173. [Google Scholar] [CrossRef]
- Yuh, E.L.; Gean, A.D.; Manley, G.T.; Callen, A.L.; Wintermark, M. Computer-aided assessment of head computed tomography (CT) studies in patients with suspected traumatic brain injury. J. Neurotrauma 2008, 25, 1163–1172. [Google Scholar] [CrossRef]
- Li, Y.; Wu, J.; Li, H.; Li, D.; Du, X.; Chen, Z.; Jia, F.; Hu, Q. Automatic detection of the existence of subarachnoid hemorrhage from clinical CT images. J. Med. Syst. 2012, 36, 1259–1270. [Google Scholar] [CrossRef]
- Li, Y.H.; Zhang, L.; Hu, Q.M.; Li, H.W.; Jia, F.C.; Wu, J.H. Automatic subarachnoid space segmentation and hemorrhage detection in clinical head CT scans. Int. J. Comput. Assist. Radiol. Surg. 2012, 7, 507–516. [Google Scholar] [CrossRef]
- Chilamkurthy, S.; Ghosh, R.; Tanamala, S.; Biviji, M.; Campeau, N.G.; Venugopal, V.K.; Mahajan, V.; Rao, P.; Warier, P. Development and validation of deep learning algorithms for detection of critical findings in head CT scans. arXiv 2018, arXiv:1803.05854. [Google Scholar]
- Ye, H.; Gao, F.; Yin, Y.; Guo, D.; Zhao, P.; Lu, Y.; Wang, X.; Bai, J.; Cao, K.; Song, Q.; et al. Precise diagnosis of intracranial hemorrhage and subtypes using a three-dimensional joint convolutional and recurrent neural network. Eur. Radiol. 2019, 29, 6191–6201. [Google Scholar] [CrossRef] [PubMed]
- Chan, T. Computer aided detection of small acute intracranial hemorrhage on computer tomography of brain. Comput. Med. Imaging Graph. 2007, 31, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Prakash, K.B.; Zhou, S.; Morgan, T.C.; Hanley, D.F.; Nowinski, W.L. Segmentation and quantification of intra-ventricular/cerebral hemorrhage in CT scans by modified distance regularized level set evolution technique. Int. J. Comput. Assist. Radiol. Surg. 2012, 7, 785–798. [Google Scholar] [CrossRef] [PubMed]
- Bhadauria, H.; Dewal, M. Intracranial hemorrhage detection using spatial fuzzy c-mean and region-based active contour on brain CT imaging. Signal Image Video Process. 2014, 8, 357–364. [Google Scholar] [CrossRef]
- Shahangian, B.; Pourghassem, H. Automatic brain hemorrhage segmentation and classification algorithm based on weighted grayscale histogram feature in a hierarchical classification structure. Biocybern. Biomed. Eng. 2016, 36, 217–232. [Google Scholar] [CrossRef]
- Muschelli, J.; Sweeney, E.M.; Ullman, N.L.; Vespa, P.; Hanley, D.F.; Crainiceanu, C.M. PItcHPERFeCT: Primary intracranial hemorrhage probability estimation using random forests on CT. NeuroImage Clin. 2017, 14, 379–390. [Google Scholar] [CrossRef]
- Kuo, W.; Häne, C.; Yuh, E.; Mukherjee, P.; Malik, J. Cost-Sensitive active learning for intracranial hemorrhage detection. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 715–723. [Google Scholar]
- Chang, P.; Kuoy, E.; Grinband, J.; Weinberg, B.; Thompson, M.; Homo, R.; Chen, J.; Abcede, H.; Shafie, M.; Sugrue, L.; et al. Hybrid 3D/2D convolutional neural network for hemorrhage evaluation on head CT. Am. J. Neuroradiol. 2018, 39, 1609–1616. [Google Scholar] [CrossRef]
- Nag, M.K.; Chatterjee, S.; Sadhu, A.K.; Chatterjee, J.; Ghosh, N. Computer-assisted delineation of hematoma from CT volume using autoencoder and Chan Vese model. Int. J. Comput. Assist. Radiol. Surg. 2018, 14, 259–269. [Google Scholar] [CrossRef]
- Kuang, H.; Menon, B.K.; Qiu, W. Segmenting Hemorrhagic and Ischemic Infarct Simultaneously From Follow-Up Non-Contrast CT Images in Patients With Acute Ischemic Stroke. IEEE Access 2019, 7, 39842–39851. [Google Scholar] [CrossRef]
- Cho, J.; Park, K.S.; Karki, M.; Lee, E.; Ko, S.; Kim, J.K.; Lee, D.; Choe, J.; Son, J.; Kim, M.; et al. Improving sensitivity on identification and delineation of intracranial hemorrhage lesion Using cascaded deep learning models. J. Digit. Imaging 2019, 32, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A.; Raman, B. Automatic segmentation of intracerebral hemorrhage from brain CT images. In Machine Intelligence and Signal Analysis; Springer: Singapore, 2019; pp. 753–764. [Google Scholar]
- Hssayeni, M.D. Computed Tomography Images for Intracranial Hemorrhage Detection and Segmentation. 2019. Available online: https://physionet.org/content/ct-ich/1.3.0/ (accessed on 25 December 2019).
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed]
- Prevedello, L.M.; Erdal, B.S.; Ryu, J.L.; Little, K.J.; Demirer, M.; Qian, S.; White, R.D. Automated critical test findings identification and online notification system using artificial intelligence in imaging. Radiology 2017, 285, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Grewal, M.; Srivastava, M.M.; Kumar, P.; Varadarajan, S. RADnet: Radiologist level accuracy using deep learning for hemorrhage detection in CT scans. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 281–284. [Google Scholar]
- Jnawali, K.; Arbabshirani, M.R.; Rao, N.; Patel, A.A. Deep 3D convolution neural network for CT brain hemorrhage classification. In Medical Imaging 2018: Computer-Aided Diagnosis; International Society for Optics and Photonics: Washington, DC, USA, 2018; Volume 10575, p. 105751C. [Google Scholar]
- Chi, F.L.; Lang, T.C.; Sun, S.J.; Tang, X.J.; Xu, S.Y.; Zheng, H.B.; Zhao, H.S. Relationship between different surgical methods, hemorrhage position, hemorrhage volume, surgical timing, and treatment outcome of hypertensive intracerebral hemorrhage. World J. Emerg. Med. 2014, 5, 203. [Google Scholar] [CrossRef]
- Strub, W.; Leach, J.; Tomsick, T.; Vagal, A. Overnight preliminary head CT interpretations provided by residents: Locations of misidentified intracranial hemorrhage. Am. J. Neuroradiol. 2007, 28, 1679–1682. [Google Scholar] [CrossRef]
- Chollet, F. Keras. 2015. Available online: https://keras.io (accessed on 1 March 2019).
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).